Abstract
Steroid hormones are known to freely partition into lipid bilayers. As a case study, we investigated the behavior of the steroid hormone cortisone in a model lipid bilayer. First, we looked at energy barriers involved in the partitioning of a single molecule into a bilayer using umbrella sampling molecular dynamics simulations. A rather wide well of −4.5 kcal/mol was observed in the interfacial region between the lipid headgroup and tailgroup. Next, using two unconstrained molecular dynamics simulations with cortisone initially positioned at distinct locations within a bilayer, we studied the preferred location and orientation of the molecule. Finally, we observed how cortisone molecules could spontaneously insert and localize in a bilayer from bulk solution. The three independent approaches produced a converged picture of how cortisone behaves in a model lipid bilayer.
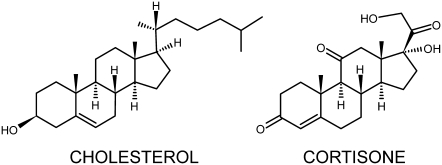
Steroid hormones like progesterone, testosterone, cortisone (Fig. 1), etc. play a critical role in regulating many life processes by acting as intercellular signaling molecules (1). These molecules are produced by steroidogenesis, a process by which various steroids are produced through a series of transformations starting from cholesterol. In the classic genomic pathway of steroid action, steroids enter a cell through the plasma membrane, bind to specific steroid receptors and the complex transported into the nucleus where it regulates gene transcription (1). However, much attention has been given recently to the rather less well understood rapid effects of steroids that include the association with membranes and modulation of neuroreceptors by circulating steroids (2–4). Intriguingly, steroids like cortisone, cortisol, and progesterone have been shown to act as noncompetitive inhibitors of nicotinic acetylcholine receptors, which are membrane-bound ion channels (5). The apparent interaction site is located on a lipid-exposed transmembrane helix. Membrane-bound γ-aminobutyric acid receptors are also modulated by corticosteroids like cortisone and cortisol (6). Specific binding sites are thought to exist for various steroids in the membrane (7).
FIGURE 1.
Structure of cholesterol and cortisone. The hydrophobic tail of cholesterol is replaced by polar groups in cortisone.
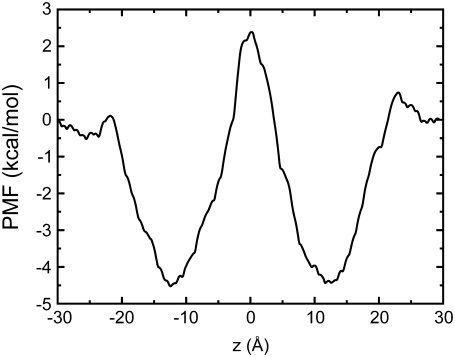
To study how a steroid interacts with a membrane-bound protein, it is important to understand how it partitions and localizes in a lipid bilayer environment. This could provide insights into how protein-steroid interactions could be initiated. In this study, we employed three independent approaches to study interactions between the steroid cortisone and a model 1-palmitoyl-2-oleoyl-sn-glycero-3 phosphocholine bilayer. First, we used an umbrella sampling (8) molecular dynamics (MD) protocol to generate biased distributions of cortisone positions along a reaction coordinate normal to the bilayer (chosen as z axis). This was then used to derive a free energy profile that indicates energy barriers along the chosen pathway. Next we ran an unconstrained MD simulation with cortisone initially positioned in the middle of the bilayer (z = 0 Å) and another with the molecule initially positioned roughly in the headgroup-tailgroup interface region (z = 10 Å). Finally, to study how cortisone partitions into a bilayer, we placed multiple copies of the molecule at random locations in bulk solution and observed cortisone-lipid interactions as it associated, partitioned, and localized in the bilayer. The umbrella sampling protocol used is detailed in Supplementary Material, Data S1. Fig. 2 shows the PMF for a cortisone molecule partitioning into the bilayer. A rather broad well of −4.5 kcal/mol exists in a region that corresponds to the interface between the headgroup and tailgroup of the lipid bilayer. The depth of the well is comparable to results obtained for steroid hormones partitioning into an implicit solvent model (9). In an implicit membrane model, steroid hormones like estradiol, testosterone, and progesterone were shown to prefer an orientation parallel to the membrane normal (9). However, these steroids have a tail that is less polar than cortisone (see Fig. S1 in Data S1). The preference for the interfacial region between the polar and the hydrophobic regions of the bilayer can be attributed to the distribution of polar groups on the cortisone molecule. Almost all hydroxyl and ketone groups of cortisone are located on one side of the long axis of the fused-ring system (Fig. 1) and associate with the polar headgroup of the lipids while the hydrophobic side of the molecule prefers the lipid tails. Thus, the molecule is amphipathic and orientates itself to maximize favorable contacts in the heterogenous bilayer interface region. The presence of polar groups in the cortisone tail introduces a barrier to free diffusion across the hydrophobic core of the membrane, resulting in a peak in the PMF in the middle of the bilayer. Steroid hormones, in general, are believed to freely diffuse across a membrane partly due to their close resemblance to cholesterol (Fig. 1), but recent evidence suggests that they could also enter cells through receptor-mediated endocytosis (10).
FIGURE 2.
PMF of cortisone in a bilayer. The value z = 0 Å corresponds to the center of the bilayer. Free energy minima exist at ∼±12 Å.
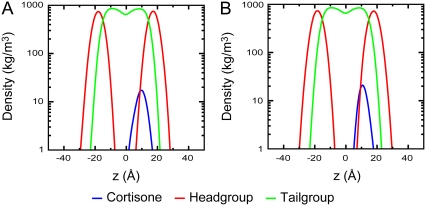
To elucidate where cortisone localizes in a bilayer, two 50-ns unconstrained MD simulations were run. Cortisone was initially placed in the middle of the bilayer (z = 0 Å) in one simulation and in the headgroup-tailgroup interface region (z = 10 Å) in the other. In both cases, the molecule was initially placed with the fused ring roughly parallel to the membrane normal in an orientation that cholesterol is thought to assume in a bilayer. Fig. 3 shows the partial densities of the various groups in the simulation box. It is clearly evident that, in both cases, the cortisone molecule spends most of the time in the region in the headgroup-tailgroup interface with a peak observed at ∼11–12 Å, which is also the location of a minimum in the PMF (Fig. 2).
FIGURE 3.
Partial densities of cortisone, lipid headgroup, and tailgroup atoms from two 50-ns MD simulations. Cortisone was initially placed at (A) the center of the bilayer (z = 0 Å), and (B) near the headgroup-tailgroup interface (z = 10 Å).
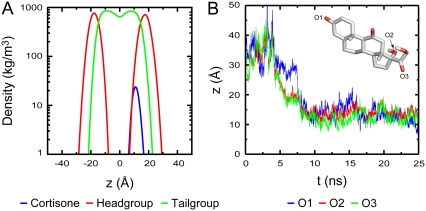
Observing spontaneous insertion into a bilayer in an unconstrained MD simulation is generally a difficult problem due to limited sampling. Despite this, we attempted to study spontaneous insertion by placing six molecules of cortisone at random positions and orientations in the bulk solution and simulated the system for 25 ns. One of the six cortisone molecules spontaneously partitioned into the bilayer after 5 ns. A second molecule did so after 20 ns while the rest remained in solution. The location of the cortisone that partitioned into the bilayer first is shown in Fig. 4 A, while that for the second is provided in the Fig. S1 in Data S1. After spontaneous insertion, the molecule adopts an orientation and location remarkably similar to the results we presented above, confirming that placing the molecule within the bilayer did not bias the results presented. Fig. 4 B gives an indication of how cortisone orients itself. In an attempt to keep the two out-of-plane methyl groups and hydrophobic regions of cortisone buried in the lipid tails, oxygen O3 (green) is always nearer to the bilayer surface compared to O2 (red), while the oxygen at the other end O1 (blue) has much more freedom (Fig. 4 B). Hence, unlike cholesterol where the hydrophobic tail is buried deep in the lipid tails attaining an orientation roughly parallel to the membrane normal, cortisone prefers an orientation that is parallel to the bilayer surface where its polar groups have maximal exposure to the lipid headgroups while keeping the hydrophobic region buried in the lipid tails.
FIGURE 4.
(A) Partial densities of the spontaneously inserted cortisone molecule, lipid headgroup, and tailgroup after full insertion of cortisone. (B) The z coordinate of three oxygen atoms, O1 (blue), O2 (red), and O3 (green), in cortisone tracked over the length of the simulation. The fluctuations indicate that the cortisone is still quite mobile. Inset shows a cortisone molecule with the tracked oxygen atoms.
Insertion takes place in a sequence of steps. The polar groups of cortisone, which replaces the hydrophobic tail in cholesterol (Fig. 1), first associate with the lipid headgroups (Fig. 5 A); the main interaction is between the cortisone tail hydroxyl group and the lipid phosphate oxygens. The molecule is then pulled deeper into the bilayer (Fig. 5 B) where the polar groups interact with the choline, glycerol, and phosphate group from three lipid molecules. It then flips to embed its hydrophobic region into the nonpolar region of the bilayer (Fig. 5 C) while allowing all five polar groups to interact with the lipid headgroups. Finally, the cortisone molecule sinks into the bilayer, finds its optimal location and orientation (Fig. 5 D) where the hydrophobic side and out of plane methyl groups interact with the nonpolar lipid tails, and remains there for the remainder of the 25 ns simulation.
FIGURE 5.
Snapshots illustrating the spontaneous association and insertion of a cortisone molecule into a lipid bilayer. For clarity, water molecules and the lower leaflet of the bilayer have been omitted. Lipids atoms are represented as spheres (C, white; O, red; N, blue; and P, green). Cortisone is shown in stick representation. (A) The polar groups from the cortisone tail associate with lipid headgroups. (B) The cortisone molecule is pulled into the bilayer. (C) The molecule flips and the hydrophobic region is embedded into the bilayer. (D) Cortisone in its preferred location and orientation.
In this study, we used three independent approaches to investigate the orientation and localization of a cortisone molecule in a lipid bilayer. A converged picture of cortisone localizing in the headgroup-tailgroup interfacial region emerged. In the nicotinic acetylcholine receptor, Thr422, a residue that is thought to associate with steroids (11), is located on a lipid-facing transmembrane helix that resides in this region suggesting that a steroid could insert into a bilayer, laterally diffuse in the bilayer, and inhibit the receptor at the putative site. In conclusion, the results presented here provide insights into the nongenomic pathway of steroid action (4) via membrane interactions.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this letter, visit www.biophysj.org.
Supplementary Material
Acknowledgments
Computations were performed on the clusters of the UK National Grid Service and the Oxford Supercomputing Centre. We thank our colleagues Henry Chew and Jussi Aittoniemi for helpful discussions.
This work was supported by the Oxford University Clarendon Fund, the UK Overseas Research Students Awards Scheme (R.V.), and the Wellcome Trust (P.C.B.). P.C.B. is a Research Councils UK fellow.
Editor: Edward H. Egelman.
References
- 1.Beato, M. 1989. Gene regulation by steroid hormones. Cell. 56:335–344. [DOI] [PubMed] [Google Scholar]
- 2.Falkenstein, E., H. C. Tillmann, M. Christ, M. Feuring, and M. Wehling. 2000. Multiple actions of steroid hormones—a focus on rapid, nongenomic effects. Pharmacol. Rev. 52:513–556. [PubMed] [Google Scholar]
- 3.Schumacher, M. 1990. Rapid membrane effects of steroid hormones: an emerging concept in neuroendocrinology. Trends Neurosci. 13:359–362. [DOI] [PubMed] [Google Scholar]
- 4.Losel, R. M., E. Falkenstein, M. Feuring, A. Schultz, H.-C. Tillmann, K. Rossol-Haseroth, and M. Wehling. 2003. Nongenomic steroid action: controversies, questions, and answers. Physiol. Rev. 83:965–1016. [DOI] [PubMed] [Google Scholar]
- 5.Nievas, G. A. F., F. J. Barrantes, and S. S. Antollini. 2007. Conformation-sensitive steroid and fatty acid sites in the transmembrane domain of the nicotinic acetylcholine receptor. Biochemistry. 46:3503–3512. [DOI] [PubMed] [Google Scholar]
- 6.Johnston, G. A. R. 1996. GABAA receptor pharmacology. Pharmacol. Ther. 69:173–198. [DOI] [PubMed] [Google Scholar]
- 7.Towle, A. C., and P. Y. Sze. 1983. Steroid binding to synaptic plasma membrane: differential binding of glucocorticoids and gonadal steroids. J. Steroid Biochem. 18:135–143. [DOI] [PubMed] [Google Scholar]
- 8.Torrie, G. M., and J. P. Valleau. 1977. Nonphysical sampling distributions in Monte Carlo free-energy estimation—umbrella sampling. J. Comput. Phys. 23:187–199. [Google Scholar]
- 9.Oren, I., S. J. Fleishman, A. Kessel, and N. Ben-Tal. 2004. Free diffusion of steroid hormones across biomembranes: a simplex search with implicit solvent model calculations. Biophys. J. 87:768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H. C., and R. V. Farese. 1999. Steroid hormones: interactions with membrane-bound receptors. Curr. Biol. 9:R478–R481. [DOI] [PubMed] [Google Scholar]
- 11.Garbus, I., A. M. Roccamo, and F. J. Barrantes. 2004. Identification of threonine 422 in transmembrane domain αM4 of the nicotinic acetylcholine receptor as a possible site of interaction with hydrocortisone. Neuropharmacology. 43:65–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.