Abstract
Objective
Lamin B receptor (LBR) is an integral protein of the inner nuclear membrane. Recent studies have demonstrated that genetic deficiency of LBR during granulopoiesis results in hypolobulation of the mature neutrophil nucleus, as observed in human Pelger-Huët anomaly (PHA) and mouse ichthyosis (ic). In this study we have utilized differentiated promyelocytes (EPRO cells) that were derived from the bone marrow of homozygous and heterozygous ichthyosis mice to examine changes to the expression of nuclear envelope proteins and heterochromatin structure that result from deficient LBR expression.
Materials and Methods
Wildtype (+/+), heterozygous (+/ic) and homozygous (ic/ic) granulocytic forms of EPRO cells were analyzed for the expression of multiple lamins and inner nuclear envelope proteins by immunostaining and immunoblotting techniques. The heterochromatin architecture was also examined by immunostaining for histone lysine methylation.
Results
Wildtype (+/+) and heterozygous (+/ic) granulocytic forms revealed ring-shaped nuclei and contained LBR within the nuclear envelope; ic/ic granulocytes exhibited smaller ovoid nuclei devoid of LBR. The pericentric heterochromatin of undifferentiated and granulocytic ic/ic cells was condensed into larger spots and shifted away from the nuclear envelope, compared to +/+ and +/ic cell forms. Lamin A/C, which is normally not present in mature granulocytes, was significantly elevated in LBR-deficient EPRO cells.
Conclusions
Our observations suggest roles for LBR during granulopoiesis which may involve augmenting nuclear membrane growth, facilitating compartmentalization of heterochromatin and promoting down-regulation of lamin A/C expression.
Category: Granulopoiesis
Key words: Lamin B receptor, neutrophil, lamin A/C, heterochromatin
Introduction
The granulocyte nucleus undergoes profound structural changes during the post-mitotic phase of terminal differentiation. Ultrastructural studies of human bone marrow granulopoiesis document significant modifications of nuclear shape, simultaneous with accumulation of peripheral heterochromatin.[1] Recent studies have demonstrated that sufficient cellular levels of a single nuclear envelope integral membrane protein (lamin B receptor, LBR) are necessary for these changes in nuclear architecture.[2] Inadequate levels of LBR during granulopoiesis lead to hypolobulated granulocyte nuclei in humans (Pelger-Huët anomaly, PHA[3]) and in mice (ichthyosis, ic [4]), with concomitant redistribution of heterochromatin.
The interphase nuclear envelope segregates nuclear and cytoplasmic functions, and facilitates intranuclear compartmentalization. Current concepts of the nuclear envelope[5–10] visualize a multi-tiered structure with a specialized inner nuclear membrane contiguous via nuclear pores to the outer nuclear membrane-endoplasmic reticulum system, but otherwise separated by a perinuclear space. Internal to the inner nuclear membrane is a meshwork layer of intermediate filaments (lamins) interacting directly or indirectly with an underlying layer of peripheral heterochromatin. The lamin network is thought to convey structural stability to the nuclear envelope and link it to the cytoskeleton.[11, 12] Integral proteins of the inner nuclear membrane are believed to tie together the various layers (membrane-lamina-heterochromatin) and may play a role in anchoring interphase chromosomes to the nuclear periphery.[10, 13, 14]
Our previous studies have employed the human leukemic cell line (HL-60) to examine changes in the nuclear envelope and heterochromatin composition, as a model for the changes during normal human granulopoiesis.[15, 16] However, HL-60 cells are an imperfect model, since various cytoplasmic and cell function features of normal peripheral blood neutrophils are not observed in the granulocytic forms of HL-60 cells (for discussion, see[17]). More faithful cell models for normal granulopoiesis are mouse bone marrow-derived cell lines: promyelocytes[18] (MPRO) and erythroid-myeloid-lymphoid/early promyelocytes (EML/EPRO cells).[19] These cell lines exhibit many properties of normal murine granulocytes, including upregulated secondary granule transcripts[20] and respiratory burst, chemotaxis, phagocytosis and upregulation of the cell surface antigen Mac-1.[17, 21] Both MPRO and EML/EPRO cells are derived from cells that have been transfected with a dominant negative form of retinoic acid receptor-α. MPRO cells grow in medium containing GM-CSF; EML cells require SCF. The previous article[22] describes the generation and properties of EML cells from murine ichthyosis bone marrow and development of EPRO cell lines. Ichthyosis (ic) is a frame shift mutation in LBR that changes amino acids 365–385, resulting in a stop codon at residue 386; homozygous mutants reveal no detectable LBR protein.[4] The present article describes the properties of EPRO nuclei during ATRA-induced granulocyte differentiation, employing cells from three different genotypes: wildtype (+/+), heterozygous ichthyosis (+/ic) and homozygous ichthyosis (ic/ic).
Materials and Methods
Cell lines
Cultivation of the EML cells and generation of the EPRO subfraction were performed as described earlier.[22] For most studies, EPRO cells were induced to differentiate to mature granulocytes by addition of 10 µM ATRA for four days.
Western Blot analysis
Undifferentiated and differentiated (4 days of ATRA treatment) EPRO-+/+, EPRO-+/ic and EPRO-ic/ic cells were washed in phosphate-buffered saline (PBS) containing the protease inhibitors Complete mini (Roche Diagnostics, Indianapolis, IN) and 7.5 µl/ml saturated phenylmethylsulphonyl fluoride (PMSF, Sigma-Aldrich Inc., St Louis, MO), and lysed in 1 × Laemmli sample as previously described.[16] Lysates were pushed 6–12 times through a #26 needle to shear DNA. Total cell extracts (10 µl, corresponding to 3 × 105 cells) and molecular weight markers (MagicMark XP, Invitrogen, Carlsbad, CA) were electrophoresed in 4–20% precast gels (Bio-Rad, Hercules, CA) and electroblotted. All immune reactions were carried out in 10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.05% Tween-20 (TBST) with 5% dried milk at RT with washing steps in TBST.
Immunofluorescence analysis
Undifferentiated and differentiated (4 days of ATRA treatment) EPRO-+/+, EPRO-+/ic and EPRO-ic/ic cells were counted and diluted in PBS. Fixation and staining methods have been previously described.[16, 23, 24] In brief, 1 × 105 cells were centrifuged onto fresh polylysine-coated slides for 5 min, fixed in 4% formaldehyde (PFA) in PBS for 15 min, permeabilized with 0.1% Triton X-100, washed in PBS and blocked with 5% normal donkey serum in PBS for 30 min. Alternatively, cells were fixed in anhydrous methanol (−20°C, 10 min) and washed in PBS. Fixed cells were incubated with antibodies for 1 hour at 37°C, with the secondary antibody including 4,6 diamidino-2-phenylindole (DAPI, Sigma-Aldrich, Inc.). Washed slides were mounted in Slow-Fade Antifade Kit (Molecular Probes, Invitrogen). Images were collected on a Zeiss 510 Meta laser confocal microscope, using a 100x objective.
Antibodies
The following antibodies were used for immunoblot analysis and/or immunofluorescence analysis: Polyclonal guinea pig serum against lamin B receptor;[3] polyclonal guinea pig serum against emerin;[25] goat anti-lamin B1 and goat anti-lamin A/C (Santa Cruz Biotechnology, Santa Cruz, CA; catalogue # sc-6216 and sc-3215); mouse monoclonal anti-lamin B2 (clone X223) and mouse monoclonal anti-Lamin A/C (clones X67 and X167) (Progen, Heidelberg, Germany; catalogue # 65147C, 65147A and 65147B, respectively); rabbit anti-histone H3, rabbit anti-trimethyl H3K9 and rabbit anti-trimethyl H4K20 (Upstate, Charlottesville, VA; catalogue # 07–690, 07–442 and 07–463); mouse anti-HP1 α, mouse anti-HP1 β and mouse anti-HP1 γ (Chemicon International, Temecula, CA; catalogue # MAB3584, MAB3448 and MAB3450); monoclonal anti-γ-tubulin (clone GTU-88, Sigma; catalogue # T6557). As kind gifts, we received rabbit anti-C-Nap1 from Eric Nigg (MPI Biochemistry, Martinsried, Germany), rabbit anti-phosphorylated H3S10 and phosphorylated H4/H2AS1 (denoted H3(S10)phos and rabbit H4/H2A(S1)phos) from David Allis, (Rockefeller University, NY; H3(S10)phos is also available from Upstate) and mouse anti-LAP2β from Amos Simon (Tel-Aviv University, Israel). All secondary antibodies (FITC-, Cy3- and Cy5-conjugated and minimally cross-reactive) were obtained from Jackson ImmunoResearch Laboratory (West Grove, PA). Primary and secondary antibody dilutions followed suggestions by the supplier.
Image J analysis
For nuclear size quantification of EPRO cells, images of DAPI-stained undifferentiated EPRO +/+, +/ic and ic/ic nuclei were acquired by conventional fluorescence microscopy. Using Image J software, nuclei boundaries were traced, areas measured in pixels2 and converted into µm2 (400 nuclei for each genotype). For quantification of the size and location of pericentric heterochromatin, undifferentiated and granulocytic EPRO-+/+, -+/ic and -ic/ic cells were treated with anti-HP1γ and anti-lamin B1 antibodies for staining of heterochromatic spots and the nuclear envelope, respectively. Confocal images were acquired, spots in 200 nuclei for each genotype (undifferentiated and granulocytic forms) were counted and associated with one of the following attributes: small diameter (<0.75 µm), medium (0.75 – 1.5 µm) or large (> 1.5 µm); touching the nuclear envelope or located in the interior of the nucleus.
Results
The level of LBR in EPRO cell lines
EPRO (early promyelocytes) cell lines (+/+, +/ ic and ic/ic) were obtained from the parent EML cells by treatment with IL-3, SCF and ATRA for 3 days. Subsequent granulocytic differentiation occurred over a 5 day period in the presence of GM-CSF and 10 µM ATRA. Since apoptotic cell death became evident by day 5, most cell analyses were performed on cells differentiated for 4 days with ATRA. The location and amounts of LBR in the different cell lines and during granulocytic differentiation were monitored by confocal immunofluorescence and by immunoblotting of total cell extracts.
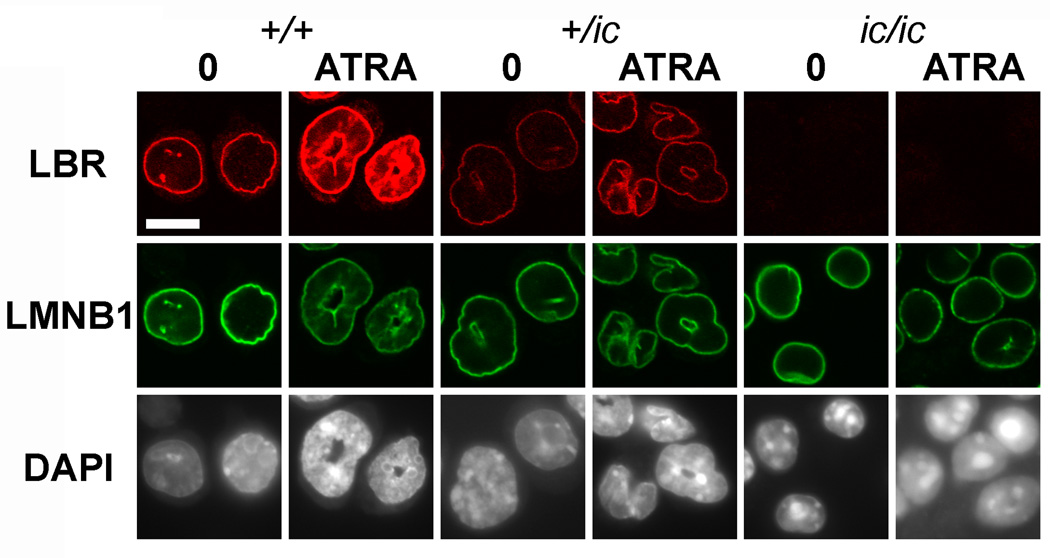
Immunostaining with guinea pig anti-LBR revealed the changing levels of LBR among the different cell lines and during ATRA-induced granulocytic differentiation (Figure 1). Also shown in Figure 1 are confocal immunostaining results with goat anti-lamin B1, and DNA localization revealed by DAPI staining (not confocal). The confocal detector gain was set for the undifferentiated (day 0) EPRO-+/+ cells and remained unchanged, during image data collection of the undifferentiated and granulocytic forms of the +/+, +/ic and ic/ic cell lines. Thus, a qualitative impression of the levels of LBR and lamin B1 can be obtained, comparing the different frames for each row of Figure 1, which are representative images of many immunostained cells. Comparing only the undifferentiated cell forms, a reduction in the nuclear envelope levels of LBR can be seen, progressing from +/+ to +/ic to ic/ic. Granulocytic differentiation by treatment with 10 µM ATRA for 4 days resulted in an evident increase of LBR in the nuclear envelope of +/+; but no LBR was apparent in granulocytic ic/ic cells. The levels of lamin B1 within the nuclear envelope appear to remain relatively unchanged, comparing the undifferentiated and granulocytic forms of the three cell lines. Furthermore, the confocal data demonstrated clear co-localization of LBR and lamin B1 staining in the EPRO-+/+ and -+/ic undifferentiated and granulocytic cell forms.
Figure 1.
Confocal immunostaining of undifferentiated and granulocytic EPRO cells with anti-LBR and anti-lamin B1. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ ic; homozygous ichthyosis, ic/ ic. Cell states: 0, undifferentiated; ATRA, granulocytic forms on day 4. Stains: anti-LBR (red); anti-LMNB1 (lamin B1, green); DAPI (DNA, uncolored). Fixation: PFA. Scale bar: 10 µm.
Nuclear shape and internal architecture differences are also apparent in the image data of Figure 1, especially comparing the anti-lamin B1 and DAPI images. Most apparent is the absence of ring-shaped nuclei in the ATRA-induced granulocytic forms of EPRO-ic/ic cells. In addition, the nuclei of undifferentiated and granulocytic ic/ic cells appear smaller in diameter than the corresponding forms of the +/+ and +/ic genotypes. Table I presents a tabulation of area measurements (using Image J) from projections of undifferentiated EPRO cells. Assuming that these nuclei can be approximated to "equivalent spheres", we estimate that ic/ic nuclei have a surface area that is ~40–50% of the surface area of +/+ cells. An additional architectural difference pertaining to the DAPI-bright regions, corresponding to AT-rich pericentric heterochromatin,[26–28] can be observed in Figure 1. These differences were quantitated, demonstrating that the heterochromatic spots are fewer in number and larger in size in the EPRO-ic/ic cells, compared to the -+/+ and -+/ic forms (Table II).
TABLE I.
AREAS OF UNDIFFERENTIATED EPRO NUCLEI* (µm)2
| +/+ | +/ic | ic/ic | |
|---|---|---|---|
| Average +/− S.D. | 109.2 +/− 32.4 | 97.9 +/− 25.4 | 54.8 +/− 16.0 |
| Mode | 87.0 | 60.9 | 37.2 |
| Relative surface area¥ | 1.0 | 0.90 | 0.50 |
| Relative surface area§ | 1.0 | 0.70 | 0.43 |
Number of nuclei counted for each genotype: 400.
Relative surface areas of "equivalent spheres" were calculated from the following formulas: 1) area of circle = πr2; 2) surface area of a sphere = 4 πr2. Measurements employed: ¥ from Average; § from Mode.
TABLE II.
CENTROMERIC HETEROCHROMATIN: "SPOT" SIZE AND LOCATION
| +/+ | +/ic | ic/ic | ||||
|---|---|---|---|---|---|---|
| 0 | ATRA | 0 | ATRA | 0 | ATRA | |
| SIZE§ | ||||||
| SMALL <0.75 µm | 75.5 | 83.1 | 72.6 | 73.5 | 28.4 | 19.4 |
| MEDIUM 0.75–1.5 µm | 22.8 | 16.1 | 25.5 | 25.0 | 45.5 | 48.3 |
| LARGE >1.5 µm | 1.6 | 0.8 | 1.8 | 1.5 | 26.2 | 32.1 |
| LOCATION | ||||||
| INTERIOR | 28.0 | 15.8 | 34.1 | 32.3 | 56.9 | 67.7 |
| Touching Nuclear Envelope | 72.0 | 84.2 | 65.9 | 67.7 | 43.1 | 32.3 |
| Spots/Nucleus¥ | 5.0 | 5.4 | 6.0 | 6.5 | 3.7 | 4.3 |
| N (total spots counted) | 105 | 1071 | 1188 | 1285 | 737 | 851 |
% of total counted
Average number of centromeric "spots" per nucleus in a 1 µm thick confocal slice.
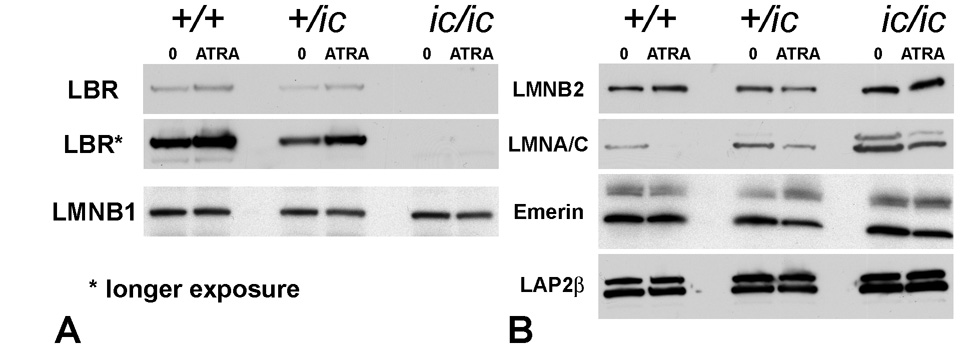
Immunoblot results from total cell extracts of undifferentiated and ATRA-treated EPRO (+/+, +/ic and ic/ic) cell lines are presented in Figure 2. Total cell extracts from the same number of cells were loaded in each lane, in agreement with densitometry of the ECL film intensities of rabbit anti-H3 (example data shown in Figure 5). Therefore, an approximate comparison of band intensities for any specific antigen should reflect the average total cellular content of that protein. Figure 2A shows very clearly that ic/ic undifferentiated and granulocytic forms possess negligible amounts of LBR, even after prolonged film exposure. The average amount of LBR in +/ic cell extracts appears discernibly lower than in +/+ cells, comparing between undifferentiated cells or between granulocytic forms. Furthermore, an increase in the cellular level of LBR can be visualized for both +/+ and +/ic, comparing granulocytic forms with undifferentiated cells of the same genotype. Also of interest, due to the relevance to Figure 1, the average cell levels of lamin B1 do not reveal any systematic or significant changes in cellular content, comparing cells of the different genotypes and extent of differentiation.
Figure 2.
Immunoblotting of total cell extracts from undifferentiated and granulocytic EPRO cells with antibodies to nuclear envelope components. (A) anti-LBR and anti-lamin B1. Rows: LBR, anti-LBR; LBR*, anti-LBR (prolonged exposure); LMNB1, anti-lamin B1. (B) anti-lamin B2, anti-lamin A/C, anti-emerin and anti-LAP2β. Rows: LMNB2, anti-lamin B2; LMNA/C, antilamin A/C; emerin, anti-emerin; LAP2β, anti-LAP2β. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ ic; homozygous ichthyosis, ic/ ic. Cell states: 0, undifferentiated; ATRA, granulocytic forms on day 4.
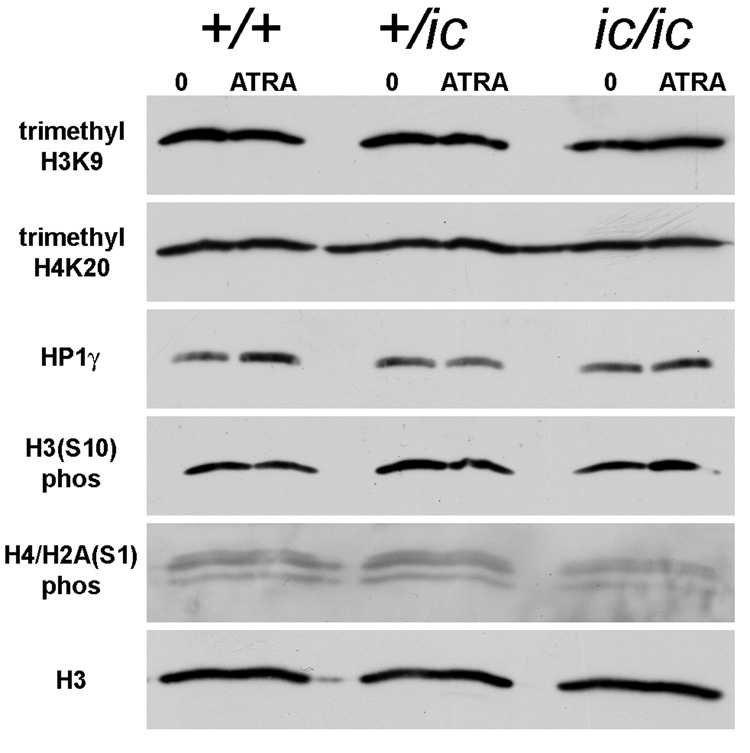
Figure 5.
Immunoblotting of total cell extracts from undifferentiated and granulocytic EPRO cells with antibodies to condensed chromatin markers. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ ic; homozygous ichthyosis, ic/ ic. Cell states: 0, undifferentiated; ATRA, granulocytic forms on day 4. Antibody specificities are listed in the left margin.
Approximate estimates of the changes in LBR amounts per cell were obtained from densitometric scans of the ECL photographic films; employing line scan features of ImageJ software (see Materials and methods). The apparent decrease in LBR, comparing undifferentiated +/ic to undifferentiated +/+ cells was ~0.6 fold. For EPRO-+/+ cells, the approximate increase in LBR after 4 days of ATRA treatment was ~1.5 fold; for EPRO-+/ic cells the increase was ~1.7 fold.
Other nuclear envelope proteins in EPRO cell lines
It was of interest to determine whether other nuclear envelope proteins might change in cellular content in correlation with the disappearance of LBR, comparing EPRO-+/+, -+/ic and -ic/ic cells in undifferentiated and granulocytic ATRA-treated cell states. Immunoblot results are shown in Figure 2B. As with lamin B1, mouse monoclonal anti-lamin B2 reveals no systematic and significant changes in amounts, comparing the different cell states (possibly a slight increase in ic/ic cells). By contrast, mouse monoclonal anti-lamin A/C reveals systematic and significant changes. Comparing only the undifferentiated EPRO cells, lamin A/C increases in cellular amount progressing from +/+ to +/ic to ic/ic. Adding together lamin A+C and estimating the relative increase (from scanned films using ImageJ), we calculate that +/ic contains ~2.5 fold and ic/ic contains ~6.2 fold more lamin A/C than +/+ undifferentiated EPRO cells. Lamin C is the major isoform in all of the examined EPRO cells lines. In addition, in all the cell genotypes, there is a reduction of lamin A/C resulting from the ATRA-induced granulocytic differentiation; i.e., relative amounts (induced/uninduced) equal to ~0.2 (+/+), ~0.5 (+/ic) and ~0.6 (ic/ic). Similar results were noted with blots employing goat antisera against lamin A/C (data not shown). Also shown in Figure 2B, guinea pig anti-emerin and mouse monoclonal anti-LAP2β both indicated relatively constant levels of these nuclear envelope proteins in the various cell states.
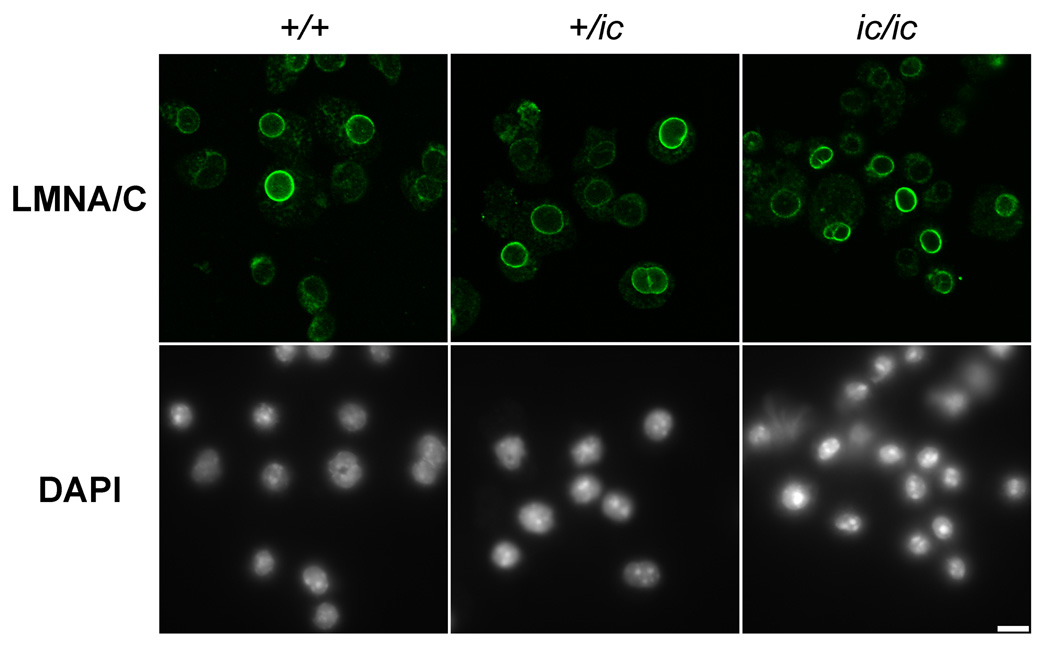
Confocal immunostaining experiments on EPRO cells were generally consistent with the immunoblotting experiments (described above). Fixation conditions had to be optimized for the various antibodies. Goat, guinea pig and mouse monoclonal anti-lamin A/C did not stain well after PFA fixation, giving high background staining. Methanol fixation worked well for the goat and guinea pig antisera, but not the mouse monoclonal anti-lamin A/C. Figure 3 presents images of undifferentiated EPRO +/+, +/ic and ic/ic fixed with methanol and stained with goat anti-lamin A/C. Immediately apparent is the variability of nuclear envelope staining, comparing cell-to-cell. Similar cell-to-cell variation in nuclear envelope staining was observed with granulocytic forms of EPRO cells (data not shown). Methanol fixation was also necessary when staining with mouse monoclonal anti-lamin B2 (data not shown). Several points were clear: 1) anti-lamin B2 co-localized with anti-lamin A/C at the nuclear periphery; 2) cell-to-cell variations in anti-lamin B2 staining were observed; 3) no systematic change in anti-lamin B2 staining was observed, comparing undifferentiated and granulocytic cell forms. Mouse monoclonal anti-LAP2β also stained poorly after PFA fixation, requiring methanol fixation. As with lamin A/C and lamin B2, LAP2β revealed cell-to-cell variation of intensity of nuclear envelope staining, with no systematic changes comparing the different cell forms (Figure 4).
Figure 3.
Confocal immunostaining of undifferentiated EPRO cells with goat anti-lamin A/C. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ ic; homozygous ichthyosis, ic/ic. Stains: anti-LMNA/C (lamin A/C, green); DAPI (DNA, uncolored). Fixation: methanol. Scale bar: 10 µm.
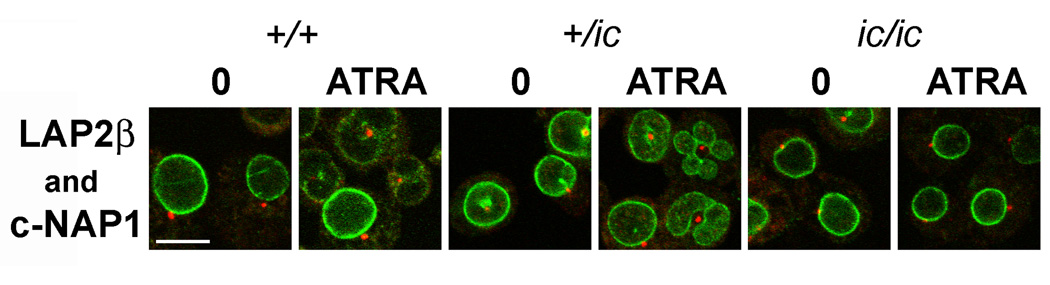
Figure 4.
Confocal immunostaining of undifferentiated and granulocytic EPRO cells with anti-LAP2β and anti-C-Nap1. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ic; homozygous ichthyosis, ic/ ic. Cell states: 0, undifferentiated; ATRA, granulocytic forms on day 4. Stains: anti-C-Nap1 (red); anti-LAP2β (green); DAPI (DNA, uncolored). Fixation: methanol. Scale bar: 10 µm.
Heterochromatin markers in EPRO cell lines
Previous studies from our laboratory have explored the presence of various heterochromatic markers in normal human and mouse peripheral blood granulocytes and in differentiating MPRO cells.[23] The conclusion of these studies was that numerous repressive histone lysine methylations were present in normal granulocytes and in MPRO cells; but only negligible amounts of HP1 proteins were observed. In the case of normal mouse granulocytes and MPRO cells, co-localization of staining was seen for me3H3K9, me3H4K20, HP1γ and DAPI-bright pericentric heterochromatin. In fact, mature mouse neutrophils frequently possess several nuclear "drumsticks" (tiny lobes) containing the DAPI-bright heterochromatin stained with these heterochromatin markers.
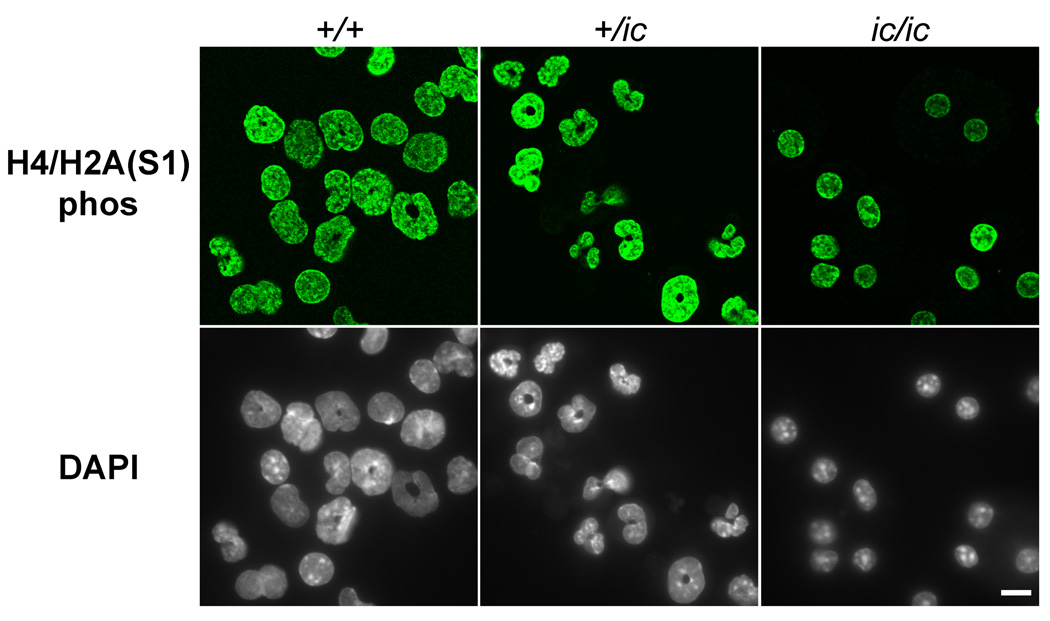
It was therefore of interest to explore the quantities and locations of heterochromatin markers in the EPRO cell lines with varying genotypes, in undifferentiated and granulocytic cell states. Figure 5 presents several immunoblots, performed to estimate the amounts of various heterochromatic markers in the undifferentiated and ATRA-treated EPRO-+/+, -+/ic and -ic/ic cell forms. Total cell extracts from the same number of cells were loaded in each lane, in agreement with the stain intensities of anti-H3 (shown at the bottom of Figure 5). It is clear that me3H3K9, me3H4K20 and HP1γ are present in all cell states, with no obvious changes in amounts. Also shown are two phosphorylated histone "mitotic" markers,[29–31] H3(S10)phos and H4/H2A(S1)phos, which also reveal no significant or systematic changes when comparing the various cell forms.
Confocal immunostaining images of selected heterochromatic markers (Figure 6) indicate that there is good co-localization of HP1γ, me3H4K20 and DAPI in the regions of pericentric heterochromatin, much as previously observed in MPRO cells and normal murine granulocytes.[23] In other experiments (data not shown), me3H3K9 showed good co-localization with HP1γ in the DAPI-bright regions. Furthermore, Figure 6 illustrates the considerable consolidation of pericentric heterochromatic regions within EPRO-ic/ic nuclei and the shift of these regions toward the center of the nucleus. Quantization of both of these structural changes in pericentric heterochromatin is presented in Table II. Neither of the repressive histone methylation epitopes nor HP1γ showed any significant or systematic change in staining intensity, comparing the various EPRO genotypes and differentiation states. Immunostaining with anti-HP1α and HP1β revealed some "spotted" staining of the nucleoplasm, with only partial co-localization to the DAPI-bright regions and no systematic differences among the different EPRO cell forms (data not shown). The phosphorylated histone "mitotic" markers, H3(S10)phos and H4/H2A(S1)phos, revealed extremely strong staining of mitotic chromosomes. In the case of H3(S10)phos, most EPRO cells (in all states) did not stain; a few showed spots that co-localized with DAPI-bright regions (data not shown). By contrast, H4/H2A(S1)phos yielded comparably strong nuclear staining in all the EPRO cell forms (Figure 7, shows only the granulocytic forms)‥
Figure 6.
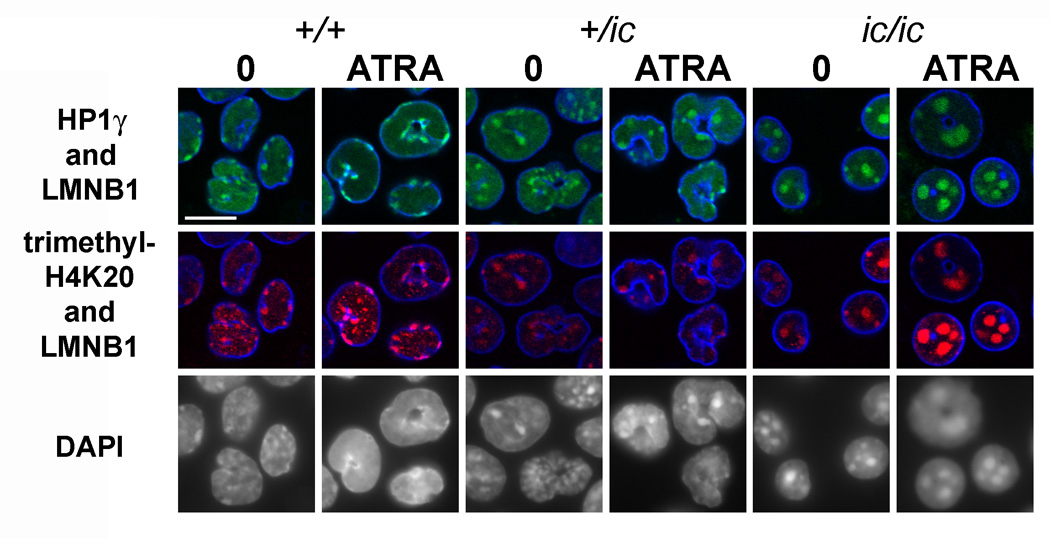
Confocal immunostaining of undifferentiated and granulocytic EPRO cells with anti-HP1γ, anti-me3H4K20 and anti-lamin B1. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ ic; homozygous ichthyosis, ic/ ic. Cell states: 0, undifferentiated; ATRA, granulocytic forms on day 4. Stains: anti-HP1γ (green); anti-me3H4K20 (red); anti-LMNB1 (lamin B1, blue); DAPI (DNA, uncolored). Fixation: PFA. Scale bar: 10 µm.
Figure 7.
Confocal immunostaining of differentiated EPRO cells with rabbit anti-H4/H2A(S1)phos. Genotypes: wildtype, +/+; heterozygous ichthyosis, +/ ic; homozygous ichthyosis, ic/ ic. Stains: anti-H4/H2A(S1)phos (green); DAPI (DNA, uncolored). Fixation: PFA. Scale bar: 10 µm. *
Centrosome location in EPRO cell lines
Previous studies from our laboratory have argued that microtubule integrity is important for lobulation of the granulocyte nucleus,[24] and that the centrosomal region can be found deep within the crevasses between the lobes of human granulocytes or in the central hole of murine granulocytes.[32] In the present study, immunostaining was performed on EPRO-+/+, -+/ic and -ic/ic cells, undifferentiated and ATRA-treated, employing two antibodies against different components of the centrosomal region, γ-tubulin and C-Nap1.[33] Figure 4 presents confocal slices merging anti-C-Nap1 (red) with anti-LAP2β (green). As previously observed for MPRO cells[32], the centrosomal region is adjacent to the convex nuclear envelope in the undifferentiated EPRO-+/+, -+/ic and -ic/ic cells; but buried deep within the central holes of the toroidal nuclear envelope in ATRA-treated EPRO-+/+ and -+/ic cells. However, the ATRA-treated EPRO-ic/ic cells retain the ovoid nuclear shape with the centrosomal region adjacent to the convex nuclear envelope, consistent with the lack of granulocytic nuclear shape differentiation. Identical results were obtained with anti-γ-tubulin (data not shown).
Discussion
During granulopoiesis in adult bone marrow, lamin B receptor (LBR, an integral membrane protein of the nuclear envelope) is necessary for normal differentiation of nuclear shape and heterochromatin distribution.[2] LBR resides in the inner nuclear membrane of the interphase nucleus, possessing a highly positively charged N-terminus (~200 amino acid residues, believed to interact with lamin B, HP1 and chromatin) and a C-terminus (~400 amino acid residues, with 8 putative transmembrane segments and sterol reductase activity).[34] At present, the mechanism by which LBR controls granulocyte nuclear shape and heterochromatin distribution remains largely speculative.[2, 24] Even so, the necessity for sufficient amounts of LBR is well documented from analysis of single gene mutations in human Pelger-Huët anomaly (PHA)[3] and murine Ichthyosis (ic).[4] The present study attempts to define some of the consequences to nuclear architecture and composition that occur during granulopoiesis in situations of LBR deficiency. Observations were made on bone marrow-derived EPRO cells[20, 21, 22] from wildtype (+/+), heterozygous ichthyosis (+/ic) and homozygous ichthyosis (ic/ic) adult mice. These EPRO cells were further differentiated with ATRA to yield mature granulocytic forms for microscopic and biochemical analyses.
Comparing ic/ic cells to corresponding +/+ and +/ic cell forms, there were several major nuclear architectural differences: 1) ic/ic granulocytes did not possess the characteristic ring-shaped nuclei, but appeared spherical or slightly indented; 2) ic/ic undifferentiated cell forms possessed smaller nuclei than the wildtype or heterozygous forms; 3) pericentric heterochromatin in ic/ic cells coalesced into fewer and larger nuclear spots, with a higher percentage shifted from peripheral to central nuclear locations, compared to the wildtype (+/+) or heterozygous (+/ic) cell forms. Thus, the absence of sufficient LBR produces profound effects on EPRO nuclear shape, size and heterochromatin distribution.
Immunochemical studies (i.e., confocal immunostaining and immunoblotting) also resulted in a number of significant observations. As expected from our earlier investigation,[4] ic/ic cells revealed no indication of any LBR protein in undifferentiated or granulocytic forms. Undifferentiated heterozygous +/ic cells had discernibly less LBR (~60%) than the wildtype +/+ cells, and both revealed an increase in LBR amount per cell (~1.5 to 1.7 fold) following granulocytic differentiation with ATRA. This increase in LBR during in vitro granulopoiesis of EPRO cells agrees with our previous observations with MPRO cells (unpublished) and with differentiating HL-60 cells (although the increase in LBR in HL-60 cells appears to be ~3 to 4 fold).[16, 35] It is notable that bone marrow is the body tissue with the highest expression of LBR mRNA.[36] An unexpected observation was the apparent increase on immunoblots of lamin A/C, increasing systematically from +/+ to +/ic (~2.5 fold) to ic/ ic (~6.2 fold). Furthermore, for all three genotypes, ATRA-induced granulocytic differentiation resulted in a decline in the level of lamin A/C. Thus, there is a general correlation between an increase in cellular LBR content and a decrease in lamin A/C, as seen after ATRA treatment or progressing from ic/ic to +/ic to +/+ genotypes. Absence of lamin A/C in mouse granulocytic (and lymphocytic) forms has been clearly documented in earlier studies,[37] consistent with our studies on differentiating HL-60 cells[16] and peripheral human blood granulocytes (unpublished). Another surprising observation was the relative uniformity and constancy of staining and immunoblotting by anti-lamin B1 with the various EPRO cell forms. This observation was surprising to us because of prior documentation of low amounts of lamin B1 in differentiating human HL-60 cells[16, 24] and human peripheral blood granulocytes.[23] However, this observation of clearly detectable lamin B1 in EPRO cells is consistent with our published images on MPRO cells and on mouse peripheral blood granulocytes.[23] Collectively, these observations argue for some differences in the nuclear envelope composition, comparing human and mouse granulocytes.
Speculations on the role of LBR in determining granulocyte nuclear shape have been previously summarized.[2, 24] Basically, it is suggested that LBR attaches the deformable lamin-depleted nuclear envelope to the underlying heterochromatin. Furthermore, it is postulated that the shape changes are powered by microtubule motors attached to the nuclear envelope and pulling towards the juxtanuclear centrosomal region to generate invaginations in the nuclear envelope. The present intriguing observation that ic/ic undifferentiated EPRO cells possess smaller nuclei than their wildtype (+/+) counterparts (Table I), suggests additional speculation. The smaller ic/ic nuclei probably possess a reduced nuclear envelope surface area (~40–50%), compared to +/+ cells. This would be consistent with our prior observations that during granulocytic differentiation of HL-60 cells, there is an increase of LBR content and an increase of nuclear envelope surface area (lobulation plus envelope-limited chromatin sheets, "ELCS").[15, 16, 35] A key question is whether the additional LBR promotes growth of the nuclear envelope.
There is increasing evidence from other laboratories that LBR stimulates nuclear envelope production. Over-expression of heterologous LBR in yeast cells (which have neither endogenous LBR nor lamin B) produces large amounts of membrane stacks, affiliated with and independent from the nuclear envelope.[38] Over-expressing LBR and portions of LBR in HeLa cells yields production of nuclear envelope associated membrane stacks.[39] Furthermore, these authors presented evidence that the C-terminal transmembrane region of LBR is responsible for the membrane overproduction, while the N-terminal portion is responsible for chromatin attachment (possibly mediated by importin β). Based upon these studies, we speculate that LBR has an inherent capability to promote membrane growth, when situated in the nuclear envelope or in the endoplasmic reticulum. The bifunctionality of LBR (i.e., the lamina-heterochromatin binding capability of the N-terminal portion and the membrane building function of the C-terminal portion) would insure membrane growth within the nuclear envelope. Defining the function of LBR in this manner, formulates a functional significance that justifies the evolutionary stability of this chimeric molecule throughout vertebrate evolution.[8] It is still not clear what role the sterol reductase property of LBR[40] may play in membrane production or cholesterol biosynthesis, since there is now clear evidence of enzymatic redundancy with the cytoplasmic enzyme DHCR14 (TM7SF2, SR-1).[41] This proposed integrated function of LBR (i.e., promoting growth of the inner nuclear membrane around heterochromatin) is consistent with the observation that LBR is among the earliest of the nuclear envelope proteins to associate with decondensing mitotic chromosomes during nuclear envelope reformation.[42, 43] It is clear, however, that other proteins must possess similar capabilities, since normal appearing nuclear envelopes can form around the chromatin of LBR-deficient ic/ic nuclei.
It is important to determine the basis of the observed negative correlation of nuclear LBR and lamin A/C content. Clearly, this negative correlation is not absolute; many cell types have both LBR and lamin A/C co-existing within the same nuclear envelope. Furthermore, confined to hematologic cell types (normal and malignant), the published literature is confusing with regards to amounts of lamin A/C (see a recent comprehensive review[44]). Even so, present studies from our laboratory and our earlier results with HL-60 cells,[16] stimulate some speculative thoughts. Evidence for direct in vivo interactions between LBR and lamin A/C is controversial: 1) co-immunoprecipitation of LBR and lamin A/C has been described;[45] 2) a single lamin A mutation has been reported to result in LBR moving from the nuclear envelope to the endoplasmic reticulum;[46] 3) but, lamin A null cells appear to have normal retention of LBR in the nuclear envelope.[47] It seems necessary to consider a more indirect coupling between LBR and lamin A/C. A recent study of the amounts of lamin A/C in various hematologic malignancies provided evidence for epigenetic silencing at the lamin A/C promoter region by CpG island methylation in a subset of leukemias and lymphomas.[48] There is increasing evidence that gene silencing occurs at the nuclear envelope (see recent reviews [10, 13]) promoted, in part, by integral proteins of the nuclear envelope. Thus, it is tempting to speculate that increased LBR content may augment repression of the lamin A/C genes and/or facilitate sequestration of the epigenetically repressed lamin A/C genes to the peripheral nuclear heterochromatin.
Similar considerations may apply to the repositioning of pericentric heterochromatin in the EPRO cells, comparing +/+, +/ic and ic/ic genotypes. The reduced amount of LBR content in the undifferentiated cell types results in a shift of centromeres away from proximity to the nuclear envelope. This conclusion is based upon published studies demonstrating that the DAPI-bright AT-rich pericentric heterochromatic regions of mouse cells are enriched in me3H3K9, me3H4K20 and HP1,[26–28] also observed in MPRO cells and normal mouse granulocytes[23] and the present analysis of these regions in undifferentiated and ATRA-treated EPRO cells (Table II). Comparing undifferentiated EPRO cells, the percentage of pericentric heterochromatic regions adjacent to the nuclear envelope declines with decreased LBR: +/+, 72%; +/ic, 66%; ic/ic, 43%. The shift is even more striking, comparing ATRA-treated EPRO cells: +/+, 84%; +/ic, 68%; ic/ic, 32%. It seems reasonable to speculate that LBR interacts with pericentric heterochromatin, sequestering it to close proximity with the nuclear envelope. This association could be promoted indirectly by LBR-HP1 interactions[49] and/or by direct binding between LBR and methyl lysine-modified nucleosomes.[50] In parallel to the repositioning of these regions, the apparent number of pericentric "spots" per nucleus (confocal slice) declines slightly and their size increases. For example, the percentage estimate of spots (>1.5 µm diameter) per nucleus in ATRA-treated cells is: +/+, 1%; +/ic, 2%; ic/ic, 32%. Coalescence of centromeric heterochromatin has been previously described in terminally differentiating mouse myotube cells,[51] with evidence presented for central roles in the aggregation process due to increased levels of methylated DNA binding proteins, MeCP2 and MBD2, and of heterochromatic DNA methylation. However, no shift of the centromeric heterochromatin towards the nuclear envelope was reported. A more recent study of mutant mouse ES cells lacking DNA methylation has challenged this postulated central role of DNA methylation and methylated DNA binding proteins in heterochromatin aggregation, suggesting instead that histone methylation and H1 binding to centromeric heterochromatin are the principal factors.[52] It is unclear whether this disagreement is a consequence of examining and comparing different types of mouse cells. Clearly, both sets of explanations for centromeric heterochromatin condensation must be explored in granulopoietic cell systems. We suggest that during normal granulopoietic nuclear differentiation, LBR pulls pericentric heterochromatin towards the nuclear envelope simultaneously with heterochromatin aggregation, promoted in part by chromatin epigenetic modifications. Thus, there may be a balance of forces tugging on heterochromatin to achieve normal chromatin compartmentalization in the granulocyte nucleus.
Why is the mouse granulocyte nucleus ring-shaped;[53] whereas the LBR-deficient granulocytic form is ovoid? We have speculated[32] that the ring-shape may be related to the frequently described "rosette" arrangement of chromosomes at the metaphase plate.[54, 55] During mitosis in the bone marrow of normal mice, the telocentric heterochromatin should situate near the central "hub" of the mitotic "rosettes" and be pulled towards the polar centrioles. We suggest that post-mitotic nuclear reformation kinetics may be influenced by this central distribution of centromeric heterochromatin, which might associate with LBR to facilitate membrane growth in the vicinity of the centrosomes creating an annulus. In the absence of LBR, centromeric heterochromatin would have no particular affinity to the nuclear periphery and might have a weakened influence on the final nuclear shape and/or the kinetics of nuclear envelope reformation.
The development of hematopoietic cell lines, such as EML/EPRO cells, possessing mutations in various nuclear envelope-associated proteins, should prove extremely useful in determining the role that these proteins may play in the determination of granulocytic nuclear shape, heterochromatin distribution and control of genetic expression. The cell lines employed in the present study, derived from LBR-deficient bone marrow cells, are suitable materials for analysis of the importance of LBR to the distribution of interphase nuclear chromosome territories[56] and to the dynamics of chromatin mobility during in vitro granulopoiesis.[13, 57]
Acknowledgements
This work was supported by grants from the National Institutes of Health to D.E.O. (R15-HL075809) and to P.G. (KO1-DK60565 and R15-HL089933), a grant to H.H. from the European Commission (Contract LSHM-CT-2005-018690), and by funds from the Department of Biology, Bowdoin College. We thank Justine Johnson for providing excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bainton DF, Ullyot JL, Farquhar MG. The development of neutrophilic polymorphonuclear leukocytes in human bone marrow. J Exp Med. 1971;134:907–934. doi: 10.1084/jem.134.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma. 2007;116:227–235. doi: 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann K, Dreger CK, Olins AL, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly) Nat Genet. 2002;31:410–414. doi: 10.1038/ng925. [DOI] [PubMed] [Google Scholar]
- 4.Shultz LD, Lyons BL, Burzenski LM, et al. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet. 2003;12:61–69. doi: 10.1093/hmg/ddg003. [DOI] [PubMed] [Google Scholar]
- 5.Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- 6.D'Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cellular and molecular life sciences. 2006;63:316–332. doi: 10.1007/s00018-005-5361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruenbaum Y, Goldman RD, Meyuhas R, et al. The nuclear lamina and its functions in the nucleus. Int Rev Cytol. 2003;226:1–62. doi: 10.1016/s0074-7696(03)01001-5. [DOI] [PubMed] [Google Scholar]
- 8.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 9.Bridger JM, Foeger N, Kill IR, Herrmann H. The nuclear lamina. Both a structural framework and a platform for genome organization. Febs J. 2007;274:1354–1361. doi: 10.1111/j.1742-4658.2007.05694.x. [DOI] [PubMed] [Google Scholar]
- 10.Shaklai S, Amariglio N, Rechavi G, Simon AJ. Gene silencing at the nuclear periphery. Febs J. 2007;274:1383–1392. doi: 10.1111/j.1742-4658.2007.05697.x. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 12.Ji JY, Lee RT, Vergnes L, et al. Cell nuclei spin in the absence of lamin b1. J Biol Chem. 2007;282:20015–20026. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- 13.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 14.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 15.Olins AL, Buendia B, Herrmann H, Lichter P, Olins DE. Retinoic acid induction of nuclear envelope-limited chromatin sheets in HL-60. Exp Cell Res. 1998;245:91–104. doi: 10.1006/excr.1998.4210. [DOI] [PubMed] [Google Scholar]
- 16.Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid-and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- 17.Gaines P, Berliner N. Current Protocols in Immunology. New York: John Wiley & Sons, Inc.; 2005. Differentiation and Characterization of Myeloid Cells; p. 22F.25.21-22F.14. [DOI] [PubMed] [Google Scholar]
- 18.Tsai S, Collins SJ. A dominant negative retinoic acid receptor blocks neutrophil differentiation at the promyelocyte stage. Proc Natl Acad Sci U S A. 1993;90:7153–7157. doi: 10.1073/pnas.90.15.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 20.Lawson ND, Krause DS, Berliner N. Normal neutrophil differentiation and secondary granule gene expression in the EML and MPRO cell lines. Exp Hematol. 1998;26:1178–1185. [PubMed] [Google Scholar]
- 21.Gaines P, Chi J, Berliner N. Heterogeneity of functional responses in differentiated myeloid cell lines reveals EPRO cells as a valid model of murine neutrophil functional activation. J Leukoc Biol. 2005;77:669–679. doi: 10.1189/jlb.1004567. [DOI] [PubMed] [Google Scholar]
- 22.Gaines P, Tien CW, Olins AL, Olins DE, Shultz LD, Carney L, Berliner N. Mouse neutrophils lacking lamin B receptor expression exhibit aberrant development and lack critical functional responses. Exp Hematol. doi: 10.1016/j.exphem.2008.04.006. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olins DE, Olins AL. Granulocyte heterochromatin: defining the epigenome. BMC cell biology [electronic resource] 2005;6:39. doi: 10.1186/1471-2121-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olins AL, Olins DE. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 2004;5:30. doi: 10.1186/1471-2121-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreger CK, Konig AR, Spring H, Lichter P, Herrmann H. Investigation of nuclear architecture with a domain-presenting expression system. J Struct Biol. 2002;140:100–115. doi: 10.1016/s1047-8477(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 26.Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 27.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens JH, O'Sullivan RJ, Braunschweig U, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Hendzel MJ, Wei Y, Mancini MA, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 31.Barber CM, Turner FB, Wang Y, et al. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma. 2004;112:360–371. doi: 10.1007/s00412-004-0281-9. [DOI] [PubMed] [Google Scholar]
- 32.Olins AL, Olins DE. The mechanism of granulocyte nuclear shape determination: possible involvement of the centrosome. Eur J Cell Biol. 2005;84:181–188. doi: 10.1016/j.ejcb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worman HJ, Courvalin JC. Nuclear envelope, nuclear lamina, and inherited disease. Int Rev Cytol. 2005;246:231–279. doi: 10.1016/S0074-7696(05)46006-4. [DOI] [PubMed] [Google Scholar]
- 35.Olins AL, Herrmann H, Lichter P, Olins DE. Retinoic acid differentiation of HL-60 cells promotes cytoskeletal polarization. Exp Cell Res. 2000;254:130–142. doi: 10.1006/excr.1999.4727. [DOI] [PubMed] [Google Scholar]
- 36.Bennati AM, Castelli M, Della Fazia MA, et al. Sterol dependent regulation of human TM7SF2 gene expression: Role of the encoded 3beta-hydroxysterol Delta(14)-reductase in human cholesterol biosynthesis. 2006 doi: 10.1016/j.bbalip.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Rober RA, Sauter H, Weber K, Osborn M. Cells of the cellular immune and hemopoietic system of the mouse lack lamins A/C: distinction versus other somatic cells. J Cell Sci. 1990;95(Pt 4):587–598. doi: 10.1242/jcs.95.4.587. [DOI] [PubMed] [Google Scholar]
- 38.Smith S, Blobel G. Colocalization of vertebrate lamin B and lamin B receptor (LBR) in nuclear envelopes and in LBR-induced membrane stacks of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994;91:10124–10128. doi: 10.1073/pnas.91.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y, Cai S, Lv Q, et al. Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci. 2007;120:520–530. doi: 10.1242/jcs.03355. [DOI] [PubMed] [Google Scholar]
- 40.Holmer L, Pezhman A, Worman HJ. The human lamin B receptor/sterol reductase multigene family. Genomics. 1998;54:469–476. doi: 10.1006/geno.1998.5615. [DOI] [PubMed] [Google Scholar]
- 41.Wassif CA, Brownson KE, Sterner AL, et al. HEM dysplasia and ichthyosis are likely laminopathies and not due to 3beta-hydroxysterol Delta14-reductase deficiency. Hum Mol Genet. 2007;16:1176–1187. doi: 10.1093/hmg/ddm065. [DOI] [PubMed] [Google Scholar]
- 42.Haraguchi T, Koujin T, Hayakawa T, et al. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci. 2000;113(Pt 5):779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- 43.Ellenberg J, Siggia ED, Moreira JE, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prokocimer M, Margalit A, Gruenbaum Y. The nuclear lamina and its proposed roles in tumorigenesis: projection on the hematologic malignancies and future targeted therapy. J Struct Biol. 2006;155:351–360. doi: 10.1016/j.jsb.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Simos G, Georgatos SD. The inner nuclear membrane protein p58 associates in vivo with a p58 kinase and the nuclear lamins. Embo J. 1992;11:4027–4036. doi: 10.1002/j.1460-2075.1992.tb05496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reichart B, Klafke R, Dreger C, et al. Expression and localization of nuclear proteins in autosomal-dominant Emery-Dreifuss muscular dystrophy with LMNA R377H mutation. BMC Cell Biol. 2004;5:12. doi: 10.1186/1471-2121-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostlund C, Sullivan T, Stewart CL, Worman HJ. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 2006;45:1374–1382. doi: 10.1021/bi052156n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agrelo R, Setien F, Espada J, et al. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. J Clin Oncol. 2005;23:3940–3947. doi: 10.1200/JCO.2005.11.650. [DOI] [PubMed] [Google Scholar]
- 49.Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 50.Makatsori D, Kourmouli N, Polioudaki H, et al. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. The Journal of biological chemistry. 2004;279:25567–25573. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- 51.Brero A, Easwaran HP, Nowak D, et al. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, Bickmore WA. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J Cell Biol. 2007;177:401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biermann H, Pietz B, Dreier R, Schmid KW, Sorg C, Sunderkotter C. Murine leukocytes with ring-shaped nuclei include granulocytes, monocytes, and their precursors. J Leukoc Biol. 1999;65:217–231. doi: 10.1002/jlb.65.2.217. [DOI] [PubMed] [Google Scholar]
- 54.Allison DC, Nestor AL. Evidence for a relatively random array of human chromosomes on the mitotic ring. J Cell Biol. 1999;145:1–14. doi: 10.1083/jcb.145.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerlich D, Ellenberg J. Dynamics of chromosome positioning during the cell cycle. Curr Opin Cell Biol. 2003;15:664–671. doi: 10.1016/j.ceb.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 56.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 57.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]