Abstract
Noncoding RNAs are crucial regulators of gene expression in prokaryotes and eukaryotes, but how they affect the dynamics of transcriptional networks remains poorly understood. We analyzed the temporal characteristics of the cyanobacterial iron stress response by mathematical modeling and quantitative experimental analyses and focused on the role of a recently discovered small noncoding RNA, IsrR. We found that IsrR is responsible for a pronounced delay in the accumulation of isiA mRNA encoding the late-phase stress protein, IsiA, and that it ensures a rapid decline in isiA levels once external stress triggers are removed. These kinetic properties allow the system to selectively respond to sustained (as opposed to transient) stimuli and thus establish a temporal threshold, which prevents energetically costly IsiA accumulation under short-term stress conditions. Biological information is frequently encoded in the quantitative aspects of intracellular signals (e.g., amplitude and duration). Our simulations reveal that competitive inhibition and regulated degradation allow intracellular regulatory networks to efficiently discriminate between transient and sustained inputs.
INTRODUCTION
Nonprotein-coding RNA regulators such as microRNAs (miRNA) and short interfering RNAs (siRNA) control diverse processes in metazoa including development, cell differentiation, and cell proliferation (1,2). Recent research revealed that small noncoding RNAs (sRNAs) also play important roles in bacteria (3,4), where they are involved mainly in the modulation of stress responses. Approximately 80 sRNAs have been identified in Escherichia coli, many of which are evolutionary conserved (5). sRNA is typically <200 nucleotides in size and is either encoded in cis or in trans. Cis-encoded sRNAs are transcribed from the antisense strand of their target mRNAs and are thus perfectly complementary, whereas trans-encoded sRNAs have only limited complementarity (4). Most sRNAs inhibit gene expression employing a noncatalytic mechanism of action: basepairing with target mRNAs either interferes with ribosome binding, and thus with target mRNA translation, or induces degradation of the whole sRNA-target complex (4,6).
Previous mathematical modeling indicated that sRNA-mediated regulation might allow faster control over gene expression than transcriptional and posttranslational regulation (7). Moreover, it was shown theoretically and experimentally that small noncoding RNAs (sRNAs) efficiently suppress steady-state target mRNA accumulation if the mRNA transcription rate is low, whereas it has little impact at higher mRNA transcription rates (8,9). Levine et al. (8) argue that this phenomenon, termed a “threshold-linear response”, efficiently prevents costly and potentially harmful expression of bacterial stress proteins under normal conditions.
In this study, we set out to analyze the impact of sRNAs on the temporal regulation of gene expression. Theoretical predictions derived from mathematical modeling are confirmed by quantitative experimental analyses of the iron stress response in a cyanobacterial organism (Synechocystis sp. PCC 6803). The IsiA (iron stress induced protein A) stress response protein, which is transcriptionally induced upon iron depletion or oxidative stress (10), is controlled by a naturally occurring antisense sRNA, IsrR (iron stress repressed RNA) (11). By comparing strains expressing different levels of IsrR sRNA, we find that IsrR is responsible for a pronounced delay in isiA induction. This delay ensures that iron stress proteins are expressed in a temporally ordered manner, with the “emergency” protein IsiA accumulating only if the stress duration exceeds a critical temporal threshold. Moreover, we find that IsrR sRNA ensures a rapid decline in isiA levels once external stress triggers are removed. IsiA expression must be tightly controlled, as it reduces photosynthesis efficiency in unstressed cells and becomes highly abundant under stress conditions. Our results reveal how the IsrR sRNA ensures that isiA accumulation is restricted to severe, prolonged, and ongoing stress conditions.
MATERIALS AND METHODS
Mathematical modeling
Numerical simulations were done in MATLAB 7.3 (MathWorks, Natick, MA; codes available upon request). PottersWheel, a multiexperiment fitting toolbox for MATLAB programmed by Thomas Maiwald, University of Freiburg (www.potterswheel.de), was used for parameter estimation (see Fig. 2, A and B). The Hill coefficient nH of the red curve in Fig. 2 D was estimated by using the formula nH = log(81)/log(D90/D10), where D90 and D10 are the stimulus durations required for the target amplitude to reach 90% and 10% of the steady state, respectively (12).
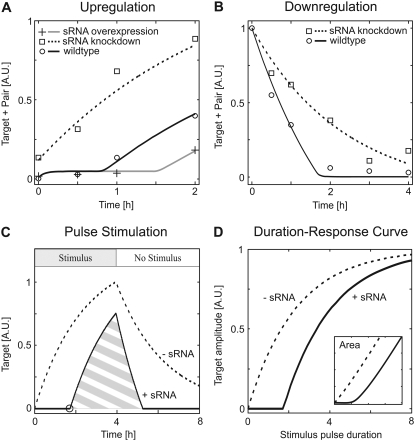
FIGURE 2.
Experimental verification of simulated dynamical behavior (A and B) and physiological relevance (C and D). (A) Regulation by sRNAs establishes a delay in target mRNA accumulation. The time course of isiA target mRNA accumulation in response to H2O2 treatment was measured in wild-type cells (circles), in cells harboring reduced amounts of IsrR sRNA (squares), and in cells overexpressing IsrR sRNA (crosses). The measurements agree well with the corresponding simulations (lines; see Table 1 for kinetic parameters). (B) The decline in target mRNA (isiA) levels is faster in wild-type cells (circles) when compared to cells depleted of IsrR sRNA (squares), as expected from corresponding simulations (lines; see Table 1 for kinetic parameters). Accumulation of isiA target mRNA was induced by iron starvation (48 h), and then isiA expression was blocked by iron readdition at t = 0 (see text). (C) Regulation by sRNAs gives rise to a sharp, spike-like time course of free target mRNA in response to step-like pulse stimulation (solid line), when compared to a corresponding system without sRNA (dashed line). In particular, target mRNA accumulation is completely suppressed until all sRNA is degraded (indicated by black circle). (D) sRNAs establish pulse filtering in gene expression. Target mRNA accumulation in response to step-like stimuli of different duration (but of constant strength) was simulated, and the time course maximum or the integral similar to the hatched area in C (inset) is plotted as a function of pulse length. Highly ultrasensitive pulse duration decoding (Hill coefficient nH ≈ 3.5) is seen in the presence (solid line) but not in the absence (dashed line) of sRNA-mediated regulation. See Table S1 in Data S1 for kinetic parameters.
Experimental
Bacterial strains and standard growth conditions were as described (11) with minor modifications (30°C; 50 μmol photons m−2s−1). Exponentially growing Synechocystis sp. PCC 6803 liquid cultures (OD750 0.7) were washed four times with iron-free medium and further grown for 48 h in fermenters supplied with a continuous stream of air. Iron pulse was achieved by the addition of 43 nmol ferric ammonium citrate per 100 mL culture at an OD750 of 0.8. RNA isolation and analysis procedures were carried out as described before (11). RNA stability was determined as described (3) with the exception that after the addition of rifampicin-arresting transcription cells were harvested on ice at different time points, followed by short-term centrifugation and resuspension in TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA was isolated with TRIzol reagent (Invitrogen) and separated on 10% polyacrylamide-urea or 1.3% agarose formaldehyde gels followed by Northern blotting. After hybridization with radiolabeled probes against either isiA or IsrR (16S or 5S ribosomal RNA (rRNA) as standard), Northern blot signals were detected and analyzed on a Personal Molecular Imager FX system with QUANTITY ONE software (Bio-Rad, Hercules, CA). For loading control of Northern blots, we used hybridization signals of rRNA as an internal standard.
RESULTS AND DISCUSSION
Mathematical model
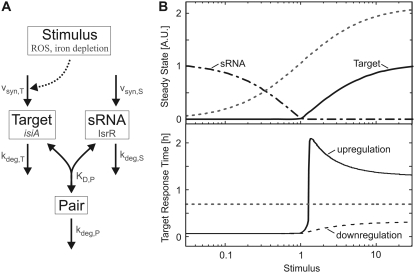
We implemented a mathematical model of sRNA-mediated regulation (schematically depicted in Fig. 1 A), which includes synthesis and degradation of target mRNA and sRNA, respectively. Additionally, we considered a reversible association reaction between the target mRNA and the sRNA and also took degradation of the resulting heteroduplex (“pair”) into account. In contrast to published models (7,8), the heteroduplex concentration was treated as a dynamic variable, mainly because our experimental analyses of the cyanobacterial iron stress response did not allow us to distinguish between free and sRNA-bound isiA mRNA. Previous studies indicated that heteroduplex association and dissociation proceed with rapid kinetics when compared to protein synthesis and degradation (13,14). We therefore applied a rapid-equilibrium assumption so that the model depicted in Fig. 1 A reduces to a two-variable system (see also Supplementary Material, Data S1):
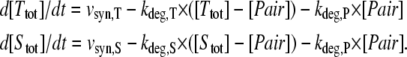 |
(1) |
Here, [Ttot] and [Stot] indicate the dynamical variables of the system which represent the total intracellular concentrations of the target mRNA and sRNA, respectively. More specifically, [Ttot] equals the sum of the concentrations of free and sRNA-bound target mRNA (i.e., [Ttot] = [Target] + [Pair]), and [Stot] = [sRNA] + [Pair] is defined similarly. Owing to the rapid equilibrium assumption, the concentration of the heteroduplex (i.e., [Pair]) is given by (see Data S1):
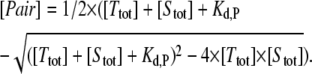 |
(2) |
Equations 1 and 2 constitute a reduced differential equation system that depends on the dynamical variables [Ttot] and [Stot] only. From the total RNA concentrations, [Ttot] and [Stot], one can calculate back to the individual concentrations (i.e., [Target], [sRNA], and [Pair]) by using Eq. 2 and the mass conservation definitions of [Stot] and [Ttot]. Numerical simulations were generally done using the units nM for intracellular concentrations and h for time. In Figs. 1 and 2, the concentrations on the y axes are given in arbitrary units (A.U.) because no absolute experimental quantification of RNA expression was available.
FIGURE 1.
Theoretical analysis of gene expression regulation by sRNAs. (A) Schematic representation of the mathematical model. The target mRNA associates with an sRNA to form a heteroduplex (pair). Molecular species and events relevant for the cyanobacterial stress response (ROSs; iron depletion; isiA and IsrR) and kinetic parameters are indicated in gray. (B) Steady-state (top) and dynamical (bottom) response to varying stimulus strength. The top graph indicates the steady-state levels of (free) sRNA (dashed-dotted line) and target mRNA (solid line), and reveals a mutually exclusive expression pattern characterized by a sharp threshold. For comparison, the gradual target mRNA dose-response curve in the absence of sRNA expression is also shown (dotted line). The bottom graph shows the response time of total target expression (i.e., sum of target and pair) as a function of the stimulus level. The solid line (upregulation) depicts the response time t50 required to reach 50% of the difference between old and new steady state upon a step-like increase in the stimulus level from no stimulation to the level indicated on the x axis. Similarly, t50 was also calculated for a step-like drop in the stimulus level from the value on the x axis to no stimulation (downregulation). The dotted line depicts the response time of a reference system with the same kinetic parameters, but without sRNA and is valid for both up- and downregulation. See Table S1 in Data S1 for kinetic parameters.
The cyanobacterial stress response protein IsiA is induced by reactive oxygen species (ROS) and by iron depletion, and its expression is further modulated by a sRNA, IsrR (Fig. 1 A). We measured the kinetics of isiA regulation to confirm the model predictions (see below). The input functions in the model were therefore chosen such that they reflect those in the cyanobacterial stress response. IsiA expression is known to be transcriptionally regulated by the iron-sensitive ferric uptake regulator repressor, whereas the activity of the promoter controlling the antagonizing sRNA, IsrR, is not affected by cellular stress (W. Hess, University of Freiburg, personal communication, 2008). The synthesis rate of target mRNA (vsyn,T) is therefore modeled to be controlled by external stimuli, as indicated in Fig. 1 A. For simplicity, it is assumed that the regulation of the isiA promoter activity occurs on a much faster timescale than downstream RNA synthesis and degradation. Thus, extracellular stimulation was simulated by a step-like change of vsyn,T from one value to another. The Michaelis-Menten equation (vsyn,T = Vmax,synT × Stimulus/(KM,synT + Stimulus)) was employed to simulate dose-response behavior (Fig. 1 B).
Theoretical analysis of steady-state and dynamical behavior
We systematically analyzed the steady-state and dynamical behavior of the sRNA circuit by numerical simulations. In the following paragraphs, we focus mainly on the scenario where the heteroduplex is much less stable than the monomeric sRNA and target species, since such behavior was recently reported for the cyanobacterial stress response (11). However, the main conclusions drawn in this work remain valid even if the sRNA merely acts as a competitive inhibitor of translation, which does not enhance target degradation (see Conclusion).
Recent work by Levine et al. (8) revealed that sRNAs establish sharp thresholds in the steady-state stimulus-response behavior of gene regulatory circuits. Similarly, our simulations also yielded an all-or-none expression pattern of target mRNA if physiologically relevant kinetic parameters were used (Fig. 1 B, top, solid line). As discussed previously (8), sRNA-mediated regulation efficiently suppresses mRNA accumulation as long as the sRNA synthesis rate exceeds that of the target mRNA (i.e., if vsyn,T < vsyn,S). In Fig. 1 B, we have vsyn,T = vsyn,S if the stimulus level equals unity and thus observe a threshold for stimulus = 1. As expected, the threshold disappeared in the absence of sRNA (dashed line) or in case the affinity of the heteroduplex was too low (not shown). Moreover, we found that sharp thresholds were typically accompanied by a mutually exclusive expression pattern of target mRNA and sRNA (solid and dashed-dotted lines in Fig. 1 B). Remarkably, our previous experimental work revealed that the isiA mRNA and its modulator, the IsrR sRNA, accumulate in a mutually exclusive manner under various stimulation conditions (11). In further experiments, we analyzed isiA and IsrR amounts for varying iron concentrations in the medium (Fig. S2 in Data S1) and found that IsrR sRNA starts to accumulate only if isiA mRNA falls below a certain level. Finally, a comparison of wild-type cells with IsrR knockdown cells reveals that IsrR completely suppresses residual isiA levels under unstressed conditions (compare circle and square at t = 0 in Fig. 2 A). Taken together, these data strongly suggest that a sharp threshold exists in the cyanobacterial iron stress response and that the system operates near this threshold under physiological conditions.
We compared the temporal dynamics of a reference system devoid of sRNA with those of the full system including sRNA-mediated regulation to derive experimentally testable predictions from the model. We focused on the time required to pass from one steady state to another (“response time”). Step-like increases in the external stimulus were applied, and the response time t50 required to reach 50% of the difference between the old and new steady state was calculated for the target mRNA. The dotted line in Fig. 1 B (bottom) reflects the response time of a reference system without sRNA and is valid for both input scenarios considered in our analysis: i), a step-like stimulus increase from no stimulation to the value indicated on the x axis (“upregulation”); and ii), a stimulus decrease from the value indicated on the x axis to no stimulation (“downregulation”). Our simulations of the full system including sRNA-mediated regulation revealed that sRNAs speed up the upregulation kinetics for subthreshold stimuli, whereas they establish a delay upon suprathreshold stimulation (solid line). Moreover, we found that sRNAs always accelerate target downregulation even for suprathreshold stimuli (dashed line).
In the subthreshold regime, the sRNA is present in excess over the target mRNA. Under these conditions, the sRNA can be assumed to be constant so that mRNA degradation via the pair intermediate behaves like a first-order decay reaction, which dominates over the much slower direct mRNA degradation step (see Data S1). The response times of mRNA up- and downregulation are known to be solely determined by the (fastest) first-order decay rate (15), which explains why sRNA-mediated regulation accelerates target regulation in both directions under subthreshold stimulation conditions. The dynamic phenomena we observed upon strong stimulation originate from nonlinear threshold effects in the sRNA circuit. The upregulation time lag arises because the residual pool of free small RNAs (sRNAs) needs to be cleared before the target mRNA can accumulate. sRNAs accelerate downregulation, since the mRNA concentration quickly falls into a subthreshold range, where sRNA-mediated regulation efficiently degrades residual mRNA. This results in an abrupt termination of the mRNA expression time course, as can be seen from the solid line in Fig. 2 B. Moreover, the initial phase of mRNA downregulation is accelerated in the presence of small noncoding RNAs (compare solid and dotted lines in Fig. 2 B), as the pair intermediate is turned over at a higher rate than the free mRNA. Taken together, our simulations suggest that IsrR sRNA delays isiA induction upon stress but speeds up isiA downregulation when the upstream trigger is removed.
Experimental verification of the simulated dynamical behavior
Quantitative experimental analyses were performed to confirm the simulations in Fig. 1 B. The impact of sRNA-mediated regulation was investigated by comparing the kinetics of isiA mRNA accumulation in mutant strains expressing different levels of IsrR sRNA. The data points shown in Fig. 2 A are densitometric quantifications of previously published measurements of isiA induction in response to hydrogen peroxide stress (11). These data revealed that the delay in isiA accumulation seen in wild-type cells can be further enhanced in cells overexpressing IsrR, whereas it is abolished in IsrR-depleted cells. The measured time courses thus agree well with the simulation result that sRNAs decelerate target mRNA induction.
We also measured the time course of isiA target mRNA downregulation after removal of the stress trigger (see Fig. S1 A in Data S1). Hydrogen peroxide could not be used as the stimulus in these experiments, since it induces irreversible oxidation of cellular components so that stress might actually persist even after hydrogen peroxide is removed. isiA accumulation was therefore induced by iron depletion (48 h), and isiA expression was subsequently downregulated by iron readdition (t = 0 in Fig. 2 B). The time course measurements after relief from iron stress were in accordance with the model predictions, as the decline in isiA levels was faster in wild-type cells when compared to knockdown cells harboring reduced levels of IsrR sRNA (Fig. 2 B).
Having established a qualitative agreement between experiments and simulations, we next asked whether our model could also quantitatively reproduce the dynamics of isiA target mRNA up- and downregulation. The model parameters were estimated from the time course data shown in Fig. 2, A and B. The trajectories of the best-fit model (solid lines in Fig. 2, A and B) and of its variants (gray and dashed lines in Fig. 2 A and B) indicate that all measurements can be accurately described by a single set of kinetic constants. Remarkably, the best-fit parameters, summarized in Table 1, suggest that the heteroduplex is rapidly degraded, whereas the single-stranded forms of isiA and IsrR are predicted to be much more stable than typical bacterial RNAs (16). A long half-life of isiA directly follows from the slow isiA downregulation kinetics in sRNA-depleted cells (Fig. 2 B; dashed line), whereas the stability of IsrR cannot be as straightforwardly deduced from the time course data. We therefore directly measured the half-life of IsrR sRNA in unstressed cells, which express negligible amounts of isiA target mRNA (11). Very little degradation of IsrR occurred within a 45-min time interval after incubation of cells with the general transcription inhibitor Rifampicin (see Fig. S1 B in Data S1). These data confirm that IsrR sRNA is stable and thus support our best-fit model.
TABLE 1.
Kinetic parameters of the best-fit model
| Figure | Fig. 2 A | Fig. 2 B |
|---|---|---|
| vsyn,T [nM h−1] | 247.2 (initial level) | 996.9 (initial level) |
| 1136 (after stimulus increase at t = 0) | 41.9 (after stimulus removal at t = 0) | |
| kdeg,T [h−1] | 0.42 | 0.42 |
| vsyn,S [nM h−1] | 516.6 (solid line) | 516.6 (solid line) |
| 706.9 (gray line) | 145.3 (dashed line) | |
| 145.3 (dashed line) | ||
| kdeg,S [h−1] | 0.35 | 0.35 |
| KD,P [nM] | 0.0045 | 0.0045 |
| kdeg,P [h−1] | 13.75 | 13.75 |
Pulse filtering properties of sRNA circuits
The above RNA measurements (Fig. 2, A and B) did not discriminate between IsrR-bound (“pair”) and free (“target”) isiA RNA. To get more direct insights into the regulation of isiA action by IsrR sRNA, we simulated time courses of free (i.e., biologically active) isiA target mRNA using the best-fit parameters. Fig. 2 C shows model responses to a step-like suprathreshold pulse stimulation and reveals that regulation by sRNAs gives rise to a sharp, spike-like time course (solid line) when compared to a system devoid of sRNA (dashed line). More specifically, target mRNA accumulation is completely suppressed until all sRNA is degraded (indicated by black circle), and the mRNA decline terminates abruptly, once the sRNA (re)starts to accumulate.
Taken together, our simulations indicate that IsrR-mediated control serves to prevent premature and unnecessarily prolonged isiA synthesis. This is consistent with the hypothesis that IsiA establishes a second line of defense against iron depletion and with the fact that its expression occurs relatively late during iron stress (17). The delay established by sRNA-mediated regulation thus enables cells to induce both early- and late-phase stress proteins in response to a single stress trigger in a temporally ordered manner. Two lines of evidence suggest that spike-like isiA expression (Fig. 2 C) is beneficial to the cellular energy budget. First, isiA becomes the most abundant transcript in cells subjected to oxidative stress (18). Second, IsiA expression saves photosynthesis in stressed cells but decreases photosynthesis efficiency and thus energy production in nonstressed cells (17). Our analyses indicate that IsrR sRNA-mediated control allows avoiding lavish isiA accumulation unless cells are subjected to severe and prolonged stress.
We sought to further confirm our hypothesis that sRNA circuits suppress short stimuli but efficiently transmit prolonged inputs. The time courses of target mRNA expression were, therefore, simulated for step-like stimulus pulses, and the time course maximum was analyzed as a function of pulse duration (Fig. 2 D). We found that the best-fit model (Table 1) indeed responds to the pulse duration in a highly ultrasensitive manner (Hill coefficient nH ≈ 3.5), whereas a much more gradual duration response is seen for a system without sRNA. A similar result was obtained when we analyzed the integral under the target mRNA time courses (Fig. 2 D, inset), thus further confirming that sRNAs establish pulse-filtering and temporal thresholds in biochemical signaling networks.
CONCLUSION
Using a combination of mathematical modeling and quantitative experimental analyses, we have shown that sRNAs establish delays and (steady-state and temporal) thresholds in gene expression. To allow a comparison of our simulations with experimental data, we analyzed the dynamical behavior of models with different sRNA expression levels for a given stimulus level and in most cases compared the wild-type model with a model devoid of sRNA. It should be noted that, from a systems theoretical point of view, the dynamical behavior of both models (±sRNA) is not directly comparable for a given stimulus level, as they differ in their steady-state dose-response curves. However, it can be seen in Fig. 1 B that sRNA-mediated regulation still establishes a delay in mRNA upregulation and accelerates mRNA downregulation if both models (±sRNA) are compared for a given suprathreshold steady-state activation level. In this context, it is important to note that the up- and downregulation response times of the model without sRNA are stimulus invariant (Fig. 1 B, bottom, dotted horizontal line).
The key finding of our work is that sRNA-mediated regulation establishes a sign-sensitive delay for suprathreshold stimulation (Fig. 2, A and B), and this conclusion does not depend on the precise dose-response behavior or on the kinetic parameters chosen. Here, the term “sign-sensitive delay” denotes that a delay is observed exclusively for mRNA upregulation but not for mRNA downregulation (19). This sign-sensitive delay is responsible for the pulse-filtering behavior shown in Fig. 2, C and D: The delay in mRNA upregulation helps to filter out short suprathreshold transients. However, ultrasensitive pulse filtering with a Hill coefficient significantly larger than unity additionally requires that no such delay and, in particular, no memory effects arise if the stimulus is removed. The experimental data presented in the work rule out that mRNA downregulation occurs with a delay (Fig. 2 B) and thus strongly support that ultrasensitive pulse filtering occurs in the cyanobacterial iron stress response. However, the mRNA downregulation kinetics might depend on the history of the system (e.g., short versus long mRNA upregulation) so that explicit pulse-stimulation experiments are required to further prove the existence of ultrasensitive pulse filtering.
In our simulations, we focused mainly on the scenario, where the heteroduplex is less stable than the single-stranded sRNA and target species. However, several bacterial sRNAs do not enhance target degradation but merely act as competitive inhibitors of translation (6). Importantly, sRNAs acting in this way still establish sharp thresholds and delays, as translation of subthreshold target mRNA levels is efficiently suppressed by sequestration into the heteroduplex (20) (Data S1). Eukaryotic miRNA action can also be described by the model scheme depicted in Fig. 1 A, as miRNAs either competitively inhibit translation or induce mRNA degradation. However, miRNAs frequently remain intact after target degradation and can guide the recognition and destruction of additional messages (1). Our numerical analysis revealed that the kinetic properties described above remain valid even if a fraction of the sRNA remains intact during the pair degradation reaction (Data S1) (8). Thus, the main results of this work apply for eukaryotic systems as well, although this remains to be confirmed in more detailed models of miRNA action (21).
In Figs. 1 and 2, we analyzed the dynamic characteristics of mRNA expression. However, the kinetic properties of the sRNA circuit may be obscured by a slow response at the level of protein expression. We therefore investigated numerically whether our conclusions regarding pulse-filtering continue to hold at the protein level. The corresponding simulation results (Fig. S5) demonstrate that for the best-fit parameters pulse filtering is preserved at the protein level even if IsiA protein is assumed to be relatively stable, with a half-life of 10 h. Up to now, studies focusing on the stability of IsiA protein are missing in the literature. However, half-life measurements of the CP43 photosynthesis protein homologous to IsiA revealed a half-life of about an hour under stress conditions (22), and a similar rapid turnover was also reported for another photosynthesis protein, D1 (23). These data suggest that IsiA protein is short lived in the experimental setup we have chosen and that the pulse-filtering property discussed here for the RNA level is observed at the level of proteins as well.
Cells face a specificity problem, as broadly overlapping signaling pathways are activated by diverse stimuli. Biological information is therefore encoded in the quantitative aspects of the signal, such as amplitude and duration (24,25). An important role for the signal duration in the initiation of cell fate decisions was described for various biological networks including mitogen-activated protein kinase signaling (24), transforming growth factor-β signaling (26), cyclic adenosine 3′:5′-cyclic monophosphate signaling (27), and nuclear factor-κB signaling (28). Previous work indicated that multistep regulation in the form of feedforward loops (29,30) and multisite phosphorylation (31,32) allows cells to discriminate transient and sustained stimuli. In this work, we identified competitive inhibition and/or regulated degradation as alternative plausible mechanisms for duration decoding (Fig. 2 D). We propose that the functional analysis of sRNAs might explain why some genes are selectively transcribed upon sufficiently long stimulation (28,33). The results are likely to be of broader relevance, because regulation by protein-protein interactions frequently involves competitive inhibition and regulated degradation as well (34–37).
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Editor: Edward H. Egelman.
References
- 1.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. [DOI] [PubMed] [Google Scholar]
- 2.Tsang, J., J. Zhu, and A. van Oudenaarden. 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 26:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axmann, I. M., P. Kensche, J. Vogel, S. Kohl, H. Herzel, and W. R. Hess. 2005. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 6:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399–404. [DOI] [PubMed] [Google Scholar]
- 5.Altuvia, S. 2007. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10:257–261. [DOI] [PubMed] [Google Scholar]
- 6.Morita, T., Y. Mochizuki, and H. Aiba. 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. USA. 103:4858–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimoni, Y., G. Friedlander, G. Hetzroni, G. Niv, S. Altuvia, O. Biham, and H. Margalit. 2007. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol. Syst. Biol. 3:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine, E., Z. Zhang, T. Kuhlman, and T. Hwa. 2007. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 5:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine, E., P. McHale, and H. Levine. 2007. Small regulatory RNAs may sharpen spatial expression patterns. PLoS Comput. Biol. 3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh, A. K., and L. A. Sherman. 2007. Reflections on the function of IsiA, a cyanobacterial stress-inducible, Chl-binding protein. Photosynth. Res. 93:17–25. [DOI] [PubMed] [Google Scholar]
- 11.Dühring, U., I. M. Axmann, W. R. Hess, and A. Wilde. 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. USA. 103:7054–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legewie, S., N. Bluthgen, and H. Herzel. 2005. Quantitative analysis of ultrasensitive responses. FEBS J. 272:4071–4079. [DOI] [PubMed] [Google Scholar]
- 13.Wagner, E. G., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361–398. [DOI] [PubMed] [Google Scholar]
- 14.Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300:1101–1112. [DOI] [PubMed] [Google Scholar]
- 15.Hargrove, J. L., M. G. Hulsey, and E. G. Beale. 1991. The kinetics of mammalian gene expression. Bioessays. 13:667–674. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA. 99:9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel, K. P., and E. K. Pistorius. 2004. Adaptation of the photosynthetic electron transport chain in cyanobacteria to iron deficiency: the function of IdiA and IsiA. Physiol. Plant. 120:36–50. [DOI] [PubMed] [Google Scholar]
- 18.Golden, S. S. 2006. Good old-fashioned (anti)sense. Proc. Natl. Acad. Sci. USA. 103:6781–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 100:11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrell, J. E. Jr. 1996. Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci. 21:460–466. [DOI] [PubMed] [Google Scholar]
- 21.Levine, E., E. Ben Jacob, and H. Levine. 2007. Target-specific and global effectors in gene regulation by MicroRNA. Biophys. J. 93:L52–L54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichacker, L., H. Paulsen, and W. Rudiger. 1992. Synthesis of chlorophyll a regulates translation of chlorophyll a apoproteins P700, CP47, CP43 and D2 in barley etioplasts. Eur. J. Biochem. 205:17–24. [DOI] [PubMed] [Google Scholar]
- 23.Li, H., and L. A. Sherman. 2000. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. Strain PCC 6803. J. Bacteriol. 182:4268–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebisuya, M., K. Kondoh, and E. Nishida. 2005. The duration, magnitude and compartmentalization of ERK MAP kinase activity: mechanisms for providing signaling specificity. J. Cell Sci. 118:2997–3002. [DOI] [PubMed] [Google Scholar]
- 25.Miller-Jensen, K., K. A. Janes, J. S. Brugge, and D. A. Lauffenburger. 2007. Common effector processing mediates cell-specific responses to stimuli. Nature. 448:604–608. [DOI] [PubMed] [Google Scholar]
- 26.Nicolas, F. J., and C. S. Hill. 2003. Attenuation of the TGF-beta-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-beta-induced growth arrest. Oncogene. 22:3698–3711. [DOI] [PubMed] [Google Scholar]
- 27.Bito, H., K. Deisseroth, and R. W. Tsien. 1996. CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 87:1203–1214. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann, A., A. Levchenko, M. L. Scott, and D. Baltimore. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 298:1241–1245. [DOI] [PubMed] [Google Scholar]
- 29.Shen-Orr, S. S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64–68. [DOI] [PubMed] [Google Scholar]
- 30.Mangan, S., A. Zaslaver, and U. Alon. 2003. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197–204. [DOI] [PubMed] [Google Scholar]
- 31.Deshaies, R. J., and J. E. Ferrell Jr. 2001. Multisite phosphorylation and the countdown to S phase. Cell. 107:819–822. [DOI] [PubMed] [Google Scholar]
- 32.Bluthgen, N., F. J. Bruggeman, S. Legewie, H. Herzel, H. V. Westerhoff, and B. N. Kholodenko. 2006. Effects of sequestration on signal transduction cascades. FEBS J. 273:895–906. [DOI] [PubMed] [Google Scholar]
- 33.Glauser, D. A., and W. Schlegel. 2006. Mechanisms of transcriptional regulation underlying temporal integration of signals. Nucleic Acids Res. 34:5175–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eissing, T., S. Waldherr, F. Allgower, P. Scheurich, and E. Bullinger. 2007. Response to bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys. J. 92:3332–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legewie, S., N. Bluthgen, and H. Herzel. 2006. Mathematical modeling identifies inhibitors of apoptosis as mediators of positive feedback and bistability. PLoS Comput. Biol. 2:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legewie, S., B. Schoeberl, N. Bluthgen, and H. Herzel. 2007. Competing docking interactions can bring about bistability in the MAPK cascade. Biophys. J. 93:2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, S. Y., and J. E. Ferrell Jr. 2007. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 128:1133–1145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.