Abstract
The peroxisome proliferator-activated receptors (PPARs), which are ligand-inducible transcription factors expressed in a variety of tissues, have been shown to perform key roles in lipid homeostasis. In physiological situations such as fasting and physical exercise, one PPAR subtype, PPARδ, triggers a transcriptional program in skeletal muscle leading to a switch in fuel usage from glucose/fatty acids to solely fatty acids, thereby drastically increasing its oxidative capacity. The metabolic action of PPARδ has also been verified in humans. In addition, it has become clear that the action of PPARδ is not restricted to skeletal muscle. Indeed, PPARδ has been shown to play a crucial role in whole-body lipid homeostasis as well as in insulin sensitivity, and it is active not only in skeletal muscle (as an activator of fat burning) but also in the liver (where it can activate glycolysis/lipogenesis, with the produced fat being oxidized in muscle) and in the adipose tissue (by incrementing lipolysis). The main aim of this review is to highlight the central role for activated PPARδ in the reversal of any tendency toward the development of insulin resistance.
1. INTRODUCTION
The modern Western lifestyle is characterized by excessive food intake and lack of physical exercise. This has led to obesity caused by a disturbance of lipid homeostasis, becoming one of the most prevalent and serious global chronic disorders. In lean organisms, fatty acids derived either from food or from hepatic lipogenesis are utilized as energy substrates by the heart and skeletal muscles. A strict physiological equilibrium between lipid availability and lipid consumption needs to be maintained to prevent development of both an impairment of insulin responsiveness and a metabolic dysfunction. In adult obesity and obesity-associated metabolic disorders (which have strict associations with type 2 diabetes, hypertension, and hyperlipidemia, and are often referred to as metabolic syndrome [1]), a disturbance of lipid homeostasis causes excess fat accumulation in various tissues (predominantly in adipose tissues, but also in other insulin-responsive organs, such as skeletal muscle and liver).
Skeletal muscle is quantitatively the largest organ in the body, and it contributes 30–40% of the resting metabolic rate in adults. It is a major site for the oxidation of fatty acids and glucose (accounting for approximately 80% of insulin-stimulated glucose uptake), and it exhibits a remarkable flexibility in its usage of fuel. One notable aspect of skeletal muscle plasticity is the specificity of its structural, biochemical, and functional adaptations to a given stimulus [2]. In view of the above features, it is perhaps not surprising that the predominant feature of type 2 diabetes is insulin resistance in skeletal muscle [3, 4].
The peroxisome proliferator-activated receptors (PPARs), which are ligand-inducible transcription factors expressed in a variety of tissues, have been shown to perform key roles in lipid homeostasis. In physiological situations such as fasting [5] and physical exercise [6], one PPAR subtype, PPARδ, triggers a transcriptional program in skeletal muscle leading to a switch in fuel usage from glucose/fatty acids to solely fatty acids, and thereby drastically increasing this tissue's oxidative capacity. In addition, recent evidence has highlighted the possibility that activating PPARδ in human subjects could increase skeletal muscle's oxidative capacity and so reverse metabolic abnormalities [7]. In mouse models, PPARδ has been shown to play a crucial role in whole-body lipid homeostasis as well as in insulin sensitivity, and to be predominantly active in skeletal muscle (as an activator of fat burning [8, 9]) but also in the liver (where it can activate glycolysis/lipogenesis, with the produced fat being oxidized in muscle [10]) and in the adipose tissue (by incrementing lipolysis [11]). Thus PPARδ activation provides a multiorgan “energy substrate-switching” phenotype that triggers tissue-specific transcriptional programs, and in which skeletal muscle plays a crucial role by reducing fat content.
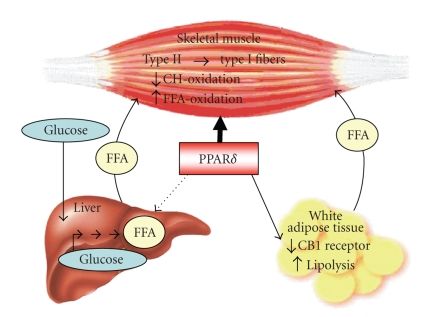
From the current data presented in the literature, a central role for activated PPARδ can be deduced in skeletal muscle, liver, and white adipose tissue (increased fat oxidation in muscle, increased carbohydrate catabolism and fat synthesis in the liver, and increased lipolysis in white adipose tissue) in the reversal of any tendency toward the development of insulin resistance. Following a brief description of the structure, the mode of activation and the action of the PPARs, this review aims to highlight the multiorgan energy switching role of PPARδ and its ultimate impact on insulin resistance. Figure 1 represents an overview of the central role played by PPARδ in the use of fatty acids as fuel by enhancing: (a) their oxidation in skeletal muscle (described in Section 3), (b) their synthesis from glucose in the liver and their subsequent release (described in Section 4), and (c) their release from the white adipose tissue (described in Section 5).
Figure 1.
Central, fuel-switching mechanisms by which PPARδ increases the use of fatty acids in skeletal muscle without provoking insulin resistance. Dotted arrow: indirect effect. For abbreviations, the reader is referred to the text.
2. PPARS, NUCLEAR RECEPTORS ACTIVATED BY FATTY ACIDS
PPARs are nuclear receptors that act as ligand-inducible transcription factors. The three known isoforms PPAR α, β (also termed δ) , and γ display tissue-specific expressions and possess different gene-regulatory profiles. PPARγ is a key regulator of adipose development and adipose insulin sensitivity [12], whereas PPARα-regulated genes are involved in hepatic lipid oxidation [13]. The PPARδ isoform, the function of which has only recently been elucidated and will be discussed in detail in this review, is predominantly expressed in skeletal muscle (where it induces fatty acid oxidation), but it is also expressed in brain, heart, liver, adipose tissue, and small intestine [14, 15].
It is generally known that PPARs heterodimerize with the retinoid X-receptor (RXR) and bind to a specific DNA sequence, (a sequence termed peroxisome proliferators response element (PPRE), that is found in a variety of genes involved in lipid and carbohydrate metabolism, inflammation, and cell proliferation and differentiation) [16, 17]. However, alternative mechanisms exist. Recently, an interplay has been reported between PPARδ and thyroid hormone receptor β (TRβ) in the activation of the gene encoding uncoupling protein 3 (UCP3) [18] in rat skeletal muscle as well as in cotransfection experiments in rat L6 myoblasts containing a reporter construct driven by the rat UCP3 promoter. Activation of UCP3 gene transcription in vivo by thyroid hormone (T3) requires the presence of fatty acids, while in the absence of fatty acids this transcription can be restored by the PPARδ agonist L165,041 [18]. The UCP3 gene promoter has been shown to contain a noncanonical thyroid hormone response element (TRE) termed TRE1 that is conserved from rodents to humans [18, 19], and this response element is also recognized by PPARs [19]. Interestingly, fatty acid responsiveness after T3 treatment was only observed in cells transfected with the rat UCP3 promoter, which suggests a species-specific regulation [18].
Dietary fatty acids and fatty acid derivatives are the natural ligands of PPARs, which display the greatest preference for monounsaturated and polyunsaturated fatty acids (MUFAs and PUFAs, resp.), as demonstrated by means of various ligand-binding assays [20, 21]. The fact that each PPAR activates a different gene program, despite their overlapping expressions, would seem to suggest ligand-specificity for each PPAR. Indeed, the structure of the ligand-binding pocket differs considerably among the various PPARs as revealed by X-ray crystal-structure analysis [21, 22]. Nevertheless, natural fatty acids can be ligands of all three PPAR isoforms. It is well known that binding of the ligand promotes a conformational change that is permissive for interactions with tissue-specific coactivator proteins, allowing nucleosome remodelling and activation of the transcription of cell type-specific target genes [21, 23]. It is, therefore, conceivable that a given fatty acid induces different conformational changes when binding to the ligand-binding pockets of the various PPAR subtypes. Given that the transcriptional activity induced by each PPAR subtype is cell type-specific [24], the different conformations induced following ligand binding may determine the cell-specificity of the different PPARs (through heterodimerization with different receptors and binding to cell-type specific cofactors). However, further research is needed to establish which natural ligands might activate each PPAR in a given cellular context.
3. ROLE OF PPARδ IN SKELETAL MUSCLE: A SWITCH TO FAT OXIDATION
3.1. PPARδ-induced fuel switching evidenced by its overexpression or ablation and by the use of synthetic ligands in rodents
In skeletal muscle, the relatively high expression of PPARδ (at 10- and 50-fold higher levels than PPARα and PPARγ, resp.; [15, 25]) as well as its preferential expression in oxidative rather than glycolytic myofibers [9], has led to the suggestion that this receptor isoform may be involved in promoting the utilization of fatty acids as fuel. Indeed, treatment of rat L6 cells with the highly specific PPARδ agonist GW0742 has been shown to increase fatty acid oxidation and induce expression of several lipid regulatory genes [26]. In addition, treatment of C2C12 cells with GW0742 [27] or with another the PPARδ-specific agonist (GW1516 [28]) induced expressions of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells [29]. However, although an in vivo magnetic resonance spectroscopy (MRS) study of rats treated with the latter agonist revealed increased lipid metabolism, no change in mitochondrial energy coupling was detected efficiency despite an increased UCP3 expression [29]. Skeletal muscle has the capacity to adapt its structure to metabolic changes, as indicated by changes in the myosin type and a subsequent shift in the number and type of muscle fibers. An increased demand for fat metabolism is reflected by a shift in fiber-type away from the fast, glycolytic form (type II) toward the slow-oxidative form (type I) [2]. PPARδ has emerged as an important stimulus in the induction of this fiber shift. Indeed, studies on transgenic mice harboring a constitutively activated form of PPARδ (VP16-PPARδ) have clearly shown an increase in skeletal muscle lipid metabolism as well as the formation of type I fibers [9]. Mice overexpressing peroxisome proliferator-activated receptor γ coactivator 1 α (PGC-1α), which activates PPARδ via a direct protein-protein interaction [30], have been shown to possess a high content of type I fibers [31]. However, experiments on mice overexpressing VP16-PPARδ have revealed that type I fiber formation can be directly stimulated by PPARδ without an induction of PGC-1α [9]. Nevertheless, in physiological situations, an upregulation of PGC-1α is likely to be involved in the early events leading to muscle-fiber shifts, since PGC-1α is a coactivator of the PPARs. Indeed, PGC-1α-null mice show clear muscle dysfunction [32], and evidence that PPARδ induces an expression of PGC-1α has come from treatment of mice with GW1516 [8] as well as from a study in which skeletal muscle C2C12 myocytes transfected with a PGC-1α promoter-driven reporter construct were treated with the same ligand [33]. A wild-type PPARδ transgene (specifically overexpressed in mouse skeletal muscle, but not constitutively activated) has been shown to promote a net increase in the number of fibers with an oxidative metabolic capability through an increase in fiber numbers in soleus and tibialis anterior muscle, while in plantaris muscle the increase was more closely related to a shift from glycolytic to more oxidative fibers [34]. Interestingly, this nonactivated PPARδ transgene failed to induce the formation of type I fibers in skeletal muscle [34]. In this case, despite its high expression, actual activation of the transgene would depend on the presence of natural ligands, such as fatty acids. As the fatty acid concentrations fluctuate, fatty regulation of PPARδ transcription is likely to be highly regulated.
This raises the question as to whether, instead of overexpression, increasing the availability of ligand (natural or artificial) would induce fuel switching as well as fiber switching toward type I fibers. Indeed, a fuel-switching role of PPARδ activation has been demonstrated by incubating rat isolated skeletal muscle strips with the agonist GW1516 for 24 hours [35]. This led to an increased use of fatty acids over glucose, as reflected by increased fatty acid oxidation and reductions in glucose oxidation, glycogen synthesis, lactate release, and glucose transport. Interestingly, this switch was independent of insulin stimulation or fiber type [35]. The PPARδ agonist GW610742 also induced this fuel switch and in addition triggered a genetic program toward initiation of muscular atrophy without compromising mitochondrial activity in rat skeletal muscle [36]. Furthermore, a somatic gene transfer of PPARδ into adult rat fibers has recently been shown to lead to activation of fuel switching as well as to a shift in the existing fiber profile toward the “slow,” oxidative phenotype, an effect that caused the number of type I fibers in the extensor digitorum longus (EDL) muscle to be tripled after 14 days [37]. From these experiments, it is clear that PPARδ rapidly programs all fiber types in skeletal muscle to oxidize fatty acids.
3.2. Evidence supporting a role for activated PPARδ in mediating fuel switching in human skeletal muscle: data from in vitro and clinical studies
Recent studies have revealed clear roles for PPARδ in the regulation of lipid and glucose metabolism in human skeletal muscle [7, 38]. PPARδ stimulates the expression of genes involved in (a) increasing of lipid oxidation (fatty acid binding protein 3 (FABP3) and CPT1) and (b) reducing carbohydrate oxidation (pyruvate dehydrogenase kinase 4 (PDK4)) in human skeletal muscle [38], as it does in rodents [5, 8–10]. These in vitro human skeletal muscle data with GW1516 support the long-known observation that an increase in fatty acid oxidation reduces the glucose utilization of isolated muscle reflecting the mutual inhibition in the metabolism of substrates involved in the glucose-fatty acid cycle [35, 36, 39]. Recent data from a clinical study indicate that this situation holds true in human subjects. A statistically significant reduction in fasting insulin levels was observed in a study performed on moderately obese men given a dose of 10 mg o.d. of GW1516 was given for 2 weeks [7]. These subjects also showed decrease fasting plasma nonesterified fatty acid (NEFA) concentrations and increased expression of CPT1b in muscle. Interestingly, GW1516 treatment slightly reduced the expression of PPARδ, showing that overexpression of the receptor itself is not necessary for the induction of PPARδ-mediated effects, the resident levels of the receptor protein being sufficient. In contrast, similar treatment with a PPARα agonist (GW590735) did not result in any change in plasma insulin levels, nor did it cause a significant increase in CPT1b mRNA [7].
3.3. PPARδ, a regulator of fuel use in skeletal muscle under physiological conditions
Since fasting promotes increased utilization of fatty acids, fasting might also regulate PPARδ expression. However, the initial reports appearing in the literature did not seem to confirm this. For instance, a 24-hour fasting period was found not to upregulate PPARδ in rat skeletal muscle [40], and our group even reported reduced levels of PPARδ (and PPARα) in gastrocnemius muscle from 48-hour-fasted rats [25]. Decreased expressions of PPARδ and PPARα upon 48 hours of fasting have recently been reported in humans, too [41]. In contrast, in mice, skeletal muscle PPARδ levels were found to be upregulated after a 24-hour fast [42]. One explanation is that studying the responses to physiological stimuli by making single time point measurements may give misleading results, due to possible transient modifications. Indeed, time course studies have shown that PPARδ actually is upregulated in fasted rat skeletal muscle, but within the first 6 hours [43]. This upregulation of PPARδ (and of PGC-1α) during fasting is, however, transient [43], downregulation (after the initial upregulation at 6 hours to around or below the control levels being evident at 48 hours). Data showing that rapid nuclear accumulations of both PGC-1α and PPARδ upon food deprivation [43], and their physical interaction within the nucleus [30], occur concomitantly with increases in fatty acid levels and an increase in the expression of myosin heavy chain Ib (MHC Ib), underline the role of PPARδ as a key regulator of fatty acid metabolism and muscle fiber switching (in concert with its coactivator, PGC-1α).
Following the nuclear accumulation of PPARδ, its target genes (such as carnitine palmitoyl transferase 1b (CPT1b), mitochondrial thioesterase I (MTE I), and UCP3) are upregulated simultaneously, and so is the rate of mitochondrial fatty acid oxidation [43]. In starved mice, the mRNA for a member of the FOXO family, FKHR (forkhead homolog in rhabdomyosarcoma), is upregulated rapidly and transiently (starting within 6 hours, peaking at 12 hours, and decreasing at 24 hours), and this is followed by an upregulation of FKHR protein and a nuclear accumulation of nonphosphorylated FKHR levels with a consequent upregulation of its target gene pyruvate dehydrogenase kinase 4 (PDK4) [44]. In mouse skeletal muscle, PDK4 has been shown to be a target of PPARδ, since it is activated in vivo by GW1516 [8]. This kinase plays an important role in the switching from glucose usage to fat usage since it phosphorylates the E1 component of the pyruvate dehydrogenase (PDH) complex, thereby downregulating carbohydrate (CH) oxidation [45]. Similarly, in human skeletal muscle, PDK4 mRNA and protein levels have been found to be elevated at both 24 hours and 48 hours of fasting, whereas at these time points the FOXO1 mRNA/protein levels were unchanged [41]. However, it is possible that regulation of the mRNA/protein levels of FOXO1 (a target of PGC-1α [46], which are transiently regulated during fasting in rats [40]) is a transient event preceding upregulation of PDK4 mRNA and protein in humans, too. If so, such transient regulatory changes may have been missed by measuring only at 24 hours and 48 hours of starvation in humans. At the 48-hour time point of starvation, PPARδ mRNA levels in the human vastus lateralis muscle were decreased [41], a finding in line with the decreased levels of PPARδ mRNA [25] and nuclear protein [43] at this time point in fasting rat gastrocnemius muscle. In the rat, this decline was preceded by a rapid rise in the mRNA and protein levels of this transcription factor, elevated levels being detected at 6 hours and 12 hours of food deprivation [43]. Unfortunately, these time points were not investigated in the human study [41]. Thus kinetic studies in the fasting situation, in rodents as well as in humans, have made it clear that the expression of PPARδ, and that of its target genes, is tightly regulated. These results offer an explanation for the rapid structural and metabolic changes favoring an increased use of lipids as fuel that occur in this condition.
The reported kinetics of the PPARδ upregulation occurring during the recovery period after exercise-imposed energy stress in humans are in line with those seen in the fasting rat; a single exhaustive bout of cycling increasing PPARδ mRNA and protein expression within 3 hours after completion of the exercise [47, 48]. Regular physical training, which elevates the levels of PPARδ and PGC-1α, leads to increases in both mitochondrial capacity and insulin sensitivity [34, 49–51], effects similar to those seen after PPARδ activation. The observations made in exercise and fasting experiments showing that PPARδ-expression kinetics are rapid, and the changes almost immediate, are supported by PPARδ acutely bringing about the fuel switching effects inpre-existing adult rat muscle fibers (as demonstrated by short-term agonist treatment [35, 36] and somatic PPARδ gene transfer [36]). Recently, a direct role for PPARδ in the suppression of glucose oxidation in fasting skeletal muscle has been shown by Nahlé et al. [52]. First, the authors demonstrated that deficiency of the fatty acid translocase CD36 (mediating muscle fatty acid uptake during fasting, [5]) blunts fasting induction of FOXO1 and PDK4 and the associated suppression of glucose oxidation in mouse skeletal muscle [52]. Next, they demonstrated that loss of PPARδ abolishes the fasting induction of muscle FOXO1 and PDK4 in vivo [52]. As the authors identified several PPRE sites in the FOXO1 promoter, these results suggest that CD36-dependent activation of PPARδ results in the trascriptional regulation of FOXO1 as well as PDK4 in fasted mice.
Taken together, the control of fuel use by activated PPARδ has emerged to be crucial for the rapid metabolic adaptation of skeletal muscle to energy stress.
4. ANTIGLYCEMIC ACTION OF PPARδ IN LIVER: CONVERTING GLUCOSE INTO FAT
At first glance, the above described PPARδ-mediated fuel switching in skeletal muscle would be expected to give rise to insulin resistance, since a preferential uptake and oxidation of fatty acids (FFAs) over glucose (mediated by PPARδ stimulation in skeletal muscle) would lead to an accumulation of blood glucose, muscle being a major player in blood-glucose homeostasis [1, 2]. However, as well as stimulating skeletal muscle fatty acid oxidation, PPARδ plays a surprising role in ameliorating hyperglycemia. It does this by increasing hepatic glucose flux through the pentose phosphate pathway and by enhancing fatty acid synthesis, the fatty acids being destined for oxidization in muscle [10]. Whereas PPARδ-deprived mice are metabolically less active and glucose intolerant, db/db mice treated with GW1516 show increased levels of CPT in muscle [10] (similar effects of this agonist being reported in [8, 9]). This effect on CPT is indicative of increased fatty acid oxidation, so it was a surprising finding that gene array analysis revealed increased expression of gene clusters involved in fatty acid synthesis and the pentose phosphate cycle in the liver [7], both pathways using glucose or its metabolites as substrates. The authors [10] suggested that PPARδ may increase glucose catabolism through these processes, with the result that peripheral insulin sensitivity may be improved [10]. Indeed, GW1516-treated db/db mice showed an improved performance in the glucose tolerance test, and also showed a tendency toward lowered fasting serum insulin levels [10].
5. ROLE FOR PPARδ IN WHITE ADIPOSE TISSUE: INCREASING LIPOLYSIS
Interestingly, adipocyte hypertrophy induced by high-fat diet was accompanied by increased cannabinoid receptor type 1 (CB1R) expression and by a decrease in PPARδ expression in adipose tissue [11]. Exercise attenuates adipocyte hypertrophy and normalizes expression of CB1R and PPARδ. Functional cross-talk between CB1R and PPARδ is established by RNA interference experiments in 3T3-L1 preadipocytes. For example, selective silencing of PPARδ by RNA interference significantly increased CB1R and increased adipocyte differentiation and adenovirus-mediated overexpression of PPARδ reduced CB1R expression [11]. The role of CB1R in obesity is well established [53]. Depletion of CB1R in knockout mice is known (in animals fed a high-fat diet) to reduce obesity through increased lipolysis [53].
The simultaneous regulation of skeletal muscle fatty acid oxidation and adipose proliferation by PPARδ underlines the powerful role this receptor may play in preventing weight gain in physiological situations.
6. CONCLUSIONS
Recent evidence shows that the fuel-switching role of PPARδ in skeletal muscle is conserved from rodents to humans. This receptor is crucial for a modulation of skeletal muscle flexibility that directs this tissue toward the use of fatty acids as fuel, an effect with rapid kinetics. It is becoming increasingly clear that even though the fat oxidation-inducing effect of PPARδ is exerted in skeletal muscle, this receptor also activates transcriptional programs in other tissues (such as adipose tissue, in which PPARδ directly inhibits proliferation and induces lipolysis), thereby channelling the fat consumed in the diet directly to muscle for oxidation. In addition, in an action that serves to prevent insulin resistance developing as a consequence of a reduction of carbohydrate oxidation in muscle, the liver is activated to form fat from glucose, thereby both allowing skeletal muscle to oxidize more fat and concomitantly lowering blood glucose levels. PPARδ is emerging as a crucial nuclear receptor for the multiorgan regulation of whole body fuel turnover, with skeletal muscle as the central organ in the burning of fat and the consequent prevention or reduction of obesity (see Figure 1). These features make PPARδ a good candidate as a central target for the future treatment of metabolic disturbances linked to obesity and insulin resistance.
ACKNOWLEDGMENTS
This work was supported in part by Grant no. MIUR-COFIN 2006 Prot 2006051517. P. de Lange and A. Lombardi contributed equally to this work.
References
- 1.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104(4):531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 2.Flück M, Hoppeler H. Molecular basis of skeletal muscle plasticity—from gene to form and function. Reviews of Physiology, Biochemistry and Pharmacology. 2003;146:159–216. doi: 10.1007/s10254-002-0004-7. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SI. Deconstructing type 2 diabetes. Cell. 1999;97(1):9–12. doi: 10.1016/s0092-8674(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 4.Zimmet P, Alberti KGMM, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414(6865):782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 5.de Lange P, Moreno M, Silvestri E, Lombardi A, Goglia F, Lanni A. Fuel economy in food-deprived skeletal muscle: signaling pathways and regulatory mechanisms. The FASEB Journal. 2007;21(13):3431–3441. doi: 10.1096/fj.07-8527rev. [DOI] [PubMed] [Google Scholar]
- 6.Fürnsinn C, Willson TM, Brunmair B. Peroxisome proliferator-activated receptor-δ, a regulator of oxidative capacity, fuel switching and cholesterol transport. Diabetologia. 2007;50(1):8–17. doi: 10.1007/s00125-006-0492-0. [DOI] [PubMed] [Google Scholar]
- 7.Risérus U, Sprecher D, Johnson T, et al. Activation of peroxisome proliferator-activated receptor (PPAR)δ promotes reversal of multiple metabolic abnormalities, reduces oxidative stress, and increases fatty acid oxidation in moderately obese men. Diabetes. 2008;57(2):332–339. doi: 10.2337/db07-1318. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y-X, Zhang C-L, Yu RT, et al. Regulation of muscle fiber type and running endurance by PPARδ . PLoS Biology. 2004;2(10, e294):1532–1539. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C-H, Olson P, Hevener A, et al. PPARδ regulates glucose metabolism and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(9):3444–3449. doi: 10.1073/pnas.0511253103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan ZC, Liu DY, Zhang LL, et al. Exercise reduces adipose tissue via cannabinoid receptor type 1 which is regulated by peroxisome proliferator-activated receptor-δ . Biochemical and Biophysical Research Communications. 2007;354(2):427–433. doi: 10.1016/j.bbrc.2006.12.213. [DOI] [PubMed] [Google Scholar]
- 12.He W, Barak Y, Hevener A, et al. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger J, Moller DE. The mechanisms of action of PPARs. Annual Review of Medicine. 2002;53(1):409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 14.Amri E-Z, Bonino F, Ailhaud G, Abumrad NA, Grimaldi PA. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. Journal of Biological Chemistry. 1995;270(5):2367–2371. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- 15.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137(1):354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 16.Michalik L, Wahli W. Peroxisome proliferator-activated receptors: three isotypes for a multitude of functions. Current Opinion in Biotechnology. 1999;10(6):564–570. doi: 10.1016/s0958-1669(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Hu E, Devine J, Beale EG, Spiegelman BM. PPARγ2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Molecular and Cellular Biology. 1995;15(1):351–357. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange P, Feola A, Ragni M, et al. Differential 3,5,3′-triiodothyronine-mediated regulation of uncoupling protein 3 transcription: role of fatty acids. Endocrinology. 2007;148(8):4064–4072. doi: 10.1210/en.2007-0206. [DOI] [PubMed] [Google Scholar]
- 19.Solanes G, Pedraza N, Iglesias R, Giralt M, Villarroya F. Functional relationship between MyoD and peroxisome proliferator-activated receptor-dependent regulatory pathways in the control of the human uncoupling protein-3 gene transcription. Molecular Endocrinology. 2003;17(10):1944–1958. doi: 10.1210/me.2002-0395. [DOI] [PubMed] [Google Scholar]
- 20.Krey G, Braissant O, L'Horset F, et al. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Molecular Endocrinology. 1997;11(6):779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 21.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ . Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 22.Xu HE, Lambert MH, Montana VG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Molecular Cell. 1999;3(3):397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 23.Surapureddi S, Yu S, Bu H, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor α-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen R, Grøntved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Molecular and Cellular Biology. 2006;26(15):5698–5714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lange P, Ragni M, Silvestri E, et al. Combined cDNA array/RT-PCR analysis of gene expression profile in rat gastrocnemius muscle: relation to its adaptive function in energy metabolism during fasting. The FASEB Journal. 2004;18(2):350–352. doi: 10.1096/fj.03-0342fje. [DOI] [PubMed] [Google Scholar]
- 26.Muoio DM, MacLean PS, Lang DB, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) α knock-out mice. Evidence for compensatory regulation by PPARδ . Journal of Biological Chemistry. 2002;277(29):26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- 27.Holst D, Luquet S, Nogueira V, Kristiansen K, Leverve X, Grimaldi PA. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochimica et Biophysica Acta. 2003;1633(1):43–50. doi: 10.1016/s1388-1981(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 28.Dressel U, Allen TL, Pippal JB, Rohde PR, Lau P, Muscat GEO. The peroxisome proliferator-activated receptor β/δ agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Molecular Endocrinology. 2003;17(12):2477–2493. doi: 10.1210/me.2003-0151. [DOI] [PubMed] [Google Scholar]
- 29.Jucker BM, Yang D, Casey WM, et al. Selective PPARδ agonist treatment increases skeletal muscle lipid metabolism without altering mitochondrial energy coupling: an in vivo magnetic resonance spectroscopy study. American Journal of Physiology. 2007;293(5):E1256–E1264. doi: 10.1152/ajpendo.00218.2007. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y-X, Lee C-H, Tiep S, et al. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113(2):159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 31.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 32.Leone TC, Lehman JJ, Finck BN, et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biology. 2005;3(4, e101):672–687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuler M, Ali F, Chambon C, et al. PGC1α expression is controlled in skeletal muscles by PPARβ, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metabolism. 2006;4(5):407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Luquet S, Lopez-Soriano J, Holst D, et al. Peroxisome proliferator-activated receptor δ controls muscle development and oxidative capability. The FASEB Journal. 2003;17(15):2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 35.Brunmair B, Staniek K, Dörig J, et al. Activation of PPAR-δ in isolated rat skeletal muscle switches fuel preference from glucose to fatty acids. Diabetologia. 2006;49(11):2713–2722. doi: 10.1007/s00125-006-0357-6. [DOI] [PubMed] [Google Scholar]
- 36.Constantin D, Constantin-Teodosiu D, Layfield R, Tsintzas K, Bennett AJ, Greenhaff PL. PPARδ agonism induces a change in fuel metabolism and activation of an atrophy programme, but does not impair mitochondrial function in rat skeletal muscle. Journal of Physiology. 2007;583(1):381–390. doi: 10.1113/jphysiol.2007.135459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lunde IG, Ekmark M, Rana ZA, Buonanno A, Gundersen K. PPARδ expression is influenced by muscle activity and induces slow muscle properties in adult rat muscles after somatic gene transfer. Journal of Physiology. 2007;582(3):1277–1287. doi: 10.1113/jphysiol.2007.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krämer DK, Al-Khalili L, Guigas B, Leng Y, Garcia-Roves PM, Krook A. Role of AMP kinase and PPARδ in the regulation of lipid and glucose metabolism in human skeletal muscle. Journal of Biological Chemistry. 2007;282(27):19313–19320. doi: 10.1074/jbc.M702329200. [DOI] [PubMed] [Google Scholar]
- 39.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in isulin sensitivity and the metabolic disturbances of diabetes mellitus. The Lancet. 1963;281(7285):785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 40.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142(10):4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 41.Tsintzas K, Jewell K, Kamran M, et al. Differential regulation of metabolic genes in skeletal muscle during starvation and refeeding in humans. Journal of Physiology. 2006;575(1):291–303. doi: 10.1113/jphysiol.2006.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holst D, Luquet S, Nogueira V, Kristiansen K, Leverve X, Grimaldi PA. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochimica et Biophysica Acta. 2003;1633(1):43–50. doi: 10.1016/s1388-1981(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 43.de Lange P, Farina P, Moreno M, et al. Sequential changes in the signal transduction responses of skeletal muscle following food deprivation. The FASEB Journal. 2006;20(14):2579–2581. doi: 10.1096/fj.06-6025fje. [DOI] [PubMed] [Google Scholar]
- 44.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochemical Journal. 2003;375, part 2:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugden MC, Howard RM, Munday MR, Holness MJ. Mechanisms involved in the coordinate regulation of strategic enzymes of glucose metabolism. Advances in Enzyme Regulation. 1993;33:71–95. doi: 10.1016/0065-2571(93)90010-b. [DOI] [PubMed] [Google Scholar]
- 46.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Watt MJ, Southgate RJ, Holmes AG, Febbraio MA. Suppression of plasma free fatty acids upregulates peroxisome proliferator-activated receptor (PPAR) α and δ and PPAR coactivator 1α in human skeletal muscle, but not lipid regulatory genes. Journal of Molecular Endocrinology. 2004;33(2):533–544. doi: 10.1677/jme.1.01499. [DOI] [PubMed] [Google Scholar]
- 48.Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. The FASEB Journal. 2005;19(11):1498–1500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- 49.Hood DA. Invited review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. Journal of Applied Physiology. 2001;90(3):1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- 50.Goodpaster BH, Katsiaras A, Kelley DE. Enhanced fat oxidation through physical activity is associated with improvements in insulin sensitivity in obesity. Diabetes. 2003;52(9):2191–2197. doi: 10.2337/diabetes.52.9.2191. [DOI] [PubMed] [Google Scholar]
- 51.Kuhl JE, Ruderman NB, Musi N, et al. Exercise training decreases the concentration of malonyl-CoA and increases the expression and activity of malonyl-CoA decarboxylase in human muscle. American Journal of Physiology. 2006;290(6):E1296–1303. doi: 10.1152/ajpendo.00341.2005. [DOI] [PubMed] [Google Scholar]
- 52.Nahlé Z, Hsieh M, Pietka T, et al. CD36-dependent regulation of muscle FoxO1 and PDK4 in the PPARδ/β-mediated adaptation to metabolic stress. The Journal of Biological Chemistry. 2008;283(21):14317–14326. doi: 10.1074/jbc.M706478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ravinet-Trillou C, Delgorge C, Menet C, Arnone M, Soubrié P. CB1 cannabinoid receptor knockout in mice leads to leanness, resistance to diet-induced obesity and enhanced leptin sensitivity. International Journal of Obesity. 2004;28(4):640–648. doi: 10.1038/sj.ijo.0802583. [DOI] [PubMed] [Google Scholar]