Abstract
Here we analyze the role of the Lhx6 lim-homeobox transcription factor in regulating the development of subsets of neocortical, hippocampal and striatal interneurons. An Lhx6 loss-of-function allele, that expresses placental alkaline phosphatase (PLAP), allowed analysis of the development and fate of Lhx6-expressing interneurons in mice lacking this homeobox transcription factor. There are Lhx6+;Dlx+ and Lhx6−;Dlx+ subtypes of tangentially migrating interneurons. Most interneurons in Lhx6PLAP/PLAP mutants migrate to the cortex, although less efficiently, and exhibit defects in populating the marginal zone and superficial parts of the neocortical plate. By contrast, migration to superficial parts of the hippocampus is not seriously affected. Furthermore, whereas parvalbumin+ and somatostatin+ interneurons do not differentiate, NPY+ interneurons are present; we suggest that these NPY+ interneurons are derived from the Lhx6−;Dlx+ subtype. Striatal interneurons show deficits distinct from pallial interneurons, including a reduction in the NPY+ subtype. We provide evidence that Lhx6 mediates these effects through promoting expression of receptors that regulate interneuron migration (ErbB4, CXCR4, CXCR7), and through promoting the expression of transcription factors either known (Arx) or implicated (bMaf, Cux2, and NPAS1) in controlling interneuron development.
Keywords: Lhx6, Dlx, GABAergic, Interneuron, NPY
Introduction
Current evidence shows that most rodent cortical interneurons are derived from progenitor domains in the prenatal subcortical telencephalon (subpallium) (reviewed in: Flames and Marin, 2005; Marin and Rubenstein, 2003; Wonders and Anderson, 2006). Ongoing studies are aimed at elucidating the precise regions within the subpallium that generate these cells, and the mechanisms that regulate their specification, migration and differentiation into distinct interneuron subtypes in pallial structures (i.e. cortex, hippocampus, parts of the amygdala, and olfactory bulb). Here, we have examined the role of the mouse Lhx6 LIM-Homeodomain transcription factor in regulating these processes.
The subpallium consists of four major subdivisions that have distinct molecular and morphological features: the lateral ganglionic eminence (LGE), medial ganglionic eminence (MGE), septum (SE) and preoptic area (POA) (Flames et al., 2007; Puelles and Rubenstein, 2003). Sulci approximate the transitions between most of these domains at early embryonic stages; however, the caudal part of the LGE and MGE lack a clear morphological demarcation, leading this region to be called the caudal ganglionic eminence (CGE), even though it has distinct LGE and MGE components. Efforts are underway to identify subdivisions within the progenitor domains of the LGE, MGE, SE and POA; the most advanced effort has used transcription factor expression patterns (Flames et al., 2007).
The MGE is thought to be the source of a large share of pallial interneurons –including the majority of interneurons that express parvalbumin and somatostatin. On the other hand, there are at least two types of calretinin+ and neuropeptide Y (NPY)+ interneurons: those expressing somatostatin are thought to derive from the MGE, and those that don’t express somatostatin are thought to derive from the CGE (Butt et al., 2005; Pleasure et al., 2000; Wonders and Anderson, 2006; Xu et al., 2004). Furthermore, there are temporal constraints that correlate with the production of specific types of interneurons and the layers that they populate (Miyoshi et al., 2007; Rymar and Sadikot, 2007).
Flames et al. (2007), using transplantation of domains within the MGE, found that its dorsal and ventral subdivisions produce different ratios of cortical interneuron subtypes. They correlate this with molecular features of the MGE to provide evidence for a transcription factor code that participates in generating distinct interneuron subtypes. Fogarty et al. (2007), using several lines of mice that express Cre in different patterns in the embryonic basal ganglia, provided evidence for the origins of different types of cortical interneurons. Ghanem et al. (2007) established that two different enhancers of the Dlx1&2locus (i12b and URE2) are expressed in distinct subsets of cortical interneurons. Differences in the expression of i12b and URE2 in the subpallium suggest why they are expressed in different subtypes of interneurons.
Here we have explored pallial interneuron development by studying the expression and function of Lhx6. Lhx6 expression begins in scattered cells in the ventricular zone of the MGE and POA, becomes broadly expressed in the subventricular zone, and is maintained in many projection neurons and interneurons (Alifragis et al., 2004; Choi et al., 2005; Flames et al., 2007; Garcia-Lopez et al., 2008; Grigoriou et al., 1998; Lavdas et al., 1999; Liodis et al., 2007; Marin et al., 2000; Sussel et al., 1999). Thus, subsets of projection neurons are Lhx6+ in the globus pallidus, ventral pallidum, bed nucleus stria terminals, medial amygdala and septum. The vast majority of MGE-derived interneurons that migrate to the pallium, and perhaps striatum, are also Lhx6+. Mature cortical interneurons (principally of the parvalbumin+ and somatostatin+ subtypes) express Lhx6 (Cobos et al., 2005; Cobos et al., 2006; Fogarty et al., 2007; Liodis et al., 2007).
Molecular regulation of interneuron specification, differentiation and migration has begun to be elucidated (reviewed in: Flames and Marin, 2005; Marin and Rubenstein, 2003; Wonders and Anderson, 2006). Lhx6 expression in the MGE is dependent on the function of the Nkx2.1 homeobox gene (Sussel et al., 1999). Nkx2.1 neonatal mutants lack somatostatin+ and NPY+ interneurons (Pleasure et al., 2000). Previous studies implicated Lhx6 function in tangential migration of immature interneurons (Alifragis et al., 2004; Liodis et al., 2007) and in the development of parvalbumin+, somatostatin+ and calretinin+ interneurons (Liodis et al., 2007).
Lhx6 expression in the subpallium is maintained in mice lacking either the Dlx1&2 or the Arx homeobox genes (Colombo et al., 2007; Marin et al., 2000; Petryniak et al., 2007). In both mutants interneuron migration to the cortex is greatly diminished, demonstrating that Lhx6 is not sufficient for migration (Anderson et al., 1997; Colombo et al., 2007; Kitamura et al., 2002). Interneuron migration is disrupted in the Dlx1&2 mutants for several reasons including: 1) immature interneurons over-express the Pak3 kinase, which contributes to premature elaboration of neurites and reduced migration (Cobos et al., 2007); 2) reduced expression of cytokine receptors, CXCR4 and RDC1 (CXCR7) (Long et al., submitted) CXCR4 function is required for the correct laminar positioning of developing interneurons (Li et al., 2008; López-Bendito et al., 2008; Stumm et al., 2007; Stumm et al., 2003) 3) Molecular changes in the MGE result in reduced Arx and Lhx6 expression (Cobos et al., 2005; Petryniak et al., 2007); and increased Olig2 expression – the latter leads to increased numbers of oligodendrocytes (Petryniak et al., 2007).
Immature interneurons are repelled from ventral migration by an unknown factor in the basal telencephalon (Marin et al., 2003). Conversely, they are attracted to the cortex (Marin et al., 2003; Wichterle et al., 2003), in part by neuregulin1/ErbB4 signaling (Flames et al., 2004). En route, semaphorin/neuropilins repulsion is implicated in targeting the interneurons to their appropriate targets (Marin et al., 2001; Tamamaki et al., 2003). Interneuron movement is stimulated by receptor tyrosine kinase signaling through trkB/BDNF and cMet/HGF (Polleux et al., 2002; Powell et al., 2001). BDNF regulates the distribution of marginal zone interneurons (Alcantara et al., 2006). Mice lacking either GDNF or GFR-alpha-l have reduced numbers of cortical interneurons (Pozas and Ibanez, 2005). Finally, there is evidence that Robo1 contributes to interneuron migration (Andrews et al., 2006), while analysis of Slit1&2 single and double mutants failed to identify a clear defect in tangential migration (Marin et al., 2003).
Herein, we explore the roles of Lhx6 in regulating cortical and hippocampal interneuron development. Previous studies have implicated Lhx6 in promoting interneuron tangential migration (Alifragis et al., 2004; Liodis et al., 2007), and in the development of interneurons expressing parvalbumin, somatostatin and calretinin (Liodis et al., 2007). Here, we have taken advantage of an Lhx6 loss-of-function allele that expresses placental alkaline phosphatase (PLAP) (Choi et al., 2005) to follow the development and fate of Lhx6-expressing interneurons in mice lacking this transcription factor. This analysis demonstrates that Lhx6−/− interneurons do migrate to the cortex, although less efficiently. However, they exhibit defects in populating the marginal zone and superficial parts of the neocortical plate. In addition, they lack expression of several markers normally expressed by cortical interneurons. Further analysis demonstrates that there are two subtypes of tangentially migrating interneurons, one Lhx6+;Dlx+ and another which is only Dlx+, suggesting that some interneurons are not dependent on Lhx6 function. By contrast, migration to superficial parts of the prenatal hippocampus does not appear to be seriously affected. While previous studies suggest that development of hippocampal interneurons follow the same general rules as for neocortical interneurons (Pleasure et al., 2000), there have been hints for differences in their development. For instance, although neonatal Dlx1/2−/− mutants lack hippocampal interneurons, their neocortex still has ~20% normal numbers (Anderson et al., 1997; Pleasure et al., 2000). As described herein, compared to Dlx1/2−/− mutants, loss of Lhx6 function has a complementary change in the relative distribution of interneurons between the neocortex and hippocampus.
Furthermore, whereas parvalbumin and somatostatin subtypes do not differentiate in the Lhx6 mutant, NPY+ interneurons are present. We suggest that late-born NPY+ interneurons are derived from the Lhx6−;Dlx+ subtype and provide evidence that Lhx6 mediates these effects through promoting expression of receptors that regulate interneuron migration (ErbB4, CXCR4, CXCR7), and through promoting the expression transcription factors either known (Arx) or implicated (bMaf, Cux2 and NPAS1) in controlling interneuron development.
Materials and Methods
Animals and Tissue Preparation
Lhx6PLAP were provided by Regeneron and genotyped as per Choi et al., 2005. Prenatal studies were performed on a C57BL/6J background (> 10 generations); postnatal studies on a CF1 background (1 generation), which may promote greater postnatal survival. Animals were treated in accordance with the protocols approved by the NICHD and UCSF animal use committee.
Embryos were immersion fixed in 4% paraformaldehyde (PFA) in phosphate buffered saline pH 7.4 (PBS) for 4–12 hours; postnatal animals were fixed by cardiac PFA perfusion; brains were post-fixed overnight, cryoprotected in a gradient of sucrose to 30%, frozen in embedding medium (OCT, Tissue-Tek, Torrance, CA) and cut using a cryostat.
In Situ Hybridization
In situ hybridization experiments (Cobos et al., 2007) were performed using digoxigenin riboprobes on 20 μm frozen sections. We thank the following people for cDNAs: Drs. Brian Condie (GAD67), Cary Lai (ErbB4), Sabine Cordes (bMAF), Thomas Jessell (ER81), Harold Cremer (Cux2), Vassilis Pachnis (Lhx6 and Lhx7), Kunio Kitamura (Arx), Thomas Lufkin (SS), Andy McMahon (Shh), Steve McKnight (NPAS1), Sam Pleasure (CXCR4), TH Rabbits (LMO3/RBTN3), ATCC (NPY and RDC1) and Image (Thrombospondin). The Dlx1 and Nkx2.1 plasmids were generated in the Rubenstein lab.
Immunohistochemistry and BrdU Labeling/Detection
Immunofluorescence and immunoperoxidase stainings were performed per Zhao et al. (2003); cryostat sections were 20 μm on slides (for embryonic ages), or 40 μm free-floating (for postnatal ages). Antibodies: rat anti-somatostatin (Chemicon, MAB354, 1/150); goat anti-choline acetyltransferase (Chemicon, AB144P, 1/250), rabbit anti-calretinin (Chemicon, AB5054, 1:2500) or (Swant, 1/2000); rabbit anti-NPY (Sigma, N9528, 1/5000); mouse anti-parvalbumin (Sigma, P3088, 1/2000); rat anti-CTIP2 (abcam, 1/1000); rabbit anti pan-DLX (kindly provided by Grace Boekhoff-Falk; Panganiban et al., 1995), 1/250), guinea pig anti-DLX2 (kindly provided by Kazuaki Yoshikawa; Kuwajima et al., 2006), 1/2000); chicken anti-GFP (1/2000, Aves labs), rabbit anti-NPY (Immunostar, 1/2000); rabbit anti-PLAP (Serotec, 1/50); rabbit anti-TBR1 (kindly provided by Robert Hevner, 1/1000); rat anti-BrdU abcam (ab6326; 1/5000); rabbit anti-NPAS1 (kindly provided by Steve McKnight, 1/1000; Erbel-Sieler et al., 2004). Secondary antibodies were: Alexa 488 goat anti-rabbit, Alexa 488 goat anti-chicken, Alexa 594 goat anti-rat, Alexa 594 goat anti-guinea pig. Immunoperoxidase staining was performed using the ABC elite or M.O.M. kit (Vector Laboratory).
Paraffin sections (4 μm thick) were treated with an antigen retrieval solution (buffer A, Electron Microscopy Sciences) using a PickCell 2100-Retriever (Electron Microscopy Sciences), for phospho-histone H3 (rabbit; Upstate, #06570, 1/500) and caspase-3 (rabbit; Cell Signaling, #9661, 1/200 to 1/400).
BrdU was injected intraperitoneally (Sigma, 100 μg/gram body weight), and detected as in Tang et al. (2007). Placental alkaline phosphatase expression was assayed as in Shah et al. (2004).
Antibody Characterization
CB
The calbindin D28k antibody was characterized by Western blot. The antibody reacts specifically to a single band at 28 kDa on Western blot of rat hippocampal cells (Kim et al., 2006). Immuno-staining of mouse embryo brain sections produces a specific staining pattern of calbindin positive neurons identical to that published previously (Jacobowitz & Abbott, 1997, Chemoarchitectonic Atlas of the Developing Mouse Brain, CRC press).
CTIP2
On immunoblot prepared from nuclear extract from Jurkat cells immunoprecipitated with anti-Sir2 antibody, the antibody detects 2 bands representing CTIP2 at about 120kD. Immunohistochemistry show expression patterns that are indistinguishable from the RNA expression patterns.
CR (Chemicon)
As described recently (Xu et al., 2008), the calretinin antibody reacts to a 32 kDa band on Western blot of rat brain specific for calretinin. Immuno-staining shows a pattern of cellular morphology and distribution in the cortex consistent with published reports (Fonseca et al., 1995).
CR (Swant)
This antibody reacts specifically with calretinin in tissue originating from human, monkey, rat and mouse. This antiserum does not cross-react with calbindin D-28k or other known calcium binding proteins, as determined by its distribution in the brain, as well as by immunoblots using brain sections. (Schwaller et al., 1994).
Cleaved caspase-3
The antibody to cleaved caspase-3 was characterized by the manufacturer on Western blot. It detects the large fragment (17/19 kDa) of activated caspase-3 resulting from cleavage adjacent to Asp 17 in apoptotic C6, NIH/3T3, and Jurkat cells. It does not recognize full-length caspase-3 or other cleaved caspases. Immuno-staining detects apoptotic cells both in culture and in tissue sections. The specificity of the staining has been confirmed by the use of blocking peptide.
ChAT
As described recently (Xu et al., 2008), the antibody to ChAT reacts to a single band of 68–70 kDa on Western blot of rat brain. Immuno-staining shows a pattern of cellular morphology and distribution in the basal telencephalon consistent with previous reports (Corcoran et al., 2004; Wozniak et al., 1989).
DLX2
DLX2 Immunoreactivity disappears in Dlx1/2 mutant brain section (Long et al., 2007). In Dlx1/2 mutants, a deletion removes most of the coding exons for both Dlx1 and Dlx2 – but does not remove exon 1 – thus an N-terminal truncated protein could be produced – however, since the guinea pig anti-DLX2 antibody is directed to the N-terminal amino acids (1–154), we conclude that there is very little of this protein is present in the Dlx1/2 mutant brain.
Pan-DLX
pan-DLX immunohistochemistry to invertebrate tissues show expression patterns that are indistinguishable from the RNA expression patterns (Panganiban et al., 1995). Immunohistochemistry to vertebrate tissues show expression patterns that are indistinguishable from the sum of the Dlx1,2,5,6 RNA expression patterns (Stuhmer et al., 2002; Stuhmer et al., 2002). In this paper, Supplemental figure 2 shows a co-labeling experiment using the DLX2 and pan-DLX antibodies on mouse brain section. This data strongly suggests that all the DLX2+ cells are also pan-DLX+ and vice versa.
GFP (Rabbit)
According to the manufacturer, this anti-GFP antibody is suited for detection of native GFP, GFP variants, and most GFP fusion proteins by western blot and immunohistochemistry analysis using transgenic mice expressing the GFP gene product. No staining was detected on brain sections from wild type mice.
GFP (Chicken)
The antibody was analyzed by western blot analysis and immunohistochemistry using transgenic mice expressing the GFP gene product (according to the manufacturer). No staining was detected on brain sections from wild type mice.
NPAS1
Lack of staining on NPAS1 knockout mice brain sections (The NPAS1 mutant mouse was generated by inserting the E. coli β-galactosidase gene into NPAS1 gene which may produce a peptide composed of NPAS1’s N-terminal 41 amino acids fused to β-galactosidase) (Erbel-Sieler et al., 2004).
NPY (Sigma)
The NPY antibody was analyzed by the manufacturer using dot blot. It shows that the antibody reacts with NPY, with no cross-reactivity with substance P, neurokinin A, neurokinin B, vasoactive intestinal peptide (VIP), calcitonin gene related peptide, calcitonin, and somatostatin. Immunostaining labels NPY-containing neurons that can be blocked by pre-incubation of the antibody with NPY.
NPY (Immunostar)
This antibody was analyzed by the manufacturerusing the Biotin-Streptavidin/HRP detection method on rat brain tissue. All staining was blocked by pre-absorption of the diluted antiserum with excess NPY (10-6 M). Cross reactivity experiments in which diluted NPY antiserum was absorbed with excess (10-6M) peptide YY, avian pancreatic polypeptide, B-endorphin, vasoactive intestinal peptide, cholecystokinin or somatostatin showed no affect in blocking the intensity of staining.
PV
As previously described (Pantazopoulos et al., 2006), the parvalbumin antibody detects a single 12 kDa band on Western blot of rabbit skeletal muscle corresponding to parvalbumin. It does not cross-react with other members of the EF-hand family of calcium binding proteins. Immunohistochemistry shows a pattern of staining in the cortex consistent with that reported previously (Liodis et al., 2007; Xu et al., 2008).
PH3
The antibody to phospho-histone H3 was characterized by the manufacturer with Western blot on acid extracts of colcemid treated Hela cells. It shows that the antibody detects a single band at 17 kDa corresponding to phospho-histone H3. Immunohistochemistry shows that the antibody labels mitotic cells.
PLAP
No staining was detected on brain sections from wild type mice.
SS
As described recently (Xu et al., 2006), the specificity of the antibody to somatostatin has been confirmed by blocking of its immunoreactivity by synthetic somatostatin peptide.
TBR1
Lack of staining on Tbr1 knockout brain section and by western blot using brain protein extract and TBR1 antibody immunostaining on brain sections match in situ hybridization data (Englund et al., 2005).
Data Collection and Analysis
Brightfield images (PLAP staining and in situ hybridization) were taken using SZX7 microscope (Olympus, Japan) and DP70 camera (Olympus, Japan). 25x magnification confocal images were taken using a Zeiss LSM 510 confocal microscope (pinhole: 2 μm). Images were manipulated with Photoshop CS2 software (Adobe). Contrast and brightness of these images were adjusted for a better visualization.
Cell Counting
We used the counting approach described in Guillery, 2002. Tissue sections were 20 μm in thickness. We only counted cells with a clear nucleus (for Table 3); when it was not possible to clearly discern the nucleus, we counted cell somata (for Table 4: counting hippocampal neurons and striatal interneurons). We measured the size of the nucleus or somata to determine whether or not there was difference between genotypes (see sizes in Supplemental Method). As no size difference was noted, we did not apply the Abercrombie correction factor. Within the neocortex and hippocampus, we counted cells within a grid (see below). For the hippocampus and striatum we determined that the Lhx6 mutation did not result in a discernable change in the size of these structures. (See supplemental method).
Table 3.
Quantification of numbers of cells that are positive for PLAP, DLX2 and both PLAP & DLX2 immunoreactivity in the E14.5 and E18.5 cortex of Lhx6PLAP/+ and Lhx6PLAP/PLAP, using confocal data from Figure 1. The numbers represent the areal density (positive cells per 1000 μm2 of the box surface area) in cortex subregions. MZ: marginal zone; CP: cortical plate; II–IV: layers II, III and IV; V: layer V; VI: layer VI; SVZ: subventricular zone; VZ: ventricular zone.
| E14.5 | +/PLAP
|
PLAP/PLAP
|
||||
|---|---|---|---|---|---|---|
| PLAP | DLX2 | PLAP+DLX2 | PLAP | DLX2 | PLAP+DLX2 | |
| MZ | 0.0 | 0.0 | 1.6 | 0.0 | 0.0 | 0.7 |
| CP | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | 0.3 |
| SVZ/IZ | 0.0 | 0.0 | 1.2 | 0.0 | 0.0 | 1.1 |
|
| ||||||
| E18.5 | +/PLAP
|
PLAP/PLAP
|
||||
| PLAP | DLX2 | PLAP+DLX2 | PLAP | DLX2 | PLAP+DLX2 | |
|
| ||||||
| MZ | 0.0 | 1.7 | 5.9 | 0.0 | 3.1 | 0.2 |
| II–IV | 0.0 | 0.2 | 1.6 | 0.0 | 0.4 | 0.4 |
| V | 0.0 | 0.1 | 0.6 | 0.0 | 0.2 | 0.6 |
| VI | 0.0 | 0.3 | 0.6 | 0.0 | 0.3 | 0.9 |
| IZ | 0.0 | 0.5 | 0.5 | 0.0 | 0.5 | 0.9 |
| SVZ | 0.0 | 0.6 | 2.9 | 0.0 | 0.7 | 2.7 |
Table 4.
Quantification of numbers of cells positive for PLAP histochemistry in different cortical layers of E18.5 Lhx6PLAP/+ and Lhx6PLAP/PLAP, using light microscopic data from Figure 2 (20 μm section plane). Boxes 1, 2 & 3 correspond to different dorsoventral regions of the neocortex as indicated in Fig. 2C&D. The numbers represent the areal density (positive cells per 1000 μm2 of the box surface area) in cortex subregions. Abbreviations: see Table 3.
| E18.5 | Box 1
|
Box 2
|
Box 3
|
|||
|---|---|---|---|---|---|---|
| +/PLAP | PLAP/PLAP | +/PLAP | PLAP/PLAP | +/PLAP | PLAP/PLAP | |
| MZ | 10.8 | 1.3 | 11.8 | 1.2 | 7.0 | 0.7 |
| II–IV | 6.3 | 1.0 | 5.6 | 1.5 | 3.7 | 0.0 |
| V | 6.3 | 3.6 | 6.9 | 2.2 | 4.2 | 1.2 |
| VI | 1.9 | 3.7 | 2.3 | 3.9 | 2.2 | 3.0 |
| IZ | 2.8 | 5.6 | 2.0 | 3.3 | 1.4 | 3.0 |
| SVZ | 5.9 | 9.3 | 4.8 | 9.0 | 4.3 | 8.8 |
Counting Cortical and Hippocampal Interneurons Labeled By Alkaline Phosphatase Histochemistry and Immunofluorescence
To count cortical cells positive for DLX2 and or PLAP at E14.5 and E18.5, we used the confocal images shown in Figure 1; boxes were drawn to delimit the approximate positions of the MZ, CP, IZ/SVZ (for E14.5 and MZ), and layer II–IV, layer V, layer VI, IZ and SVZ (for E18.5; aided by the expression of CR for MZ, CTIP2 for layer V and TBR1 for layer VI; see Fig. 3). The area for each box was as follow:
Figure 1.
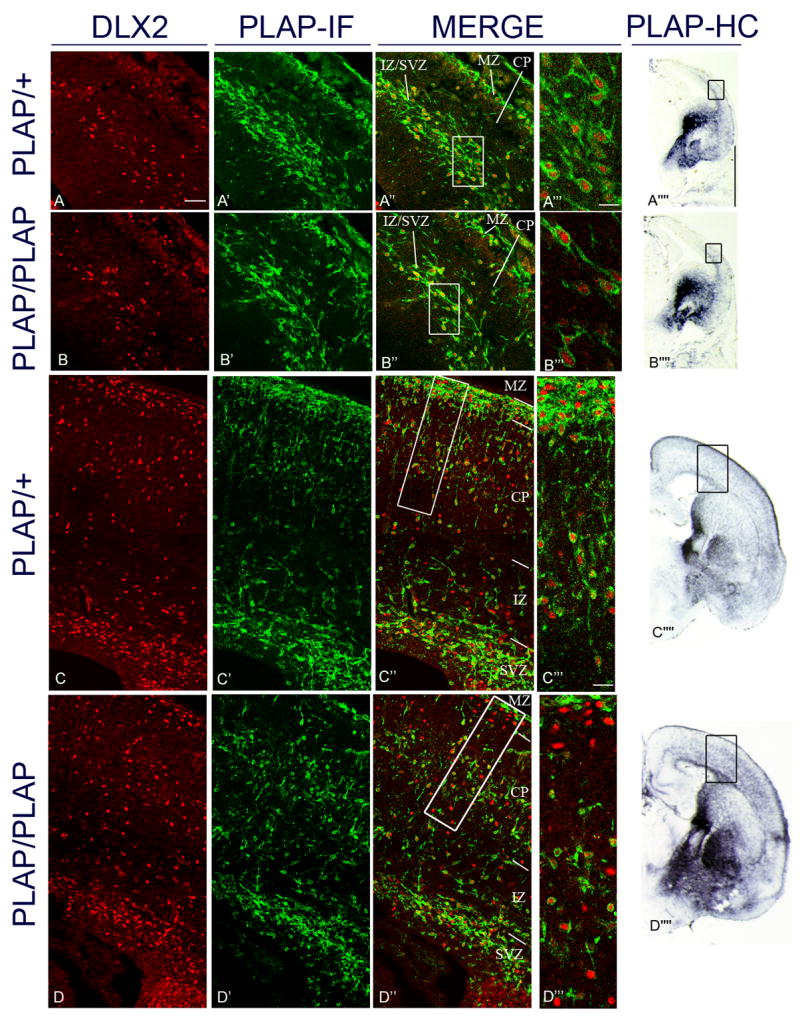
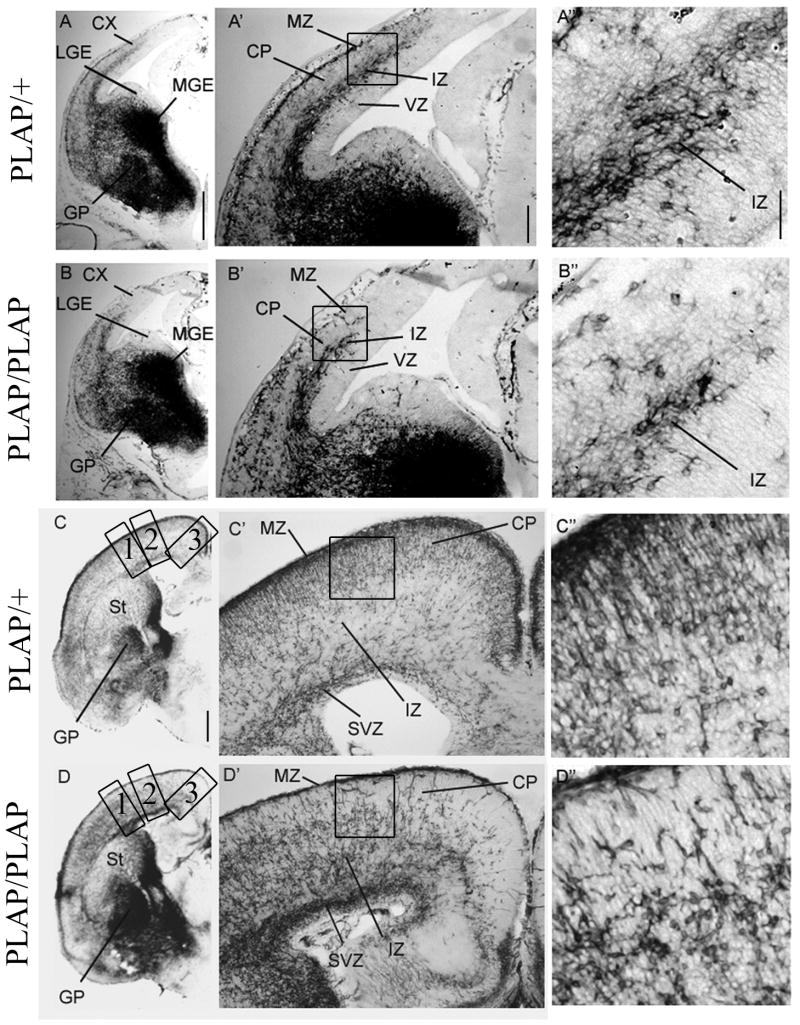
Analysis of Dlx2 and Lhx6 co-expression in the cortex of Lhx6PLAP/+ and Lhx6PLAP/PLAP at E14.5 and E18.5 by immunofluorescence of coronal sections. E14.5 data are shown in panels A–A’’’’ and B–B’’’’ and E18.5 data are shown in panels C–C’’’’ and D–D’’’’. PLAP expression was used to identify Lhx6+ cells. DLX2 immunofluorescence (red; first column); PLAP immunofluorescence (IF) (green; second column); merged (third column); higher magnification of marginal zone and cortical plate (see boxes in A″, B″, C″ and D″)(fourth column). Low magnification views of PLAP expression (histochemically-stained, HC) in telencephalic hemispheres (fifth column); boxed regions correspond to the approximate regions shown in the panels to the left. The boxes in the panels in the third column identify the regions shown at higher magnification in the fourth column. Panels C, C′ and C″ and D, D′ and D″ are composite images from the fusion of 2 images (confocal pictures; 25x objective) from the superficial region of the cortex and the deep region of the cortex.
At E14.5, the mutant has slightly reduced numbers of PLAP+ cortical interneurons compared to the heterozygote (A′ and B′), while at E18.5, the distribution of the PLAP+ cells in the mutant is very abnormal; compare with the heterozygote (C′ and D′). The mutant had very few PLAP+ cells in the superficial layers of the cortex (MZ and superficial layers of the CP). By contrast, the mutant had increased numbers of PLAP+ cells in deep regions of the CP and IZ (C′ and D′). At E14.5, Lhx6PLAP/+ and Lhx6PLAP/PLAP only exhibited interneurons that were Dlx2+;Lhx6+ (A″, A‴ and B″, B‴); whereas by E18.5, there were Dlx2 +;Lhx6− and Dlx2+;Lhx6+. Abbreviations: see Table: 3. Scale bar in A: 50 μm (for A–D, A′–D′ and A″–D″). Scale bar for A‴–B‴: 10 μm; for C‴ and D‴: 20 μm; for A’’’’ and B’’’’: 1mm; for C’’’’ and D’’’’: 1mm. See Supplemental Figure 20 for the magenta-green copy.
Figure 3.
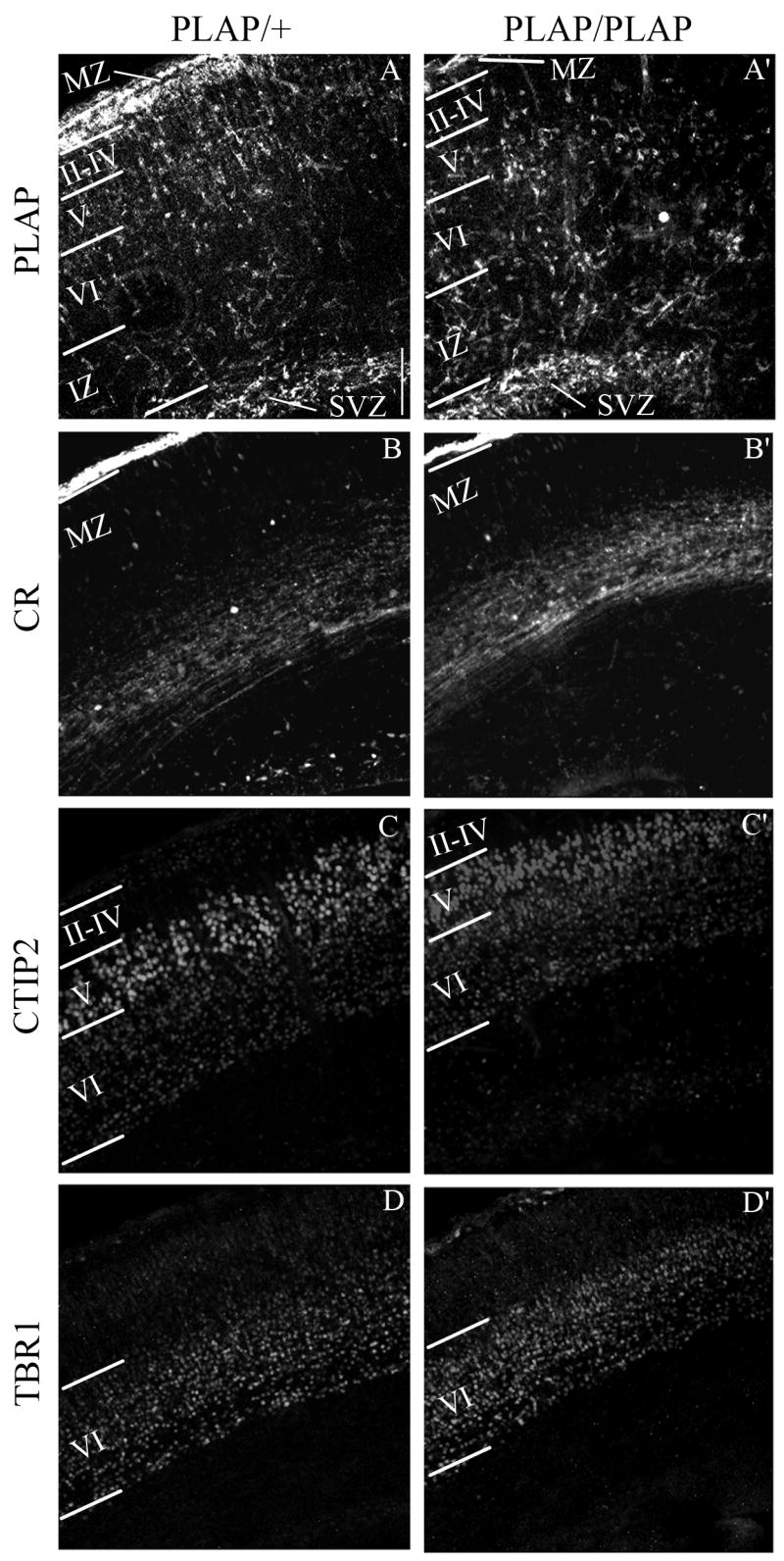
Laminar organization of cortical Lhx6+ interneurons and glutamatergic neurons in E18.5 Lhx6PLAP/+ and Lhx6PLAP/PLAP in coronal sections. The Lhx6+ interneurons were identified using rabbit anti-PLAP antibody (green) (A and A′) and the glutamatergic neurons (red) using the following antibodies: calretinin (CR) marks the neurons in the MZ (B and B′), CTIP2 identifies neurons in the layers VI and V (C and C′), and TBR1 labels the layer VI (D and D′). Abbreviations: see Table 3 and: II–IV: layers II, III and IV (cortical plate); V: layer V; VI: layer VI. Scale bar in A: 100 μm (for A–D and A′–D′).
E14.5: MZ: 6115 μm2; CP: 16163 μm2; IZ/SVZ: 19657 μm2. E18.5: MZ: 4238 μm2; layer II/IV: 8150 μm2; layer V: 19071 μm2; layer VI: 20864 μm2; IZ: 15322 μm2; SVZ: 14833 μm2.
To count E18.5 PLAP+ cortical cells, we used the images in Figure 2; boxes were drawn to delimit the MZ, layer II/IV, layer V, layer VI, IZ and SVZ, aided by the expression of CR for MZ, CTIP2 for layer V and TBR1 for layer VI; see Fig. 3. We counted three different dorso-ventral positions (see boxes in Fig. 2C and 2D). The area for each box was: MZ: 6953 μm2; layer II/IV: 11821 μm2; layer V: 15993 μm2; layer VI: 26772 μm2; IZ: 21904 μm2; SVZ: 15298 μm2. The same boxes were used for the same layer in the three different regions.
Figure 2.
Analysis of the position of Lhx6-expressing cells in the cortex and basal ganglia of Lhx6PLAP/+ and Lhx6PLAP/PLAP of coronal hemisections from E14.5 and E18.5 using alkaline phosphatase histochemical reaction to visualize the Lhx6+ cells. E14.5 data are shown in panels A–A″ and B–B″ and E18.5 data are shown in panels C–C″ and D–D″. Telencephalic left hemisphere (left column); higher magnification view of the ganglionic eminences and cortex (middle column); High magnification view of the cortex (right column; boxes in the middle column identify these regions). At E14.5, there is a retardation of the dorsal progression of the tangentially migrating PLAP+ cortical cells in the mutant (Figure 2A′, 2B′). At E18.5, the distribution of the PLAP+ cells is strongly affected in the mutant compared to the control. In the mutant, there is a large reduction in PLAP+ cells of the MZ and upper CP (figure 2C′ and 2D′), whereas there is an excess of PLAP+ cells in the lower part of the CP, the IZ and the SVZ (figure 2C′ and 2D′). There is a ventrodorsal gradient in the phenotype; the dorsomedial cortex is most affected. Abbreviations: see Table 3 and CX: cortex; GP: globus pallidus; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; St: striatum. Scale bar for A and B: 500 μm; for C and D: 500 μm; for A′–D′: 200 μm; for A″–D″: 50 μm.
To count the E18.5 cortical cells positive for pan-DLX and or GFP (Lhx6-BAC-GFP), we used the confocal images in Supplemental Fig. 1; boxes were drawn to delimit the approximate positions of the MZ, CP, IZ and SVZ. The area for each box was: MZ: 16463 μm2; CP: 105557 μm2; IZ: 60952 μm2; SVZ: 47773 μm2. Numbers shown in Table 3, Table 4 and supplemental Table 1 are the areal density (number of positive cells/surface area of the box μm2)). The values were multiplied by 1000 to make it easier to read.
Counting Cortical and Hippocampal Interneurons Labeled By In Situ Hybridization
To count cortical cells labeled by in situ hybridization, we drew a box on images of the cortex of P17 animals; the box extended from the ventricular zone to the pial surface. Three horizontal lines divided the box into three strata; these were drawn at approximately 9%, 53% and 85% from the pial surface. We counted cells in these boxes, and estimate that they correspond to different cortical layers: 0–9%: ~layer I; 9%–53% ~layers II–III; 53%–85% ~layers IV–VI.
Counting of Immunostained Striatal Interneurons
To count striatal interneurons positive for SOM, PV, NPY, or ChAT, three successive coronal sections, 40 μm thick and 240 μm apart, were selected from a region anterior to the crossing of the anterior commissure. The entire striatum on both sides of these sections was photographed. Positively labeled neurons were manually counted. The numbers for the striatum on each side of these sections were then averaged. The counting was performed exactly the same for both control and Lhx6 homozygous mutant animals. The differences between these two were statistically analyzed using the student’s t-test.
Results
Lhx6 Expression Marks a Subset of Tangentially Migrating Interneurons
Lhx6 expression in the adult neocortex primarily marks parvalbumin+ and somatostatin+ interneurons (Cobos et al., 2005; Cobos et al., 2006; Liodis et al., 2007). We compared Lhx6 and Dlx expression in the prenatal mouse cortex; we chose Dlx expression, because most (perhaps all) immature cortical interneurons express these genes (Anderson et al., 1999; Anderson et al., 1997; Cobos et al., 2007; Cobos et al., 2005; Cobos et al., 2006; Stuhmer et al., 2002b). We studied the co-expression of DLX2 and all DLX proteins (pan-distaless antibody) with marker genes expressed from the Lhx6 locus at E14.5 and E18.5 (Fig. 1; Supplemental Fig. 1). We identified Lhx6+ cells using either PLAP expressed by the endogenous Lhx6 locus (the heterozygote mouse has only one functional Lhx6 allele; (Choi et al., 2005), or green fluorescent protein (GFP) expressed from a BAC transgenic mouse (Lhx6 function is believed to be inactivated in this allele; Cobos et al., 2006; Gong et al., 2003).
We compared the numbers of Dlx+ and Lhx6+ cells in different layers of the prenatal neocortex at E14.5 and E18.5 (Table 3; Supplemental Table 1). Double-immunofluorescence labeling identified two types of cells: Dlx+;Lhx6− and Dlx+;Lhx6+ using both the Lhx6PLAP and Lhx6-GFP mice; we did not unequivocally identify Dlx−;Lhx6+ cells. Between E14.5–E18.5, there is an increase in Dlx+;Lhx6−, particularly in the subventricular zone (Fig. 1A″, C″). To examine whether Dlx+;Lhx6− cells contribute to distinct interneuron subtypes, we studied the phenotype of mice lacking Lhx6..
Lhx6PLAP/PLAP Cells Migrate to the Cortex But Show Deficits in Populating the Marginal Zone and Cortical Plate
In the Lhx6PLAP allele, PLAP replaces part of exon 2 and all of exons 3, 4, 5, and 6; this deleted region encodes amino acid residues 25 to 264, which includes a large part of the N-terminus, the entire LIM1 and LIM2 domains, and 49 out of the 62 amino acid residues of the homeodomain (Choi et al., 2005). The increased intensity of PLAP expression in the Lhx6PLAP/PLAP mutants, relative to the Lhx6PLAP/+, could primarily reflect PLAP dosage.
PLAP labels the MGE progenitors and radially migrating cells that become pallidal projection neurons and tangentially migrating cells, which contribute to the population of cortical interneurons (Fig. 2; Supplemental Figs. 3, 4). Lhx6PLAP/PLAP mice express PLAP in the MGE progenitor domains, its mantle (pallidum) and in cells tangentially migrating to the cortex at E12.5, E14.5, E16.5 and E18.5 (Fig. 2; Supplemental Figs. 3, 4). This demonstrates that cells lacking Lhx6 expression can tangentially migrate to the cortex. However, both the superficial (marginal zone) and deep (intermediate/subventricular zone) E14.5 cortical tangential migrations in the Lhx6PLAP/PLAP mutants do not advance as far dorsally (particularly in rostral regions) (Fig. 2B, B′; Supplemental Fig. 3). The PLAP+ mutant cells continue to express DLX2 (Fig. 1B–B″).
At E16.5 and E18.5, the deficit in the mutant’s deep tangential migration has recovered, whereas the marginal zone and neocortical plate defect persists (Fig. 1C′, D′; Fig. 2C′, D′; Supplemental Fig. 4). Those interneurons that remain in the superficial layers are DLX2+;PLAP− (Fig. 1D–D‴). There are very few PLAP+ cells in dorsomedial parts of the neocortex. Surprisingly, the hippocampal primordium does have PLAP+ cells in the marginal zone suggesting that neocortical and the hippocampal interneurons have different dependence on Lhx6 function (Fig. 2; Supplemental Fig. 4).
We compared the laminar distribution of PLAP+ cells in the neocortex of Lhx6+/PLAP and Lhx6PLAP/PLAP mice at E18.5 (Table 4), using the following markers: Tbr1 (subplate and layer VI), CTIP2 (layer V), and calretinin (marginal zone, subplate and intermediate zone) (Fig 3). The phenotype varied as a function of dorsoventral position, although in all regions the mutant had ~10-fold fewer PLAP+ cells in the marginal zone (MZ) and presumptive layers II–IV (Table 4). This phenotype was most severe in the dorsal neocortex. Layer V also had fewer PLAP+ cells, but the reduction was ~2–3-fold. On the other hand, presumptive layer VI, subplate, intermediate zone and subventricular zone had increased PLAP+ cells.
Molecular Defects of the Lhx6PLAP/PLAP MGE and Pallidum
Lhx6 is expressed in the progenitors in the MGE, and neurons in the globus pallidus, bed nucleus stria terminals and the ventral pallium (Flames et al., 2007; Garcia-Lopez et al., 2008; Grigoriou et al., 1998; Lavdas et al., 1999; Liodis et al., 2007; Marin et al., 2000; Sussel et al., 1999). Based on the PLAP expression, MGE development appears to be preserved in the Lhx6PLAP/PLAP mutants (Figs. 2, 4A–B; Supplemental Figs. 3, 4). Proliferation in the MGE appears normal based on the number of phospho-histone 3+ (M-phase) cells at E12.5, E14.5, E16.5 and E18.5 (Supplemental Fig. 18). In addition, we did not detect increased levels of prenatal apoptosis (caspase-3+ cells) (Supplemental Fig. 16).
Figure 4.
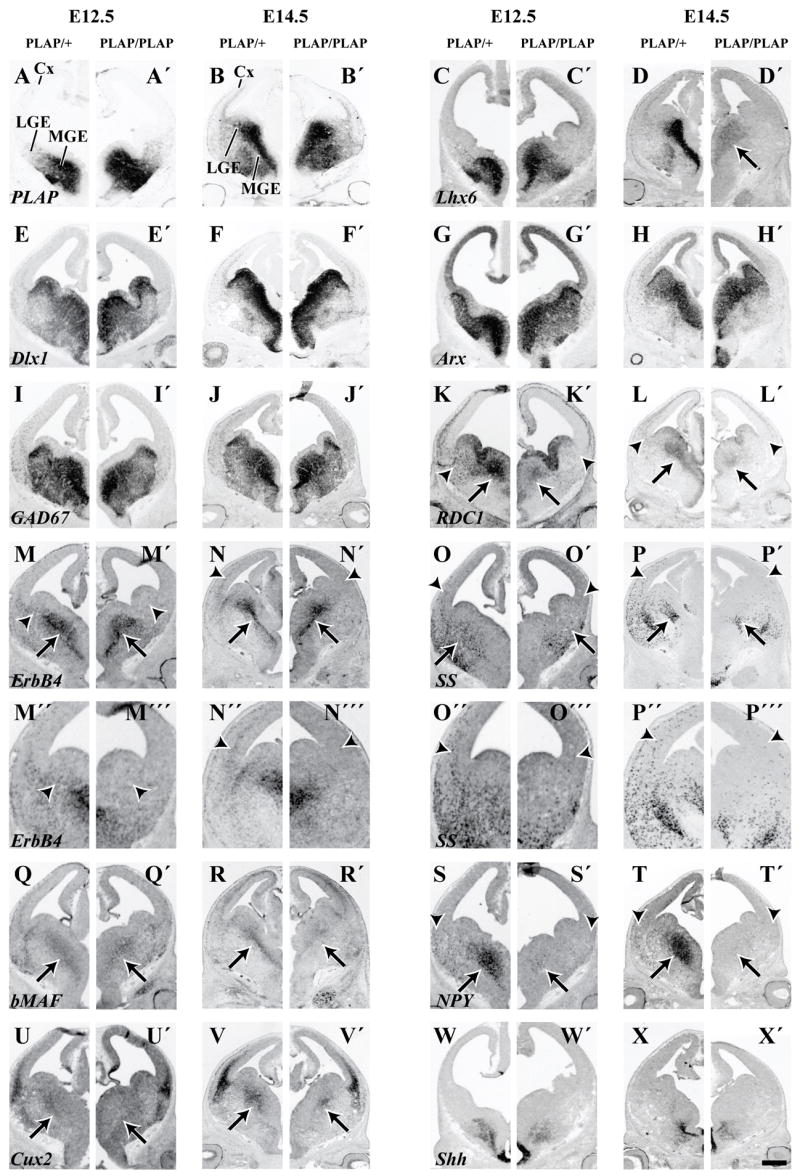
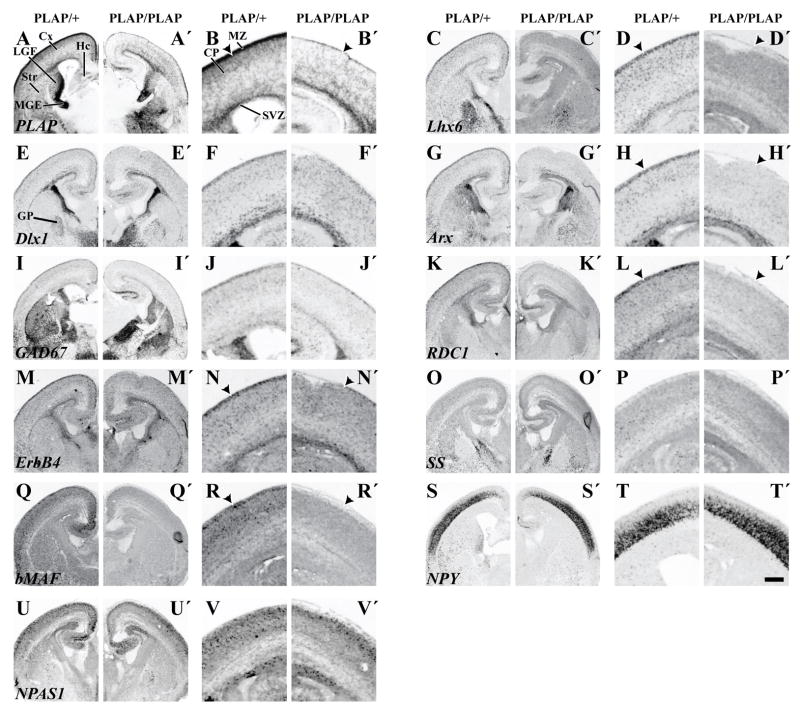
Molecular features of the basal ganglia and cortex of Lhx6 PLAP/+ and Lhx6 PLAP/PLAP at E12.5 and E14.5 shown by in situ RNA hybridization on coronal hemisections, from a middle position along the rostrocaudal dimension. PLAP histochemistry is also shown. Several markers show little or no change in expression in the MGE/LGE of Lhx6 PLAP/PLAP mutant animals; PLAP (A–B′), Dlx1 (E–F′), Arx (G–H′), GAD67 (I–J′) and Shh (W–X′). However, Lhx6 expression is reduced in the MGE at E14.5 (D, D′). ErbB4 expression is not detectable in the LGE (arrowheads; M, M′) and in cells migrating to the cortex (arrowheads; N, N′). The changes in expression of ErbB4 and SS in the LGE and ventral neocortex are shown at higher magnification (M″, M‴, N″, N‴). The same is true for RDC1 (K–L′), SS (O–P′) and NPY (S–T′). The MGE expression of ErbB4 appears relatively unaffected, while for RDC1 (K–L′), bMAF (Q–R′), NPY (S–T′) and Cux2 (U–V′) all show a dramatic reduction in expression (arrows). Abbreviations: Cx, Cortex; LGE, Lateral Ganglionic Eminence; MGE, Medial Ganglionic Eminence. Scale bar: A–X′, 500 μm.
To obtain molecular information on whether loss of Lhx6 function alters the development of pallidal neurons, we compared gene expression from Lhx6PLAP/PLAP and Lhx6PLAP/+ E12.5, E14.5, E16.5 and E18.5 brains. We studied MGE marker expression using in situ RNA hybridization for: Arx, bMaf, Cux2, CXCR4, Dlx1, ER81, ErbB4, GAD1 (GAD67), Lhx6, Lhx7, Lmo3, Nkx2.1, NPAS1, NPY, RDC1, somatostatin and thrombospondin (Figs. 4, 5; Supplemental Figs. 5, 6, 7, 8).
Figure 5.
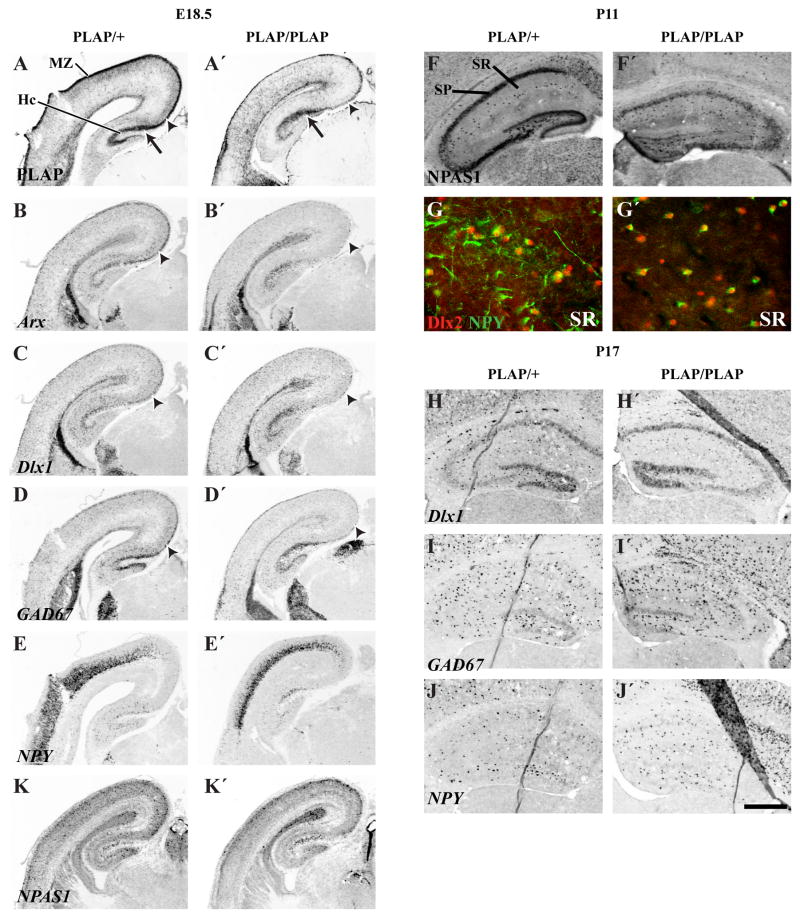
Molecular features of the basal ganglia and cortex of Lhx6 PLAP/+ and Lhx6 PLAP/PLAP at E18.5 shown by in situ RNA hybridization on coronal hemisections, from a middle position along the rostrocaudal dimension. Two-fold higher magnification views of the cortex for each specimen are shown to the right. PLAP histochemistry is also shown. PLAP expression is nearly lost in the marginal zone (B, B′; arrowheads). Lhx6 (D, D′); Arx (H, H′); RDC1 (L, L′); ErbB4 (N, N′); and bMAF (R, R′) also show a decrease in marginal zone expression. Lhx6 (D, D′); Arx (H, H′); RDC1 (L, L′); ErbB4 (N, N′); SS (P, P′) and bMAF (R, R′) also show a decrease in cortical interneuron expression. Note: most of the neocortical NPY expression is in immature pyramidal cells at this age (S, S′, T, T′). Scale bar: 500 μm: low magnification’; 250 μm high magnification.
Expression of Lhx6 RNA and PLAP activity are maintained in the SVZ domain of the MGE in the Lhx6PLAP/PLAP mutants at E12.5, E14.5, E16.5 and E18.5 (Fig. 4A–D′, 5A–D′; Supplemental Figs. 5, 7, 8). However, by E14.5, in the MZ of the Lhx6PLAP/PLAP MGE, PLAP activity is much easier to detect than Lhx6 RNA, by contrast to the Lhx6PLAP/+, where Lhx6 RNA and PLAP expression are both strong (Fig. 4B, B′, D, D′). This suggests that Lhx6 RNA expression fails to be maintained in most of the postmitotic cells that migrate from the progenitor domains into the MZ, where the neurons mature. The residual expression of Lhx6 RNA overlaps with expression of Lhx7 and Nkx2.1 (Supplemental Fig. 5), suggesting that these TFs preserve Lhx6 expression in progenitor cells.
Lack of Lhx6 function leads to reduced MGE and pallidal expression of three transcription factors (TFs) Cux2, bMaf and NPAS1 at E12.5 and E14.5 (arrows; Fig. 4Q–R′, U–V′; Supplemental Fig. 5). Cux2 and bMaf expression become restricted to a small domain in the dorsal MGE; bMaf expression is nearly lost from a thin “stream” of SVZ cells that extends from the VZ of the ventral MGE (arrow; Fig. 4Q–R′). NPAS1 expression is reduced in the dorsal MGE (arrow), but is maintained in the ventral MGE and CGE (arrowhead) (Supplemental Fig. 5).
Despite these molecular defects, expression of other TFs in MGE progenitor and postmitotic zones (including the presumptive globus pallidus), appear preserved (Arx, Dlx1, Er81, Lhx7, Lmo3 and Nkx2.1; Fig. 4E–H′; Supplemental Fig. 5).
The differential effect on TF expression suggests that Lhx6 has a selective role in controlling the differentiation of specific sets of effector genes and/or subsets of MGE-derived cells. Consistent with this idea is the maintained MGE/pallidal expression of the ErbB4 receptor tyrosine kinase and the GABA synthesizing enzyme (GAD1) (Fig. 4M–N′; I–J′). However, RNA expression of NPY is virtually eliminated and somatostatin expression is greatly reduced (arrow; Fig. 4S–T′; O–P′). Furthermore, expression of the cytokine receptors CXCR4 and RDC1 (CXCR7, CMKOR1) are greatly reduced (arrow; Fig. 4K–L′; Supplemental Figs. 5, 6). These CXCL12 (SDF1) receptors are implicated in regulating cortical interneuron migration (Li et al., 2008; López-Bendito et al., 2008; Stumm et al., 2007; Stumm et al., 2003; Y. Wang, J. Long, D. Crawford, T. Behrens, and J. Rubenstein, unpublished).
Analysis of PLAP, Arx, Dlx1 and Lhx7 expression in the E16.5 and E18.5 basal ganglia of Lhx6PLAP/PLAP mutants did not identify major anatomical or molecular defects (Fig. 5; Supplemental Figs. 4, 6, 7), except for the periventricular accumulation of PLAP+ cells. This ectopia is located in the dorsal MGE and expresses DLX2 (Supplemental Fig. 9). BrdU birthdating shows that many of these cells had their last mitosis around E13.5 (Supplemental Fig. 9).
Molecular Defects in Tangentially Migrating Lhx6PLAP/PLAP Neocortical Interneurons
To determine the molecular profile of tangentially migrating cells in the Lhx6PLAP/PLAP mutants from E12.5–E18.5, we examined expression of genes known to mark, or regulate, their development (Fig. 4; Supplemental Figs. 5, 6, 7). Cortical expression of Dlx1 and GAD67 in Lhx6PLAP/PLAP mutants at E14.5 was moderately decreased, similar to the reduction in PLAP expression (Fig. 4F, F′, J, J′; see higher magnification in Supplemental Fig. 3, 6). Furthermore, interneuron expression of Arx was greatly reduced and interneuron expression of Cux2, CXCR4, ErbB4, bMaf, NPY, RDC1, somatostatin and thrombospondin were not detectable in the Lhx6PLAP/PLAP mutants (Fig. 4; see higher magnification in Supplemental Fig. 6). Arx encodes a homeobox gene whose function is required for development of cortical interneurons and is implicated in promoting migration (Colombo et al., 2007; Kitamura et al., 2002). In the Lhx6PLAP/PLAP mutants, Arx expression is maintained in the MGE and LGE, thus the Lhx6PLAP/PLAP mutants should not fully phenocopy the Arx mutant.
ErbB4 function is linked to the attraction of interneurons into the cortex (Flames et al., 2004). While ErbB4 expression is maintained in the MGE (arrows; Fig. 4M–N′), ErbB4+ cells fail to spread into the LGE and the cortex (arrowhead; Fig. 4M′, N′; Supplemental Fig. 6). This suggests that Lhx6 is required to maintain ErbB4 expression in the immature interneurons migrating from the MGE en route to the cortex. CXCR4 and RDC1 encode cytokine receptors that transduce CXCL12 (SDF1) signaling (Li et al., 2008; López-Bendito et al., 2008; Stumm et al., 2007; Stumm et al., 2003). CXCL12 (SDF1) is an attractant for Lhx6+ interneurons (Li et al., 2008). CXCR4 regulates the laminar organization of migrating cortical interneurons. RDC1 is expressed in developing cortical interneurons (J. Long and J. Rubenstein, unpublished results). While RDC1 function in migrating cortical interneurons is not known, it regulates migration of lateral line cells in zebrafish (Dambly-Chaudiere et al., 2007; Valentin et al., 2007). CXCR4 and RDC1 expression in the Lhx6PLAP/PLAP mutant MGE are greatly reduced (arrows; Fig. 4K–L′; Supplemental Fig. 6), and expression in tangential streams to the cortex is not detectable (arrowheads; Fig. 4K′, L′; Supplemental Fig. 6).
In sum, loss of Lhx6 function results in important molecular defects in migrating interneurons that can account, at least in part, for reduced tangential migration and abnormal laminar location of those interneurons that reach the cortex. These results suggest that Lhx6 may not have a major function in regulating expression of some markers like Dlx1 and GAD67, but rather it regulates the number and types of interneurons reaching the cortex, and their position within cortical layers and regions.
Late Lhx6PLAP/PLAP Tangential Migrations in the Cortical SVZ/IZ Are Relatively Preserved
With advancing age (E16.5 and E18.5), the cortical SVZ of Lhx6PLAP/PLAP mutants showed strong expression of markers of tangentially migrating interneurons (Arx, Dlx1, ErbB4, GAD67) (Fig. 5E–J′, M–N′; Supplemental Figs. 7, 8). This result is consistent with PLAP expression (Figs. 1C′, D′, 2C–D′; Supplemental Fig. 4) and the observation that there are increasing numbers of Dlx+;Lhx6− tangentially migrating interneurons in the cortical SVZ/IZ late in gestation (Fig. 1C–C‴; Supplemental Fig. 1). To establish the effect of the mutation on interneuron subtypes and their distributions within the cortex and hippocampus, we studied late gestation and early postnatal Lhx6PLAP/PLAP mutants.
Distribution and Types of Cortical and Hippocampal Interneurons Late in Gestation and Postnatally
At E18.5, PLAP expression shows that cortical interneurons have failed to adequately populate the marginal zone and cortical plate of Lhx6PLAP/PLAP mutants (Fig. 2D–D″; Supplemental Fig. 4). Furthermore, these layers have very low expression of Arx, ErbB4 bMaf, RDC1, and somatostatin (Fig. 5G–H′, K–R′; Supplemental Fig. 8). However, marginal zone and cortical plate expression of Dlx1, GAD67 and NPAS1 are better preserved (Fig. 5E–F′, I–J′, U, U′, V, V′; Fig. 5A, A′, B, B′, C, C′, D, D′, K, K′); The NPAS1+ cells are DLX+ (Supplemental Fig. 19).
Unlike the neocortex, many molecular features of hippocampal interneurons are preserved in the E18.5 Lhx6PLAP/PLAP mutants. Interneurons, in the superficial layers of the CA fields, maintain robust expression of PLAP, Arx, Dlx1, GAD67 and NPAS1 (Figs. 5, 7; Supplemental Figs. 4, 19), with lower levels of ErbB4, and NPY (Fig. 5 and not shown). On the other hand, hippocampal expression of bMaf, RDC1 and somatostatin was not detected (Fig. 5K, K′, O, O′, Q, Q′; Supplemental Fig. 8).
Figure 7.
Hippocampal interneuron phenotypes in Lhx6 PLAP/+ and Lhx6 PLAP/PLAP at E18.5, P11 and P17. While in situ hybridization and PLAP histochemistry shows a dramatic reduction in staining in the dorsomedial cortical plate and marginal zone in E18.5 Lhx6 PLAP/PLAP animals (arrowheads; A–D′), the hippocampus maintains its PLAP staining (arrow; A, A′). Similar results are seen for Arx (B, B′), Dlx1 (C, C′), GAD67 (D, D′) and NPAS1 (K, K′) at E18.5. Note how hippocampal expression of NPY (E, E′) is maintained in the same region showing PLAP, Arx, Dlx1, GAD67 and NPAS1 expression. At P11, there is ~2-fold increase in the number of NPAS1 immunoreactive cells in hippocampal interneuron layers (easiest to see in stratum radiatum, SR) in the Lhx6 PLAP/PLAP animals (F′). NPY+ cells in the hippocampus completely co-label with Dlx2 (G), but there are more Dlx2+/NPY− cells in the Lhx6 PLAP/PLAP animals at P11 (G′). Note the reduction of NPY+ processes. Cell counting in rostral hippocampal P17 sections (H–J′): Dlx1+: 143±36 (Lhx6 PLAP/+); 156±34 (Lhx6 PLAP/PLAP) (p=0.7); GAD67+: 289±45 vs. 281±39 (p=0.7); NPY+: 141±20 vs. 196±50 (p=0.1). Abbreviations: Hc, Hippocampus; MZ, Marginal Zone; SR, Stratum Radiatum; SP, Stratum Pyramidale. Scale bar: A–E′: 700 μm; F–F′:480 μm; G–G′: 80 μm; H–J′: 500 μm. See Supplemental Figure 21 for the magenta-green copy.
Neonatal Lhx6PLAP/PLAP mutants were indistinguishable from the heterozygote or wild type littermates for at least one week (based on simple examination of their size, movements and behaviors). By P10 the mutants appeared smaller than the controls, and by P14–P18, they were ~half the size and less active than controls. No Lhx6PLAP/PLAP mutants were detected by weaning at P21.
Analysis of the postnatal neocortex and hippocampus at P11, P15 and P18 showed profound interneuron defects (Figs. 6, 7; Supplemental Figs. 12, 13; and data not shown). As reported by Liodis et al., (2007), parvalbumin immunoreactivity (at P15 and P18) was reduced to just a few cells in deep neocortical layers. Strongly positive somatostatin immunoreactive cells (at P15 and P18) were almost eliminated; weakly positive cells were detectable. Calbindin and calretinin immunoreactivity were greatly reduced (at P15 and P18); dysmorphic calbindin+ cells remained in three layers (Supplemental Figs. 12, 13).
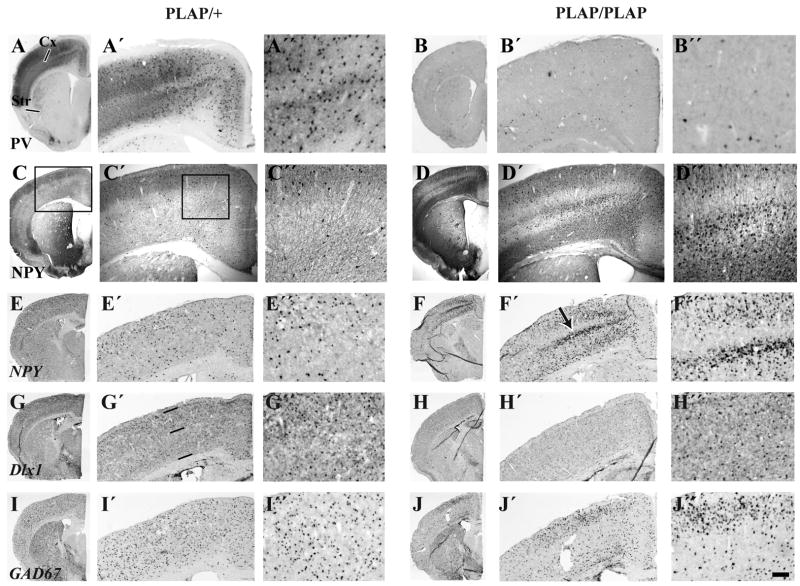
Figure 6.
Cortical interneuron defects in Lhx6 PLAP/+ and Lhx6 PLAP/PLAP in coronal hemisections from P17 animals. Parvalbumin histochemical staining in the cortex of Lhx6 PLAP/PLAP animals is severely reduced (B′, B″), while NPY histochemical staining is increased (D′, D″). The cortex was split into two zones for counting analysis of in situ hybridization signals. Zone 1 includes roughly layers I–IV, and zone 2 includes roughly layers V–VI; the zones are demarcated by three horizontal lines (G′). The in situ hybridization analysis shows that the numbers of Dlx+ and GAD67+ cells in zone 1 don’t change significantly, whereas there was a ~2–3 fold increase in zone 2 in the Lhx6 PLAP/PLAP animals at P17 (G′–H″). Comparing Lhx6 PLAP/+ and Lhx6 PLAP/PLAP: Dlx1: Zone 1 (126 vs. 146); Zone 2: 112 vs. 223); GAD67: Zone 1 (272 vs. 318); Zone 2 (133 vs. 317); NPY: Zone 1 (68 vs. 353); Zone 2 (65 vs. 237). Scale bar: A–J: 1 mm; A′–J′:370 μm; A″–J″: 150 μm.
On the other hand, cortical and hippocampal expression of Dlx1 (P18), Dlx2 (P11), GAD67 (P18) and NPY (at P11, P15 and P18) remained strong (Figs. 6 and 7; Supplemental Figs. 11, 12, 13); these aspects of the Lhx6 mutant were previously not reported. The number of Dlx1, Dlx2, GAD67 and NPY expressing neocortical cells were increased; this was easiest to quantify using in situ RNA hybridization (Fig. 6; see numbers in the legend to Fig. 6). Some of the NPY+ cells are probably not interneurons because their laminar distribution differs from Dlx1 and GAD67 expression (arrow Fig. 6F′). Double immunofluorescence labeling showed that many NPY+ cells co-express DLX2 in the neocortex and hippocampus, providing evidence that these are interneurons (Fig. 7 and Supplemental Fig. 19). Unfortunately, because postnatal expression of PLAP was too low in both the Lhx6PLAP/+ and Lhx6PLAP/PLAP mice, we could not determine whether the NPY+ interneurons were PLAP+. Thus, while early (E12.5 and E14.5) NPY expression in the MGE and migrating cells is dependent on Lhx6 (Fig. 4), later expression (P11 and P18) in cortical and hippocampal interneurons is Lhx6-independent.
Finally, like Dlx1, NPAS1 expression was maintained in the mutant hippocampus (Fig. 7; Supplemental Fig. 19). In the neocortex, NPAS1 expression was maintained in superficial layers, but was reduced in deep layers (Fig. 5 U, U′, V, V′; Supplemental Fig. 19).
Selective Postnatal Deficits of Striatal Interneurons
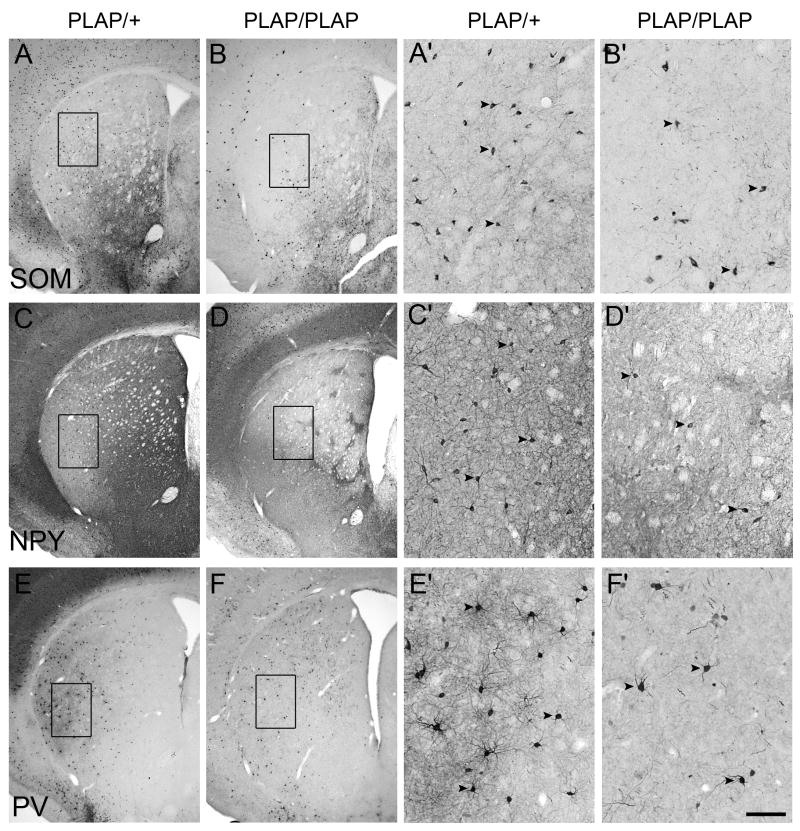
Postnatal analysis of Lhx6PLAP/PLAP mutant basal ganglia showed no overt morphological defects (Fig. 8; Supplemental Figs. 10, 15). The Lhx6PLAP/PLAP mutant continues to express parvalbumin in the globus pallidus (Supplemental Fig. 10). However, there was an abnormal distribution of calretinin, NPY, parvalbumin and somatostatin immunoreactivity in the septum, nucleus accumbens, striatum, BSTN and amygdala (Fig. 8; Supplemental Fig. 10). Some of these defects are due to reduced interneuron numbers (legend to Fig. 8) – for instance, the striatum has a ~2-fold reduction of somatostatin+ and NPY+ interneurons.
Figure 8.
Striatal interneuron phenotypes in Lhx6 PLAP/PLAP (B, B′, D, D′, F, F′) mutant as compared to Lhx6 PLAP/+ control (A, A′, C, C′, E, E′). Parts of A–F are enlarged and shown in A′–F′. Immunohistochemical analysis on coronal hemisections from P17 animals showing a ~2-fold reduction of somatotstatin (SOM) (A, A′, B, B′) and neuropeptide Y (NPY) (C, C′, D, D′) cells, non-significant reduction in parvalbumin+ interneurons (PV) (E, E′, F, F′). Somatostatin: heterozygote: 268 +/− 23; homozygote: 112 +/− 5; P = 0.0003; NPY: heterozygote: 193 +/− 36; homozygote: 104 +/− 4; P = 0.013; parvalbumin: heterozygote: 243 +/− 41; homozygote: 204 +/− 58; P = 0.4; ChAT: heterozygote: 232 +/− 20; homozygote: 190 +/− 21; P = 0.07. Three animals for each genotype were examined. Arrowheads point to selected labeled cells. Note the change in neuropile staining and interneuron distribution. Scale bar in F′: 500 μm for A–F and 100 μm for A′–F′.
Discussion
Lhx6 Dependent and Independent Tangential Migrations
Here we provide evidence that Lhx6 is expressed in a large subset of the DLX+ tangentially migrating immature interneurons. At the earliest stages of tangential migration, the vast majority of interneurons are Dlx+;Lhx6+ (Fig. 1; Supplemental Fig. 1). With progressive time, there are increasing numbers of Dlx+;Lhx6− tangentially migrating immature interneurons; this cell type migrates in both the deep and superficial streams, but is predominantly found in the IZ and SVZ. While our analyses do not discern the anatomical origins of Dlx+;Lhx6+ and Dlx+;Lhx6− cells, a parsimonious hypothesis is that the Dlx+;Lhx6+ are derived from the MGE, whereas the Dlx+;Lhx6− cells arise from the caudal LGE – the dorsal CGE (dCGE) (Flames et al., 2007). The CGE is currently implicated in producing late-born interneurons that express calretinin and NPY (Butt et al., 2005; Fogarty et al., 2007; Xu et al., 2004).
Lhx6 Is Required For ErbB4 Expression and Efficient Tangential Migration to the Cortex
Consistent with previous studies, we find that Lhx6 function participates in tangential migration to the cortex. Unlike previous studies, our analyses had the advantage of a cell autonomous marker of the Lhx6 homozygous mutant cells, as PLAP was inserted into the deleted Lhx6 allele (Choi et al., 2005). Unlike Alifragis et al., 2004, who used siRNA to reduce Lhx6 expression, we find that loss of Lhx6 does not block tangential migration to the cortex, except perhaps in a subset of cells that remain as a periventricular ectopia (Supplemental Fig. 9). Our results are similar to Liodis et al., 2007, who studied a different Lhx6 mutant. The reduced number of cortical GAD67+ cells in their Lhx6−/−mutant resembles the Lhx6PLAP/PLAP mutant, particularly in the E14.5 dorsal neocortex. As development proceeds the superficial zone is more affected (Figs. 1, 2, 3, 4; Supplemental Figs. 3, 5, 6, 7, 8). We suggest that the reduced migration is caused in part by reduced ErbB4 expression, a receptor mediating neuregulin1 attraction of tangentially migrating cortical interneurons (Flames et al., 2004).
Lhx6 Is Required For Expression of CXCL12 (SDF1) Receptors (CXCR4 and RDC1) and Cortical Plate Laminar Position
Lhx6PLAP/PLAP mutant PLAP+ cells are greatly reduced in the marginal zone and superficial layers of the E16.5 and E18.5 cortical plate. However, they are found in increased numbers in layer VI, the intermediate and subventricular zones (Table 4; Figs. 1, 2, 3). These data support a model that Lhx6 regulates the laminar position of the Lhx6PLAP+ interneurons.
On the other hand, DLX2+;Lhx6PLAP− interneurons do not show the deficit in populating superficial layers of the cortex (Fig. 1D; Table 3), providing evidence that the laminar phenotypes of Lhx6PLAP+ cells are due to autonomous molecular deficits in responding to environmental cues. The postnatal cortex (P11, P15 and P18) has interneurons (Dlx1+, Dlx2+ or GAD67+ cells) in all cortical layers (Fig. 6), but with skewing of their laminar distribution (~2-fold increase in deep layers of Dlx1+ or GAD67+ cells; see Fig. 6 legend).
Little is known about how interneurons radially migrate into the cortical plate, although there is evidence that cytokine signaling participates in this process. Mice lacking either the CXCL12 (SDF1) cytokine, or its receptor (CXCR4), show defects in interneuron laminar position (Li et al., 2008; López-Bendito et al., 2008; Stumm et al., 2007; Stumm et al., 2003). In particular, CXCR4 mutants show deficits in interneuron localization to the marginal zone, presumably because CXCR4 mediates their attraction to SDF1 secreted by the meninges.
CXCR4 expression is not detectable in immature cortical interneurons in the Lhx6PLAP/PLAP mutants (Supplemental Figs. 5, 6). Thus, this could account for the decreased number of interneurons in the marginal zone. However the laminar phenotype is more severe than in the CXCR4 mutant. We found that the Lhx6PLAP/PLAP mutants also lack detectable expression of a new SDF1 receptor named RDC1 (CXCR7; CMKOR1) (Figs. 4, 5)(Balabanian et al., 2005). RDC1 expression is down-regulated in the basal ganglia and cortical interneurons in Dlx1&2−/− mutants (Long et al., submitted). RDC1 (CXCR7) regulates the migration of lateral line cells in zebrafish (Dambly-Chaudiere et al., 2007; Valentin et al., 2007). While the function of this receptor has not been established in the development of cortical interneurons, perhaps the combined reduction in CXCR4 and RDC1 contributes to the lamination defects of the immature interneurons.
Preservation of Migration in the SVZ in Lhx6 Mutants
While the overall number of cortical PLAP+ cells is reduced in Lhx6PLAP/PLAP mutants, there are increased numbers in the SVZ at E16.5 and E18.5, where Arx and Dlx1 expression is also robust (Table 4; Figs. 1, 2, 3, 5; Supplemental Fig. 3, 6, 7, 8). Furthermore, ErbB4 expression, which is not detectable in the mutant migratory cells at E12.5 and E14.5 (Fig. 4; Supplemental Fig. 5), is present in the SVZ migratory pathway at E16.5 and E18.5 (Fig. 5, 6, 7). Thus, Lhx6 function appears to be less important for interneurons that migrate within the SVZ at E16.5 and E18.5. Note that the wild-type SVZ is enriched in migratory cells that express DLX proteins but not Lhx6 (Fig. 1; Table 3; Supplemental Fig. 1). Thus, we propose that there is a subset of SVZ cells that are independent of Lhx6 function, but require Arx, Dlx and ErbB4 function (Fig. 9). While the fate of these cells are not certain, it is known that late born calretinin+ interneurons are produced by the caudal ganglionic eminence and at least this population is not dependent on Nkx2.1 function (Butt et al., 2005; Fogarty et al., 2007; Xu et al., 2004). Thus, we examined the types of cortical and hippocampal interneurons that are present in the postnatal Lhx6PLAP/PLAP mutant.
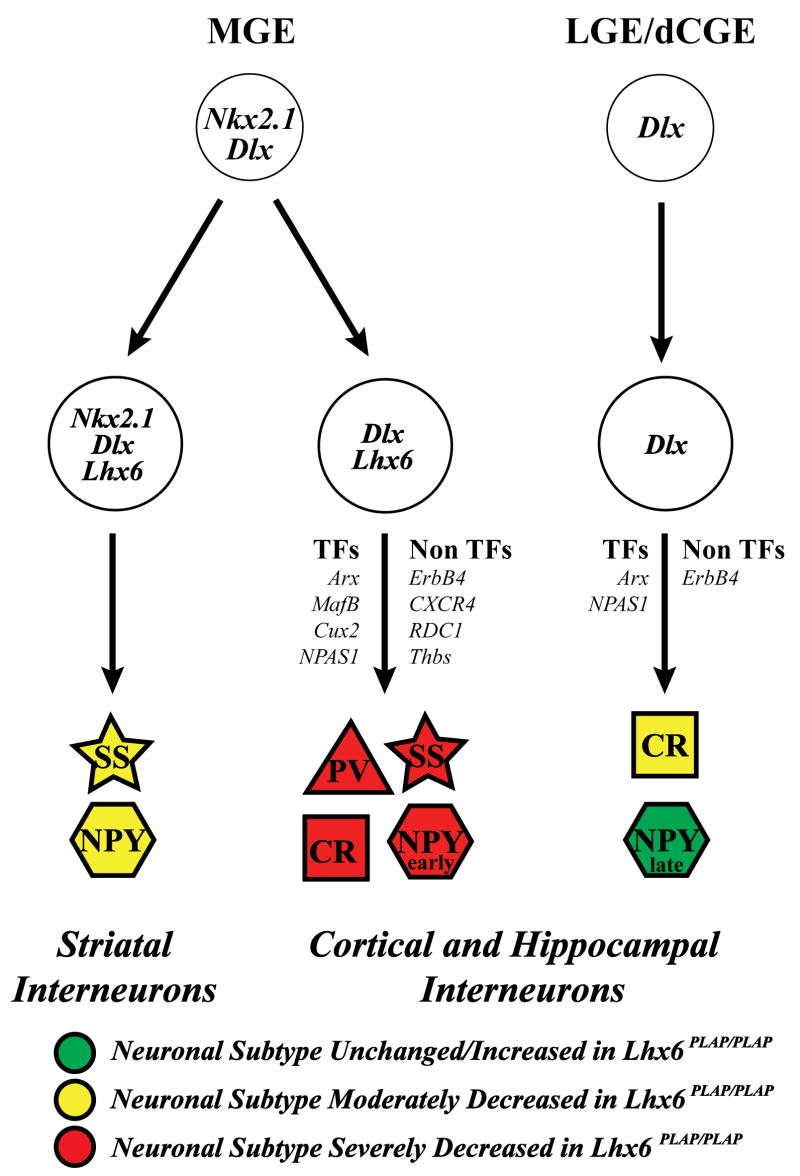
Figure 9.
Model of molecular mechanisms that control development of striatal, cortical and hippocampal interneurons. MGE precursors express Nkx2.1 and Dlx; as they differentiate, they split into Nkx2.1+ and Nkx2.1−. Nkx2.1+ differentiate into SS+ and NPY+ striatal interneurons. Nkx2.1−;Lhx6+ cells become cortical and hippocampal interneurons, through the hypothesized actions of transcription factors (Arx, bMaf, Cux2 and NPAS1) and non-transcription factors (ErbB4, CXCR4, RDC1 and Thrombospondin). These cells generate the cortical and hippocampal PV+ and SS+ interneurons, and a subset of NPY+ (early-born) and CR+ interneurons. Precursors are located in the LGE and dorsal CGE (dCGE). These neurons express Dlx genes, but do not express Nkx2.1 or Lhx6, and may not use RDC1 and CXCR4 signaling. They differentiate into a different subset of CR+ and NPY+ (late born neurons) through the hypothesized actions of the Arx and NPAS1. We postulate that the early born NPY cortical/hippocampal interneurons are Lhx6-dependent, based on the loss of NPY expression in the E12.5 and E14.5 MGE; we provided evidence that late born NPY cortical/hippocampal interneurons are Lhx6-independent (Figs. 6, 7). See Supplemental Figure 22 for the magenta-green copy.
Interneuron Subtype Defects in Postnatal Lhx6 Mutants
Lhx6PLAP/PLAP mutants survive until ~P20; the cause of death is not known. The mutant cortex shows increased apoptosis at P11 and P16 (Supplemental Fig. 17 and not shown), which may contribute to the low numbers of PLAP+ cells. We were not able to determine whether the apoptosis in the cortex was limited to the interneurons that lacked Lhx6 expression because of the low PLAP expression postnatally. Prenatally, we did not find increased apoptosis in the mutant telencephalon (Supplemental Fig. 16).
The Lhx6PLAP/PLAP cortex and hippocampus have severe deficits in parvalbumin+ and somatostatin+ cells (Fig. 6; Supplemental Figs 11, 12, 13), consistent with Liodis et al. (2007), and the expression of Lhx6-GFP in these interneurons (Cobos et al., 2006). There is also a reduction in calretinin+ interneurons in the Lhx6PLAP/PLAP mutants (as did Liodis et al., 2007). This finding was unexpected as previously we observed little co-expression of Lhx6-GFP in these cells (Cobos et al., 2006). Perhaps, Lhx6 is required in progenitors for some calretinin+ interneurons; this is likely as a subset of calretinin+ interneurons also express somatostatin (Xu et al., 2006; Gonchar et al., 2008). Alternatively, the postnatal cortical milieu does not support calretinin expression. Furthermore, given that Lhx6 is expressed in the hypothalamus, it is possible that additional physiological defects could contribute to the mutant’s phenotype.
Unlike the other interneuron subtypes, NPY+ cells were not reduced in the neocortex or hippocampus at later stages (Figs. 6, 7; Supplemental Figs. 11, 12, 13). In fact, the number of cells expressing NPY increases (~4-fold at P18; Fig. 6). However, quantifying the increase has been difficult, because we believe that some of the increase is due to expression in cells other than interneurons. None-the-less, we confirmed that in the hippocampus and superficial layers of the neocortex that most of the NPY+ cells are interneurons, based on co-expression with DLX2 (Fig. 6 and not shown).
The preservation of NPY-expressing interneurons is consistent with the observation that few Lhx6-GFP+ interneurons express NPY (Cobos et al., 2006). Of note, however, NPY expression in the MGE at E12.5 and E14.5 is not detectable in the Lhx6PLAP/PLAP mutants (Fig. 4). This suggests that early NPY expression is dependent on Lhx6, whereas late expression is Lhx6-independent. Cavanagh and Parnavelas (1990) previously presented evidence for two types of NPY+ interneurons; an early born population that decreases postnatally, and a later born population that persists. Our results suggest that the early born population depends on Lhx6, whereas the later-born NPY+ interneurons are largely Lhx6-independent. This conclusion is consistent with fate-mapping suggesting the NPY+ interneurons come from both the MGE and LGE/CGE (Butt et al., 2005; Fogarty et al., 2007).
Striatal NPY+ interneurons show the opposite result as in the cortex; the Lhx6PLAP/PLAP mutants exhibited a decrease in NPY+ cells (Fig. 8; Supplemental Fig. 10). Perhaps this reflects the loss of NPY expression in the MGE at E12.5 and E14.5 (Fig. 4). Furthermore, while parvalbumin and somatostatin expression are essentially eliminated from the pallium, there are only modest decreases in the numbers of striatal interneurons expressing these proteins. These results reflect the specificity of the genetic pathways that control the development of pallial and striatal interneurons (Fig. 9). Furthermore, these results highlight differences between Lhx6 and Lhx7(8) in the genesis of striatal interneurons. While Lhx7(8) mutants have large reduction in subcortical cholinergic neurons (Zhao et al., 2003; Mori et al., 2004; Fragkouli et al., 2005), this cell type appears normal in the Lhx6 mutants (Supplemental Fig. 15). These findings add to earlier studies on the roles of the Dlx1&2, Mash1 and Nkx2.1 transcription factors on striatal interneuron development (Marin et al., 2000).
PLAP Expression in the Absence of Lhx6 Function
Lhx6 RNA and PLAP expression is maintained in MGE progenitors of the Lhx6PLAP/PLAP mutants (Figs. 4, 5; Supplemental Figs. 4, 6, 7). Thus, Lhx6 function is not required in these cells to maintain Lhx6 RNA expression. On the other hand, while Lhx6 RNA can be detected in pallidal neurons at E12.5, its expression becomes progressively weak in pallidal neurons at E14.5, E16.5 and E18.5. Furthermore, it is not detected as MGE-derived cells tangentially migrate into the LGE/CGE and cortex at any age, whereas PLAP activity and immunoreactivity is preserved in pallidal neurons and in tangentially migrating interneurons. (Figs. 4, 5; Supplemental Figs. 4, 6, 7). Thus, perhaps Lhx6 is required to maintain its transcription in these cells, and the PLAP activity represents perdurance of the protein.
Transcription Factor Expression Defects in Lhx6PLAP/PLAP Mutants
Lack of Lhx6 function leads to reduced MGE and pallidal expression of three transcription factors, bMaf, Cux2 and NPAS1 (Fig. 4; Supplemental Fig. 5). While the function of these genes has not been established in the MGE/pallidum, they may contribute to the Lhx6PLAP/PLAP phenotype. On the other hand, MGE/pallidal expression of Arx, Dlx1, ER81, Lmo3 and Nkx2.1 appear normal (Fig. 4; Supplemental Fig. 5), and may explain why pallidal nuclei show no obvious phenotype.
As Lhx6PLAP/PLAP interneurons tangentially migrate into the LGE and cortex, they show reduced Arx, bMaf and Cux2 expression (Fig. 4; Supplemental Fig. 6). Constitutive loss of Arx function results in severe reductions in the numbers of cortical interneurons (Colombo et al., 2007; Kitamura et al., 2002). Arx expression appears normal in the MGE/pallidum of the Lhx6PLAP/PLAP mutants; therefore, Arx expression may be reduced as interneurons enter the LGE. It is unknown how reduced Arx levels in tangentially migration interneurons affect their migration.
Dlx1 expression appears less affected in Lhx6PLAP/PLAP mutants. Dlx1&2function are required for tangential migration (Anderson et al., 1997), suggesting why this process is largely normal in Lhx6PLAP/PLAP mutants. Furthermore, Dlx1 regulates the number of calretinin, somatostatin and NPY neurons (Cobos et al., 2005). Perhaps the maintenance of Dlx1 expression is related to the persistence of NPY interneurons.
NPAS1 expression is reduced in deep cortical layers, but is maintained in many cortical and hippocampal interneurons at E18.5 and P11 (Figs. 5, 7, and not shown). In fact, there are ~2-fold more NPAS1+ cells in the mutant P11 hippocampus (Fig. 7). Expression of this transcription factor is reduced in Dlx1−/− mutants (Cobos et al., 2005). Thus, we propose that NPAS1 is a marker of interneurons that are in part derived from the Lhx6-independent lineage that depend on Dlx1 function, and perhaps come primarily from the LGE/dorsal CGE. We also hypothesize that NPAS1 is expressed in NPY+ interneurons; proof of this awaits double-labeling experiments.
In sum, appear to be two major subtypes of immature pallial interneurons that differ in their origins, expression of transcription factors and fate (Fig. 9; Butt et al., 2005; Flames et al., 2007; Fogarty et al., 2007; Liodis et al., 2007; Wonders and Anderson, 2006; Xu et al., 2004). In the absence of Lhx6, some cells fail to migrate out of the MGE; many of these are born around E13.5, and remain as a periventricular ectopia, showing that Lhx6 is essential for migration of a subset of MGE-derived cells (Supplemental Fig. 9). However, most of the PLAP+ MGE-derived cells, which are fated to become pallial interneurons, migrate into the cortex and hippocampus. These cells continue to express PLAP and DLX2 in the cortex and hippocampus at E18.5 (Fig. 1 and not shown); therefore they have the potential to develop along the pathway of Dlx+;Lhx6−interneurons. Postnatally, the vast majority of mutant PLAP+ cells either lose PLAP expression and/or die (Supplemental Figs. 11, 14); thus we cannot follow their fate. We postulate that the increase in NPAS1 expression in the postnatal hippocampus may reflect a fate change of some MGE-derived cells.
Supplementary Material
Quantification of numbers of cells that are positive for GFP (Lhx6-GFP), pan-DLX and both GFP & pan-DLX immunoreactivity in the E14.5 and E18.5 cortex of Lhx6PLAP/+ and Lhx6PLAP/PLAP, using data from Supplemental Figure 1. The numbers represent the areal density (positive cells per 1000 μm2 of the box surface area) in cortex subregions. Abbreviations: see Table 3.
Table 1.
Information on RNA probes used for in situ hybridization experiments. The size of the cDNA determines the region of the RNA from which they were derived, as their 3′ end is at the terminus of the transcript; except for ErbB4, where the insert encodes amino acids 326-633.
| Plasmid Name | Source | Size Insert (nucleotides 3′ from poly-A tail) |
|---|---|---|
| Arx | Kunio Kitamura | 1000 |
| bMAF | Sabine Cordes | 451 |
| Cux2 | Harold Cremer | 1000 |
| CXCR4 | Dan Littman | 580 |
| Dlx1 | JLRR | 2800 |
| ER81 | Thomas Jessell | 2000 |
| ErbB4 | Cary Lai | 920 (corresponding to AA 326-633) |
| GAD67 | Bryan Condie | 2000 |
| Lhx6 | Vassilis Pachnis | 1500 |
| Lhx7 | Vassilis Pachnis | 1500 |
| Lmo3 | TH Rabbitts | 2000 |
| Nkx2.1 | JLRR | 2200 |
| NPAS1 | Steve McKnight | 1800 |
| NPY | Unigene MGC-57879 | 561 |
| RDC1 | ATCC MGC-18378 | 1700 |
| Shh | Andy McMahon | 640 (in 200nt from 5′ and 300nt up from 3′) |
| SS | Thomas Lufkin | 1000 |
| Thbs | ATCC 930244 | 2000 |
Table 2.
Information on the antibody reagents; details on the specificity controls are described in the Materials and Methods.
| Target | Antigen | Species | Dilution | Source |
|---|---|---|---|---|
| Calbindin D28k | Recombinant calbindin D28k | Rabbit
AB1778 |
1:1000 | Chemicon |
| CTIP2 | human CTIP2 fusion protein | Rat
AB18465 |
1:1000 | Abcam |
| Calretinin | Recombinant rat calretinin | Rabbit
AB5054 |
1:2500 | Chemicon |
| Calretinin | Recombinant human calretinin | Rabbit
7699/4 |
1:4000 | Swant |
| Cleaved Caspase-3 | RGTELDCGIETD | Rabbit
#9664 |
1:400
1:200 |
Cell Signaling |
| ChAT | Human placental ChAT | Goat
AB144P |
1:250 | Chemicon |
| DLX2 | purified MBP-DLX2 fusion protein (aa:1-154) | Guinea pig | 1:2000 | Kazuaki Yoshikawa |
| Pan-DLX | N-terminal 200 aa + protein (aa:1-154) butterfly DLX protein | Rabbit | 1:250 | Grace Panganiban |
| Rabbit GFP | GFP isolated directly from Aequorea Victoria | Rabbit
A11122 |
1:1000 | Invitrogen |
| Chicken GFP | purified recombinant GFP | Chicken
GFP-1020 |
1:1000 | Aves lab. |
| NPAS1 | Mouse NPAS1 peptide (aa: 139-594) | Rabbit | 1:1000 | Steve McKnight |
| NPY | Synthetic porcine NPY | Rabbit
N9528 |
1:5000 | Sigma |
| NPY | synthetic porcine NPY | Rabbit
22940 |
1:2000 | Immunostar |
| PV | Purified frog muscle PV | Mouse
P 3088 |
1:2000 | Sigma |
| PH3 | Human histone H3 peptide (aa: 7-20) | Rabbit
06-570 |
1:500 | Upstate |
| PLAP | Purified human PLAP | Rabbit
AHP537HT |
1:50 | AbD Serotec |
| SS | Synthetic 1-14 cyclic SS | Rat
MAB354 |
1:150 | Chemicon |
| TBR1 | TBR1 peptide (aa:1-20) | Rabbit | 1:1000 | Robert Hevner |
Acknowledgments
This work was supported by the research grants to JLRR from: Nina Ireland, Larry L. Hillblom Foundation, NIMH RO1 MH49428-01 and K05 MH065670; PF: Swiss National Science Foundation (PA00A-117463 and PBGEA-112882); HW from Intramural Research Program of the National Institute of Child Health and Human Development. We thank Regeneron for providing the Lhx6 mutant mice.
Abbreviations
- BSTN
bed nucleus striata terminalis
- CA
CA fields
- CB
calbindin
- CC
corpus callosum
- CGE
caudal ganglionic eminence
- dCGE
dorsal caudal ganglionic eminence
- ChAT
choline acetyltransferase
- CP
cortical plate
- DB
diagonal band
- CR
calretinin
- DG
dentate gyrus
- GFP
green fluorescent protein
- GP
globus pallidus
- Hc
Hippocampus
- IZ
intermediate zone
- LGE
lateral ganglionic eminence
- MZ
marginal zone
- MGE
medial ganglionic eminence
- NAc
nucleus accumbens
- NPY
neuropeptide Y
- PH3
phospho histone-3
- PLAP
placental alkaline phosphatase
- POA
preoptic area
- PV
parvalbumin
- SE
septum
- SP
Stratum Pyramidale
- SR
Stratum Radiatum
- SS
somatostatin
- STR
striatum
- SVZ
subventricular zone
- TF
transcription factor
- Thbs
Thrombospondin
- VZ
ventricular zone
References
- Alcantara S, Pozas E, Ibanez CF, Soriano E. BDNF-modulated spatial organization of Cajal-Retzius and GABAergic neurons in the marginal zone plays a role in the development of cortical organization. Cereb Cortex. 2006;16(4):487–499. doi: 10.1093/cercor/bhi128. [DOI] [PubMed] [Google Scholar]
- Alifragis P, Liapi A, Parnavelas JG. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J Neurosci. 2004;24(24):5643–5648. doi: 10.1523/JNEUROSCI.1245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9(6):646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278(5337):474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133(11):2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280(42):35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48(4):591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cavanagh ME, Parnavelas JG. Development of neuropeptide Y (NPY) immunoreactive neurons in the rat occipital cortex: a combined immunohistochemical-autoradiographic study. J Comp Neurol. 1990;297(4):553–563. doi: 10.1002/cne.902970408. [DOI] [PubMed] [Google Scholar]
- Choi GB, Dong HW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Swanson LW, Anderson DJ. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46(4):647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54(6):873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nature neuroscience. 2005;8(8):1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(Suppl 1):i82–88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Colombo E, Collombat P, Colasante G, Bianchi M, Long J, Mansouri A, Rubenstein JL, Broccoli V. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J Neurosci. 2007;27(17):4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran JP, So PL, Maden M. Disruption of the retinoid signalling pathway causes a deposition of amyloid beta in the adult rat brain. Eur J Neurosci. 2004;20(4):896–902. doi: 10.1111/j.1460-9568.2004.03563.x. [DOI] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25(1):247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci U S A. 2004;101(37):13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Gelman DM, Pla R, Rubenstein JLR, Puelles L, Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007 doi: 10.1523/JNEUROSCI.2750-07.2007. In Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marin O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44(2):251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Flames N, Marin O. Developmental mechanisms underlying the generation of cortical interneuron diversity. Neuron. 2005;46(3):377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27(41):10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M, del Rio JA, Martinez A, Gomez S, Soriano E. Development of calretinin immunoreactivity in the neocortex of the rat. J Comp Neurol. 1995;361(1):177–192. doi: 10.1002/cne.903610114. [DOI] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur J Neurosci. 2005;21(11):2923–38. doi: 10.1111/j.1460-9568.2005.04141.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez M, Abellan A, Legaz I, Rubenstein JL, Puelles L, Medina L. Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J Comp Neurol. 2008;506(1):46–74. doi: 10.1002/cne.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27(19):5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, Burkhalter A. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Frontiers Neuroanatomy. 2008;1(3):1–11. doi: 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125(11):2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. J Comp Neurol. 2002;447(1):1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Jacobowitz DM, Abbott LC. Chemoarchitectonic atlas of the developing mouse brain. Boca Raton; CRC Press: 1998. 1 v. (various pagings) p. [Google Scholar]
- Kim JH, Lee JA, Song YM, Park CH, Hwang SE, Kim YS, Kaang BK, Son H. Overexpression of calbindin-D28K in hippocampal progenitor cells increases neuronal differentiation and neurite outgrowth. FASEB J. 2006;20(1):109–111. doi: 10.1096/fj.05-4826fje. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, Matsuo M, Kamijo S, Kasahara M, Yoshioka H, Ogata T, Fukuda T, Kondo I, Kato M, Dobyns WB, Yokoyama M, Morohashi K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32(3):359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Kuwajima T, Nishimura I, Yoshikawa K. Necdin promotes GABAergic neuron differentiation in cooperation with Dlx homeodomain proteins. J Neurosci. 2006;26(20):5383–5392. doi: 10.1523/JNEUROSCI.1262-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci. 1999;19(18):7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Adesnik H, Li J, Long JE, Nicoll R, Rubenstein JLR, Pleasure SJ. Regional distribution of cortical interneurons and development of inhibitory tone is regulated by Cxcl12/Cxcr4 signaling. J Neurosci. 2008;28(5):1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27(12):3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JLR. Dlx1&2 and Mash1 Transcription Factors Control Striatal Patterning and Differentiation Through Parallel and Overlapping Pathways. doi: 10.1002/cne.21854. Submitted to Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27(12):3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G, Sánchez-Alcañiz JA, Pla R, Borrell V, Picó E, Valdeolmillos M, Marín O. Chemokine Signaling Controls Intracortical Migration and Final Distribution of GABAergic Interneurons. Journal of Neuroscience. 2008 doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20(16):6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Plump AS, Flames N, Sanchez-Camacho C, Tessier-Lavigne M, Rubenstein JL. Directional guidance of interneuron migration to the cerebral cortex relies on subcortical Slit1/2-independent repulsion and cortical attraction. Development. 2003;130(9):1889–1901. doi: 10.1242/dev.00417. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. Cell Migration in the Forebrain. Annu Rev Neurosci. 2003 doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Marin O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001;293(5531):872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27(29):7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, Kitanaka J, Takemura M, Misawa H, Ikawa M, Okabe M, Wanaka A. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur J Neurosci. 2004;19(12):3129–41. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- Panganiban G, Sebring A, Nagy L, Carroll S. The development of crustacean limbs and the evolution of arthropods. Science. 1995;270(5240):1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- Pantazopoulos H, Lange N, Hassinger L, Berretta S. Subpopulations of neurons expressing parvalbumin in the human amygdala. J Comp Neurol. 2006;496(5):706–722. doi: 10.1002/cne.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak M, Potter G, Rowitch D, Rubenstein J. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007 doi: 10.1016/j.neuron.2007.06.036. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28(3):727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Polleux F, Whitford KL, Dijkhuizen PA, Vitalis T, Ghosh A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 2002;129(13):3147–3160. doi: 10.1242/dev.129.13.3147. [DOI] [PubMed] [Google Scholar]
- Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30(1):79–89. doi: 10.1016/s0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Pozas E, Ibanez CF. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic neurons. Neuron. 2005;45(5):701–713. doi: 10.1016/j.neuron.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26(9):469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9(20):2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Rymar VV, Sadikot AF. Laminar fate of cortical GABAergic interneurons is dependent on both birthdate and phenotype. J Comp Neurol. 2007;501(3):369–380. doi: 10.1002/cne.21250. [DOI] [PubMed] [Google Scholar]
- Schwaller B, Buchwald P, Blumcke I, Celio MR, Hunziker W. Characterization of a polyclonal antiserum against the purified human recombinant calcium binding protein calretinin. Cell Calcium. 1993;14(9):639–648. doi: 10.1016/0143-4160(93)90089-o. [DOI] [PubMed] [Google Scholar]
- Shah NM, Pisapia DJ, Maniatis S, Mendelsohn MM, Nemes A, Axel R. Visualizing sexual dimorphism in the brain. Neuron. 2004;43(3):313–319. doi: 10.1016/j.neuron.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Stuhmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002a;129(1):245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- Stuhmer T, Puelles L, Ekker M, Rubenstein JL. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex. 2002b;12(1):75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- Stumm R, Kolodziej A, Schulz S, Kohtz JD, Hollt V. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. J Comp Neurol. 2007;502(3):382–399. doi: 10.1002/cne.21336. [DOI] [PubMed] [Google Scholar]
- Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, Hollt V, Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci. 2003;23(12):5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126(15):3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Fujimori K, Nojyo Y, Kaneko T, Takauji R. Evidence that Sema3A and Sema3F regulate the migration of GABAergic neurons in the developing neocortex. J Comp Neurol. 2003;455(2):238–248. doi: 10.1002/cne.10476. [DOI] [PubMed] [Google Scholar]
- Tang X, Falls DL, Li X, Lane T, Luskin MB. Antigen-retrieval procedure for bromodeoxyuridine immunolabeling with concurrent labeling of nuclear DNA and antigens damaged by HCl pretreatment. J Neurosci. 2007;27(22):5837–5844. doi: 10.1523/JNEUROSCI.5048-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17(12):1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Alvarez-Dolado M, Erskine L, Alvarez-Buylla A. Permissive corridor and diffusible gradients direct medial ganglionic eminence cell migration to the neocortex. Proc Natl Acad Sci U S A. 2003;100(2):727–732. doi: 10.1073/pnas.242721899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wozniak DF, Stewart GR, Finger S, Olney JW. Comparison of behavioral effects of nucleus basalis magnocellularis lesions and somatosensory cortex ablation in the rat. Neuroscience. 1989;32(3):685–700. doi: 10.1016/0306-4522(89)90290-x. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24(11):2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506(1):16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499(1):144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A. 2003;100(15):9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of numbers of cells that are positive for GFP (Lhx6-GFP), pan-DLX and both GFP & pan-DLX immunoreactivity in the E14.5 and E18.5 cortex of Lhx6PLAP/+ and Lhx6PLAP/PLAP, using data from Supplemental Figure 1. The numbers represent the areal density (positive cells per 1000 μm2 of the box surface area) in cortex subregions. Abbreviations: see Table 3.