Abstract
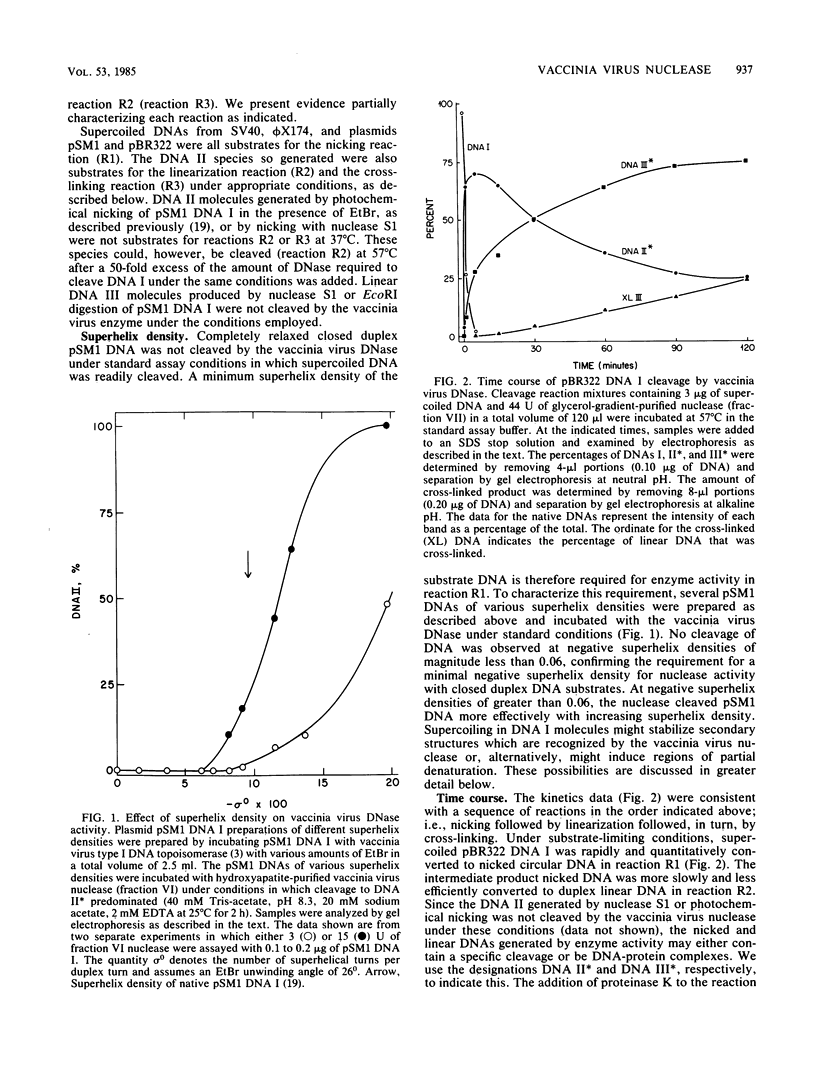
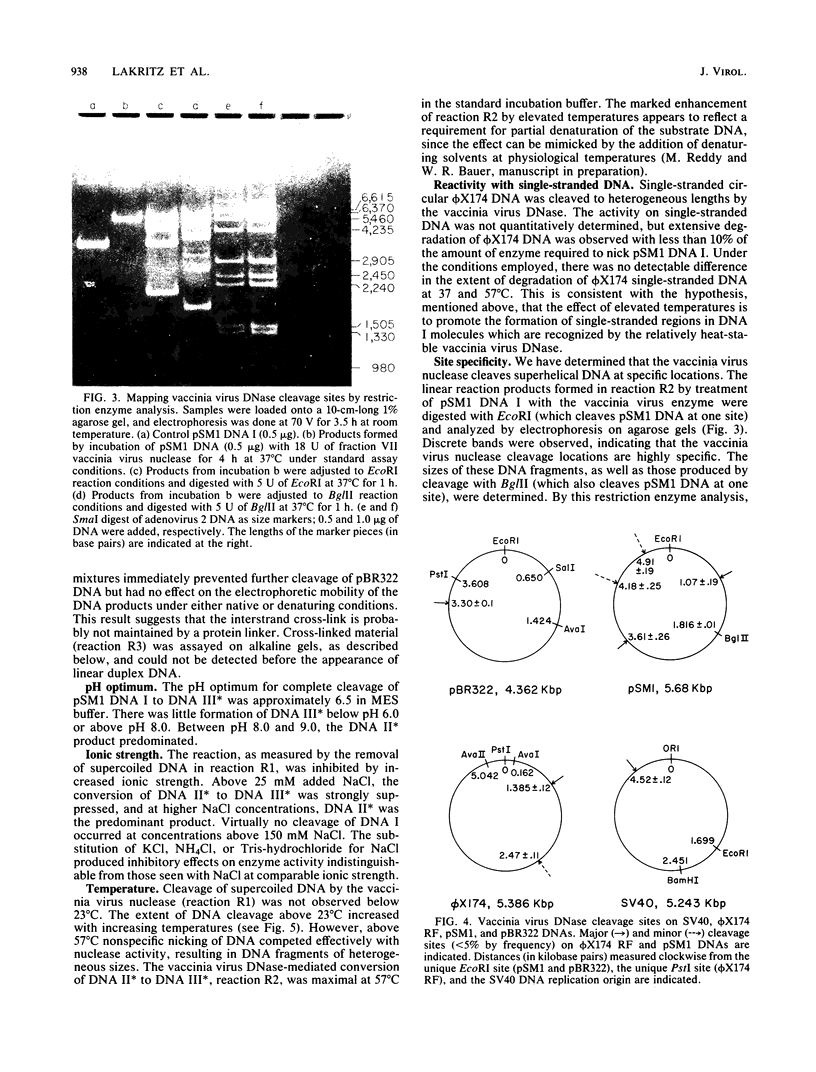
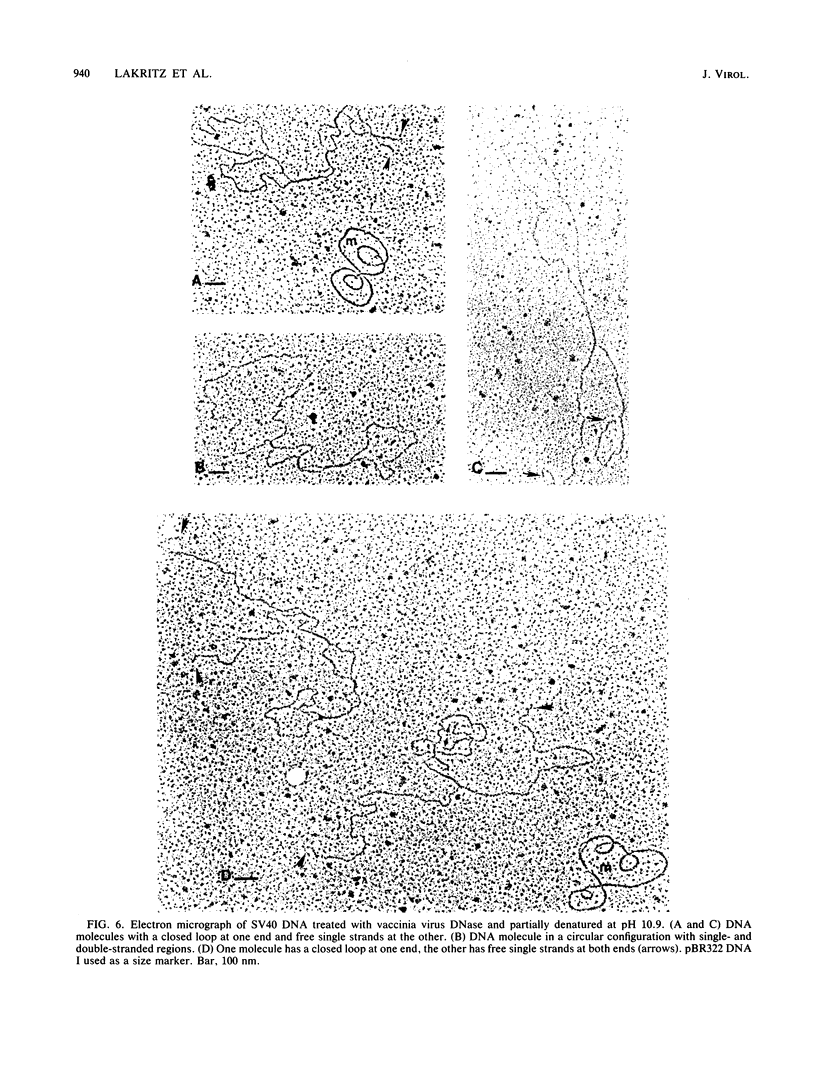
Multiple DNA-dependent enzyme activities have been detected in highly purified preparations of a single-strand-specific nuclease from vaccinia virus. These enzyme preparations were extensively purified and characterized by using superhelical DNAs as substrates. In particular, the nuclease activity was monitored by the extent of conversion of supercoiled closed duplex DNA (DNA I) to nicked circular DNA (DNA II), which could subsequently be converted to duplex linear DNA (DNA III) by prolonged incubation with the enzyme. DNA species which were not substrates for the enzyme included relaxed closed duplex DNA, DNA II which had been prepared by nuclease S1 treatment or by photochemical nicking of DNA I, and DNA III. With plasmid pSM1 DNA as substrate, the extent of cleavage of DNA I to DNA II was found to increase with superhelix density above a threshold value of about -0.06. The linear reaction products were examined by gel electrophoresis after restriction enzyme digestion of the DNAs from plasmids pSM1 and pBR322 and of the viral DNAs from bacteriophage phi X174 (replicative form) and simian virus 40, and the map coordinate locations of the scissions were determined. These products were further examined by electron microscopy and by gel electrophoresis under denaturing conditions. Electron micrographs taken under partially denaturing conditions revealed molecules with terminal loops or hairpins such as would result from the introduction of cross-links at the cutting sites. These species exhibited snapback renaturation. The denaturing gel electrophoresis experiments revealed the appearance of new bands at locations consistent with terminal cross-linking. With pSM1 and pBR322 DNAs, this band was shown to contain DNA that was approximately twice the length of a linear single strand. The terminal regions of the cross-linked linear duplex reaction products were sensitive to nuclease S1 but insensitive to proteinase K, suggesting that the structure is a hairpin loop not maintained by a protein linker. A similar structure is found in mature vaccinia virus DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baroudy B. M., Venkatesan S., Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982 Feb;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Bauer W. R., Ressner E. C., Kates J., Patzke J. V. A DNA nicking-closing enzyme encapsidated in vaccinia virus: partial purification and properties. Proc Natl Acad Sci U S A. 1977 May;74(5):1841–1845. doi: 10.1073/pnas.74.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W. R. Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown D. R., Reinberg D., Schmidt-Glenewinkel T., Roth M., Zipursky S. L., Hurwitz J. DNA structures required for phi X174 A-protein-directed initiation and termination of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):701–715. doi: 10.1101/sqb.1983.047.01.081. [DOI] [PubMed] [Google Scholar]
- DeLange A. M., Futcher B., Morgan R., McFadden G. Cloning of the vaccinia virus telomere in a yeast plasmid vector. Gene. 1984 Jan;27(1):13–21. doi: 10.1016/0378-1119(84)90234-8. [DOI] [PubMed] [Google Scholar]
- Foglesong P. D., Bauer W. R. Effects of ATP and inhibitory factors on the activity of vaccinia virus type I topoisomerase. J Virol. 1984 Jan;49(1):1–8. doi: 10.1128/jvi.49.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte M. A., Fangman W. L. Yeast chromosomal DNA molecules have strands which are cross-linked at their termini. Chromosoma. 1979 Apr 30;72(2):131–150. doi: 10.1007/BF00293230. [DOI] [PubMed] [Google Scholar]
- Funnell B. E., Inman R. B. Comparison of partial denaturation maps with the known sequence of simian virus 40 and phi X174 replicative form DNA. J Mol Biol. 1979 Jun 25;131(2):331–340. doi: 10.1016/0022-2836(79)90079-2. [DOI] [PubMed] [Google Scholar]
- Garon C. F., Barbosa E., Moss B. Visualization of an inverted terminal repetition in vaccinia virus DNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4863–4867. doi: 10.1073/pnas.75.10.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Meyer T. F. Gene-II protein of bacteriophage fd in enzymatic replication of viral duplex DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):59–62. doi: 10.1101/sqb.1979.043.01.010. [DOI] [PubMed] [Google Scholar]
- Goddard J. M., Cummings D. J. Mitochondrial DNA replication in Paramecium aurelia. Cross-linking of the initiation end. J Mol Biol. 1977 Jan 15;109(2):327–344. doi: 10.1016/s0022-2836(77)80037-5. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Winston T. P., Hodnett J. L., Legerski R. J., Nees D. W., Wei C. F., Robberson D. L. The extracellular nuclease from Alteromonas espejiana: an enzyme highly specific for nonduplex structure in nominally duplex DNA. Gene Amplif Anal. 1981;2:169–203. [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Mickel S., Arena V., Jr, Bauer W. Physical properties and gel electrophoresis behavior of R12-derived plasmid DNAs. Nucleic Acids Res. 1977;4(5):1465–1482. doi: 10.1093/nar/4.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981 Dec;27(2 Pt 1):391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Mulder C., Delius H. Specificity of the break produced by restricting endonuclease R 1 in Simian virus 40 DNA, as revealed by partial denaturation mapping. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3215–3219. doi: 10.1073/pnas.69.11.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortín J., Enjuanes L., Viñuela E. Cross-links in African swine fever virus DNA. J Virol. 1979 Sep;31(3):579–583. doi: 10.1128/jvi.31.3.579-583.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pogo B. G., Dales S. Two deoxyribonuclease activities within purified vaccinia virus. Proc Natl Acad Sci U S A. 1969 Jul;63(3):820–827. doi: 10.1073/pnas.63.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogo B. G., O'shea M. T. Further characterization of deoxyribonucleases from vaccinia virus. Virology. 1977 Mar;77(1):56–66. doi: 10.1016/0042-6822(77)90405-6. [DOI] [PubMed] [Google Scholar]
- Pogo B. G. Terminal crosslinking of vaccinia DNA strands by an in vitro system. Virology. 1980 Jan 30;100(2):339–347. doi: 10.1016/0042-6822(80)90525-5. [DOI] [PubMed] [Google Scholar]
- Pritchard A. E., Cummings D. J. Replication of linear mitochondrial DNA from Paramecium: sequence and structure of the initiation-end crosslink. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7341–7345. doi: 10.1073/pnas.78.12.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Zipursky S. L., Weisbeek P., Brown D., Hurwitz J. Studies on the phi X174 gene A protein-mediated termination of leading strand DNA synthesis. J Biol Chem. 1983 Jan 10;258(1):529–537. [PubMed] [Google Scholar]
- Rosemond-Hornbeak H., Moss B. Single-stranded deoxyribonucleic acid-specific nuclease from vaccinia virus. Endonucleolytic and exonucleolytic activities. J Biol Chem. 1974 May 25;249(10):3292–3296. [PubMed] [Google Scholar]
- Rosemond-Hornbeak H., Paoletti E., Moss B. Single-stranded deoxyribonucleic acid-specific nuclease from vaccinia virus. Purification and characterization. J Biol Chem. 1974 May 25;249(10):3287–3291. [PubMed] [Google Scholar]
- Rosen J., Ohtsubo H., Ohtsubo E. The nucleotide sequence of the region surrounding the replication origin of an R100 resistance factor derivative. Mol Gen Genet. 1979 Mar 27;171(3):287–293. doi: 10.1007/BF00267583. [DOI] [PubMed] [Google Scholar]
- Sadowski P., McGeer A., Becker A. Terminal cross-linking of DNA catalyzed by an enzyme system containing DNA ligase, DNA polymerase, and exonuclease of bacteriophage T7. Can J Biochem. 1974 Jun;52(6):525–535. doi: 10.1139/o74-077. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Shishido K. Relationship between S1 endonuclease-sensitivity and number of superhelical turns in a negatively-twisted DNA. FEBS Lett. 1980 Mar 10;111(2):333–336. doi: 10.1016/0014-5793(80)80821-0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. Relationship between superhelical density and cruciform formation in plasmid pVH51. J Biol Chem. 1982 Jun 10;257(11):6292–6295. [PubMed] [Google Scholar]
- Stellwagen N. C. Accurate molecular weight determinations of deoxyribonucleic acid restriction fragments on agarose gels. Biochemistry. 1983 Dec 20;22(26):6180–6185. doi: 10.1021/bi00295a022. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Szostak J. W. Replication and resolution of telomeres in yeast. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1187–1194. doi: 10.1101/sqb.1983.047.01.134. [DOI] [PubMed] [Google Scholar]
- Tattersall P., Ward D. C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976 Sep 9;263(5573):106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- Weiss B. Terminal cross-linking of DNA strands by an enzyme system from Escherichia coli infected with bacteriophage T4. Proc Natl Acad Sci U S A. 1970 Mar;65(3):652–659. doi: 10.1073/pnas.65.3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A., Langeveld S. A., Teertstra R., van Arkel G. A., Weisbeek P. J. Enzymatic properties of the bacteriophage phi X174 A protein on superhelical phi X174 DNA: a model for the termination of the rolling circle DNA replication. Nucleic Acids Res. 1981 May 11;9(9):2037–2053. doi: 10.1093/nar/9.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]