Abstract
Binding of cell surface glycans by influenza hemagglutinin controls viral attachment and infection of host cells. This binding is a three-way interaction between viral proteins, host glycans, and viral glycans; many structural details of this interaction have been difficult to resolve. Here, we use a series of 100-ns molecular dynamics simulations to further analyze available crystallographic data on hemagglutinin-ligand interactions. Based on our simulations, we predict that the viral glycans contact the host glycans within 1–2 residues of the ligand-binding site. We also predict that the glycan-glycan interactions contain both stabilizing and destabilizing components. These predictions suggest a structural means to explain why changes to viral glycosylation alter the efficiency and selectivity of ligand binding. We also predict that the proximity of these interactions to the ligand-binding pocket will impact the binding affinity of small glycomimetic ligands analogous to the influenza neuraminidase inhibitors currently in clinical use.
Influenza hemagglutinin binds host cell glycans to mediate viral attachment and ultimately entry into cells. Hemagglutinin itself is heavily glycosylated (1), and changes in viral glycosylation alter the specificity and efficiency of host-cell attachment (2–4), but the structural basis for this remains unknown. It is also unknown whether the glycan-ligand interaction occurs close to the ligand-binding site on the protein or distal to it. The specificity of ligand binding and cell attachment by influenza is challenging to predict because it involves a complex interaction between hemagglutinin on the virion surface, multivalent target-cell glycans, and glycans covalently attached to hemagglutinin.
Altering any of these three components can affect efficiency of infection (2–6). Modification of viral glycans by producing influenza in different host cell types has been shown to alter efficiency and specificity of infection in different target cell types. Truncation of viral glycans near the ligand-binding site has a complex effect: truncation of fowl plague virus glycans increased viral binding to host receptors, while deletion of similarly located glycans in human H1N1 influenza decreased binding to α-2,6 sialyllactose ligands but did not change binding to α-2,3 ligands. These differences are particularly important for considering interspecies transmission of influenza; hemagglutinin glycosylation varies with host cell type, and the cells that influenza infects in the human upper respiratory tract, the human lower respiratory tract, and the avian respiratory tract can be differentiated based on expression of α-2,6 vs. α-2,3 sialyllactose-containing glycans (7–9).
A structural explanation for these biochemical data is challenging because only a subset of the viral glycans have been resolvable via x-ray crystallography, and the ligands used in structural studies represent a fragment of the cell-surface receptors in the respiratory tract. Even those short fragments are quite flexible and have been difficult to resolve in many crystallographic datasets (10,11). It therefore remains unknown whether glycan-glycan interaction occurs near the protein-binding pocket and whether motions of the viral glycan affect binding pocket conformation. Molecular dynamics simulations provide a means to analyze this glycoprotein flexibility using existing structures as a starting point. In this report, we use a series of 100-ns molecular dynamics simulations to investigate the interaction between viral glycans of differing length, host ligands, and the hemagglutinin protein. Our analysis suggests that viral glycans contact the host ligands within 1–2 residues of the terminal sialic acid and therefore may affect binding near the binding pocket rather than distant from it.
We have simulated the binding of trisaccharide ligands by hemagglutinin with two different glycosylation patterns: a core sequence comprising only those residues that were crystallographically ordered (1,10) and a longer full glycan detected in mammalian cell culture at the N165 site (see Scheme 1 in Supplementary Material, Data S1) (12,13). Crystal structures of A/Aichi/68 H3N2 hemagglutinin bound to α2,3-sialyllactose (10) formed the starting point for all simulations. Each simulation was run for 100 ns using GROMACS 4 and the AMBER03 and GLYCAM force fields (14–16); see supporting information for additional details.
Glycan and protein motion was measured by scoring residues in the hemagglutinin-ligand complex using root mean-squared fluctuation (RMSF) during the simulation. The protein as a whole did not substantially drift from the crystal structure (RMSF 1.9 Å), and RMSF values show agreement with crystallographic B-factors (Fig. S1 in Data S1). Consistent with their crystallographically ordered state, the core glycans maintain a relatively stable conformation throughout the simulation. RMSF values for core glycans at the N165 glycosylation site were 1.9 Å, compared with 1.4 Å for protein residues in the binding pocket. Glycans at other sites did not interact with the ligand in our simulations. The Gal2 and Glc3 residues of the ligand display increased flexibility, with RMSF values of 2.2 and 4.0 Å for Gal2 and Glc3, respectively, after rigid-body alignment to the sialic acid positions (91st and 98th percentiles among all residues in the complex). Simulations of hemagglutinin with longer glycan chains show greater flexibility in the distal residues; RMSF values are given in Tables S1–S2 in Data S1.
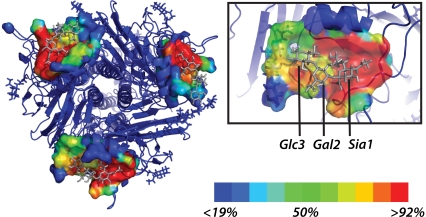
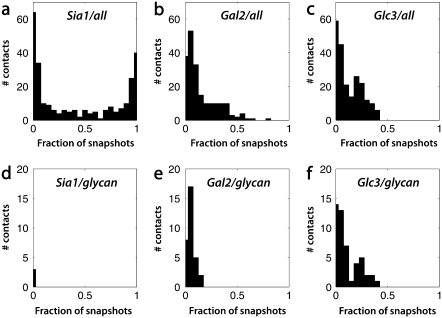
This predicted glycan flexibility allows additional contacts between the ligand and the glycoprotein beyond those visualized in the crystal structure. We therefore assess each contact by the fraction of simulation snapshots in which it is present rather than using a single static structure (Fig. 1, and Fig. S2 in Data S1). As shown in Fig. 2, a and d, the binding pocket for the terminal sialic acid is relatively stable in our simulations and is composed almost entirely of protein residues.
FIGURE 1.
Ligand-binding pocket for core glycan simulations. Atoms with contact frequency >10% are rendered in surface form and colored by contact frequency. A single binding pocket is enlarged in the inset.
FIGURE 2.
Distribution of glycan-ligand contacts by ligand residue. (a–c) Contact frequency histograms between the ligand and all glycoprotein atoms. (d–f) Histograms for contact between ligand and viral glycans only.
The proximal D-galactose residue (Gal2) of the ligand has a much more diverse set of interactions, with many residues contacting but few in constant contact (Fig. 2, b and e). The core viral glycan residues contact Gal2 in up to 20% of snapshots. This more broadly distributed contact histogram suggests the importance of dynamic heterogeneity in hemagglutinin-ligand interactions.
The distal D-glucose (Glc3) of the ligand shows a similarly diverse set of interactions, but it interacts much more strongly with the core glycan sequences (up to 40% of simulation time (Fig. 2, c and f). Similar results were obtained from simulations of the longer full glycans (Figs. S3 and S4 in Data S1). These simulation data suggest that the viral glycans contact the host glycan extensively within two residues of the terminal sialic acid. The diversity of contacts also argues that multiple structures are required to best explain the effect of viral glycosylation on ligand binding.
In addition to estimating equilibrium contact frequency (a value analogous to KD), we performed kinetic analysis of the simulation data to estimate contact dissociation timescales. In our simulations, some contacts between ligand and glycoprotein were transient, while others were much longer-lived—we quantitate this via autocorrelation functions for each contact. Analysis of the time autocorrelation functions (Fig. S5 in Data S1) for glycan-ligand contacts shows that 40–50% of encounters are transient in our simulations, decaying with time constant τ of 0.05–0.09 ns. The remaining ∼50% of encounters result in more stable association between viral glycans and ligand (τ 26–56 ns). This slowed decay suggests an energetically favorable interaction. While each individual glycan-glycan contact is relatively short-lived on experimental timescales, repeated encounters may stabilize the overall hemagglutinin-ligand interaction.
These simulation results have several important consequences for understanding the interaction between the cell type in which influenza virus was grown and the cell type that it will infect. Our simulations extend the available crystallographic information to help explain functional and biochemical data in this regard. Based on our simulations, we predict that: 1), many viral glycan-host glycan interactions occur within 1–2 residues of the terminal sialic acid; and 2), these interactions may include steric or entropic penalties of ligand binding and energetically favorable contacts that stabilize binding. Truncation of H1N1 influenza glycans showed a differential effect on binding of α-2,3 vs. α-2,6-linked glycans (3); our simulations suggest that this effect, if present in H3N2 hemagglutinin, is detectable using trisaccharide ligands. Simulations are now underway to compare α-2,3 and α-2,6 sialyllactose in complex with hemagglutinin bearing either truncated glycans or the full sequence; these simulations enable specific prediction of glycan effects on experimental binding assays.
The ΔΔG of ligand binding as the length of the viral glycan extends depends on three main factors: an increased entropic penalty from the restriction of glycan configurational entropy, the change in binding energy from favorable glycan-ligand interactions, and a possible enthalpic penalty if ligand binding increases the occupancy of high-energy glycan conformations. Accurately estimating these quantities requires extensive sampling to estimate the relative probabilities of different conformations. Future work will utilize methods such as free-energy perturbation to quantitatively predict the effects of glycan length on ligand-binding affinity and specificity.
The Aichi/2/68 strain used here is the earliest isolate from the 1968 epidemic that marked the emergence of H3N2 influenza in humans. While the 1968 epidemic was relatively mild compared with 1918, it nonetheless serves as a model for a newly emergent influenza subtype. At such times, existing vaccines may be of little use and effective antivirals thus particularly helpful in therapy. Moreover, the Aichi isolate binds both α-2,3 and α-2,6 sialyllactose in solution (KD of 3.2 and 2.1 mM, respectively (10)) and binds α-2,3 gangliosides in plate-based assays (17). Since the biochemical data for H1N1 suggest that changes to hemagglutinin glycosylation affect the binding of these two ligands differently, a hemagglutinin that binds both ligands (and may be somewhat intermediate between avian- and human-adapted forms) is of particular interest.
While biochemical data have shown that the process of ligand binding by influenza hemagglutinin depends on both ligand-protein interactions and ligand-glycan interactions, they do not demonstrate whether these two sets of interactions are coupled. Our results suggest that ligand-glycan interaction can occur quite close to the sialic-acid-binding site and thus any conformational bias induced by the glycan could affect protein-ligand interactions. We therefore predict that the ligand-protein and ligand-glycan interactions are coupled rather than completely separable.
Our predictions carry particular importance for the design of glycomimetic ligands for hemagglutinin, including inhibitors analogous to the neuraminidase inhibitors currently in clinical use. If viral glycan-host glycan interactions occurred distant from the sialic-acid binding site, viral glycosylation would affect ligand binding in a manner independent from small-molecule binding. Since we predict that contact can occur close to the sialic acid site, however, viral glycosylation and glycan motion can have a substantial effect on small-molecule binding.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank K. Branson, V. Voelz, R. Brandman, and A. Laederach for many helpful discussions and National Science Foundation award No. CNS-0619926 for computer resources.
P.M.K. was supported by a fellowship from the Berry Foundation. This work was also supported by the National Science Foundation Center on Polymer Interfaces and Macromolecular Assemblies, and dissemination support was provided by the National Institutes of Health SimBIOS Center.
Editor: Kathleen B. Hall.
References
- 1.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 289:366–373. [DOI] [PubMed] [Google Scholar]
- 2.Marinina, V. P., A. S. Gambarian, N. V. Bovin, A. B. Tuzikov, A. A. Shilov, B. V. Sinitsyn, and M. N. Matrosovich. 2003. Mol. Biol. (Mosk.). [The effect of losing glycosylation sites near the receptor-binding region on the receptor phenotype of the human influenza virus H1N1]. 37:550–555. [PubMed] [Google Scholar]
- 3.Gambaryan, A. S., V. P. Marinina, A. B. Tuzikov, N. V. Bovin, I. A. Rudneva, B. V. Sinitsyn, A. A. Shilov, and M. N. Matrosovich. 1998. Effects of host-dependent glycosylation of hemagglutinin on receptor-binding properties of H1N1 human influenza A virus grown in MDCK cells and in embryonated eggs. Virology. 247:170–177. [DOI] [PubMed] [Google Scholar]
- 4.Ohuchi, M., R. Ohuchi, A. Feldmann, and H. D. Klenk. 1997. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J. Virol. 71:8377–8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569. [DOI] [PubMed] [Google Scholar]
- 6.Yamada, S., Y. Suzuki, T. Suzuki, M. Q. Le, C. A. Nidom, Y. Sakai-Tagawa, Y. Muramoto, M. Ito, M. Kiso, T. Horimoto, K. Shinya, T. Sawada, M. Kiso, T. Usui, T. Murata, Y. Lin, A. Hay, L. F. Haire, D. J. Stevens, R. J. Russell, S. J. Gamblin, J. J. Skehel, and Y. Kawaoka. 2006. Hemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 444:378–382. [DOI] [PubMed] [Google Scholar]
- 7.Shinya, K., M. Ebina, S. Yamada, M. Ono, N. Kasai, and Y. Kawaoka. 2006. Avian flu: influenza virus receptors in the human airway. Nature. 440:435–436. [DOI] [PubMed] [Google Scholar]
- 8.van Riel, D., V. J. Munster, E. de Wit, G. F. Rimmelzwaan, R. A. Fouchier, A. D. Osterhaus, and T. Kuiken. 2006. H5N1 Virus attachment to lower respiratory tract. Science. 312:399. [DOI] [PubMed] [Google Scholar]
- 9.van Riel, D., V. J. Munster, E. de Wit, G. F. Rimmelzwaan, R. A. Fouchier, A. D. Osterhaus, and T. Kuiken. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauter, N. K., J. E. Hanson, G. D. Glick, J. H. Brown, R. L. Crowther, S. Park, J. J. Skehel, and D. C. Wiley. 1992. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and x-ray crystallography. Biochemistry. 31:9609–9621. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, M. B., S. Sabesan, J. J. Skehel, and D. C. Wiley. 1997. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by x-ray crystallography. Virology. 232:19–31. [DOI] [PubMed] [Google Scholar]
- 12.Mir-Shekari, S. Y., D. A. Ashford, D. J. Harvey, R. A. Dwek, and I. T. Schulze. 1997. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J. Biol. Chem. 272:4027–4036. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, K., H. Geyer, R. Geyer, W. Doerfler, and H. D. Klenk. 1990. The oligosaccharides of influenza virus hemagglutinin expressed in insect cells by a baculovirus vector. Virology. 174:418–429. [DOI] [PubMed] [Google Scholar]
- 14.Hess, B., C. Kutzner, D. van der Spoel, and E. Lindahl. 2008. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4:435–447. [DOI] [PubMed] [Google Scholar]
- 15.Kirschner, K. N., A. B. Yongye, S. M. Tschampel, and J. Gonzalez-Outeiriño. 2007. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29:622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan, Y., C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang, and P. Kollman. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24:1999–2012. [DOI] [PubMed] [Google Scholar]
- 17.Matrosovich, M. N., A. S. Gambaryan, S. Teneberg, V. E. Piskarev, S. S. Yamnikova, D. K. Lvov, J. S. Robertson, and K. A. Karlsson. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 233:224–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.