Abstract
Eukaryotic proteins containing a C-terminal CAAX motif undergo a series of posttranslational CAAX-processing events that include isoprenylation, C-terminal proteolytic cleavage, and carboxyl methylation. We demonstrated previously that the STE14 gene product of Saccharomyces cerevisiae mediates the carboxyl methylation step of CAAX processing in yeast. In this study, we have investigated the subcellular localization of Ste14p, a predicted membrane-spanning protein, using a polyclonal antibody generated against the C terminus of Ste14p and an in vitro methyltransferase assay. We demonstrate by immunofluorescence and subcellular fractionation that Ste14p and its associated activity are localized to the endoplasmic reticulum (ER) membrane of yeast. In addition, other studies from our laboratory have shown that the CAAX proteases are also ER membrane proteins. Together these results indicate that the intracellular site of CAAX protein processing is the ER membrane, presumably on its cytosolic face. Interestingly, the insertion of a hemagglutinin epitope tag at the N terminus, at the C terminus, or at an internal site disrupts the ER localization of Ste14p and results in its mislocalization, apparently to the Golgi. We have also expressed the Ste14p homologue from Schizosaccharomyces pombe, mam4p, in S. cerevisiae and have shown that mam4p complements a Δste14 mutant. This finding, plus additional recent examples of cross-species complementation, indicates that the CAAX methyltransferase family consists of functional homologues.
INTRODUCTION
Proteins that contain a C-terminal CAAX motif (in which C is a cysteine, A is an aliphatic amino acid, and X is one of several amino acids) are found in a wide variety of eukaryotes ranging from yeast to mammals. In all species examined to date, CAAX proteins undergo an ordered series of posttranslational modifications at their C termini: isoprenylation, proteolytic cleavage, and carboxyl methylation, which are collectively referred to in this article as CAAX processing (reviewed in Clarke, 1992; Zhang and Casey, 1996). Three well-characterized proteins that undergo CAAX processing in Saccharomyces cerevisiae are Ras1p, Ras2p, and the mating pheromone a-factor (Hrycyna et al., 1991; Clarke, 1992). In S. cerevisiae, the first CAAX modification, isoprenylation, is mediated by the Ram1p/Ram2p farnesyltransferase or by the Ram2p/Cdc43p geranylgeranyltransferase (Finegold et al., 1991; He et al., 1991). This is followed by the proteolytic cleavage of the last three amino acids (AAX) by the Rce1p and/or the Ste24p/Afc1p proteases (Boyartchuk et al., 1997; Fujimura-Kamada et al., 1997; Tam and Michaelis, unpublished observations). Finally, the prenylated CAAX protein from which the AAX tripeptide has been removed is carboxylmethylated by the prenylcysteine carboxyl methytransferase Ste14p (Hrycyna and Clarke, 1990). This study focuses on Ste14p, the carboxyl methyltransferase of yeast. Because Ste14p mediates the last step of CAAX processing, determining the intracellular localization of Ste14p will provide insight into the intracellular site of CAAX processing, which is currently not known.
The expression of an enzymatically active recombinant Ste14p fusion protein in Escherichia coli and the demonstration that a Δste14 mutant lacks carboxyl methyltransferase activity provided evidence that Ste14p mediates this enzymatic activity in yeast (Hrycyna et al., 1991). Carboxyl methylation is important for several aspects of the biogenesis and activity of the mating pheromone a-factor (Sapperstein et al., 1994). A MATa Δste14 strain has a sterile phenotype (i.e., mating does not occur) as a result of combined defects in a-factor transport, receptor recognition, and stability (Sapperstein et al., 1994). The unmethylated a-factor produced in a Δste14 mutant fails to be exported, suggesting that the methyl group of a-factor may be an essential determinant for recognition of a-factor by its transporter Ste6p (Sapperstein et al., 1994). Methylation is also important for the interaction of a-factor with its receptor, because an in vitro synthesized a-factor species lacking its methyl group is inactive (Marcus et al., 1991). In addition, methylation seems to play a role in the intracellular metabolic stability of a-factor, because a-factor biosynthetic intermediates inside the cell are highly unstable in a ste14 mutant (Sapperstein et al., 1994). Likewise, in a macrophage cell line, the CAAX protein RhoA was found to have a decreased half-life when methylation was inhibited (Backlund, 1997). Interestingly, STE14 is not an essential gene despite its role in the modification of essential proteins such as Ras1p and Ras2p. Consistent with this observation, there are no significant cellular defects associated with unmethylated Ras1p or Ras2p, although there is a slight delay in Ras2p maturation and a subtle defect in Ras2p membrane localization (Hrycyna et al., 1991; Sapperstein et al., 1994).
Ste14p was the first CAAX methyltransferase to be cloned and sequenced and is predicted to contain multiple membrane spans (Blair, 1979; Wilson, 1985; Sapperstein et al., 1994). Recently, Imai et al. (1997) have described the cloning of CAAX methyltransferases from Schizosaccharomyces pombe and Xenopus laevis, mam4p and Xmam4p, respectively. These proteins share significant amino acid similarity with Ste14p, are similar to Ste14p in their hydropathy profiles, and have been shown to function as CAAX methyltransferases in vitro (Imai et al., 1997). Mutations in mam4p result in a sterile phenotype in S. pombe, just as mutations in Ste14p result in a sterile phenotype in S. cerevisiae. Enzymatic activities analogous to that of Ste14p have been described in a number of mammalian systems (Stephenson and Clarke, 1990; Perez-Sala et al., 1991; Pillinger et al., 1994), and the cloning of the first mammalian CAAX methyltransferase, designated pcCMT, has recently been reported (Dai et al., 1998).
In this study we have raised antibodies to Ste14p that, together with an in vitro methyltransferase activity assay, have permitted us to characterize the localization of Ste14p in S. cerevisiae. We demonstrate by immunofluorescence and subcellular fractionation that Ste14p is localized to and is active on the endoplasmic reticulum (ER) membrane, despite its lack of known signals for ER localization or retrieval. Importantly, these results imply that the processing of CAAX proteins occurs at the ER membrane. Consistent with this view, we have recently determined that the gene products involved in the preceding step of CAAX processing, Rce1p and Ste24p, are localized to the ER membrane (Schmidt and Michaelis, unpublished observations). Surprisingly, we find here that Ste14p constructs that are epitope tagged with the triply iterated epitope from influenza hemagglutinin (HA) at the N terminus, at the C terminus, or internally are mislocalized, emphasizing the importance of caution when interpreting results based on tagged membrane proteins. In addition, we show that mam4p, the S. pombe CAAX methyltransferase, complements a Δste14 mating defect. This result together with other transcomplementation studies indicates that the CAAX methyltransferases comprise a family of functional homologues.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions
The S. cerevisiae strains used in this study are listed in Table 1. Complete (YEPD), synthetic (SD), and synthetic dropout (SC-Leu, SC-Ura, SC-Leu-Ura) media were prepared as described previously (Michaelis and Herskowitz, 1988), except that dropout media lacked cysteine. All experiments were performed at 30°C. Yeast transformations were performed either by the lithium acetate method (Ito et al., 1983) or by the method of Elble (Elble, 1992).
Table 1.
Yeast strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| SM1058 | MATa trp1 leu2 ura3 his4 can1 | Michaelis and Herskowitz, 1988 |
| SM1068 | MATα lys1 | Michaelis and Herskowitz, 1988 |
| SM1188 | MATa trp1 leu2 ura3 his4 can1 ste14-3::TRP1 | Sapperstein et al., 1994 |
| SM2544 | MATa trp1 leu2 ura3 his4 can1 ste6Δ4(738)(-368nt—843) | Loayza and Michaelis, 1998 |
| SM2905 | MATa trp1 leu2 ura3 his4 can1 [CEN URA3 OCH1::HA] | Transformant of SM1058 with pO-Nhe |
| Harris and Waters, 1996 | ||
| SM2915 | MATa trp1 leu2 ura3 his4 can1 ste6Δ4(738)(-368nt—843) [CEN URA3 STE6::HA] | Transformant of SM2544 with pSM1085 |
| SM2926 | [CEN URA3] | Transformant of SM1188 with pRS316 |
| SM3041 | MATa trp1 leu2 ura3 his4 can1 [CEN URA3] | Transformant of SM1058 with pRS316 |
| SM3185 | [CEN URA3 STE14] | Transformant of SM1188 with pSM1237 |
| SM3187 | [CEN URA3 STE14::HA Q3] | Transformant of SM1188 with pSM1248 |
| SM3189 | [CEN URA3 STE14::HA I239] | Transformant of SM1188 with pSM1250 |
| SM3431 | [CEN URA3 STE14::HA I226] | Transformant of SM1188 with pSM1260 |
| SM3495 | [2μ URA3 STE14] | Transformant of SM1188 with pSM1317 |
| SM3583 | [CEN URA3 mam4] | Transformant of SM1188 with pSM1334 |
| SM3584 | [2μ LEU2 STE14::HA Q3] | Transformant of SM1188 with pSM1335 |
| SM3585 | [2μ LEU2 STE14::HA I239] | Transformant of SM1188 with pSM1336 |
| SM3586 | [2μ LEU2 STE14::HA I226] | Transformant of SM1188 with pSM1337 |
All strains used in this study are isogenic to SM1188 unless otherwise indicated.
Plasmid Constructions
To analyze Ste14p expressed at CEN levels, we constructed plasmid pSM1237 that contains the STE14 coding sequence preceded by 503 bp of 5′-noncoding sequence and 677 bp of 3′-untranslated sequence. This plasmid is essentially the same as pSM186 (Sapperstein et al., 1994) except that the 5′-noncoding sequence was extended to 503 bp from 66 bp. This step was necessary because previous experiments showed that expressing STE14 with only 66 bp of 5′-upstream noncoding sequence resulted in the production of Ste14p from two aberrant translational start sites (Romano and Michaelis, unpublished observations). The expanded 5′-noncoding region was PCR amplified from pSM187 (Sapperstein et al., 1994) and subcloned into pSM186 to generate pSM1237 (CEN URA3 STE14).
A LEU2 version of pSM1237, pSM1316, was constructed in vivo by homologous recombination (Ma et al., 1987; Oldenburg et al., 1997) using a PvuI fragment from pSM1237 that was cotransformed with PvuII-gapped pRS315 (CEN LEU2) (Sikorski and Hieter, 1989) into SM1188 (Δste14-3) and selecting for Leu+ transformants. pSM1316 (CEN LEU2 STE14) was used to generate a 2μ plasmid containing STE14, pSM1317 (2μ URA3 STE14), by cotransforming a PvuI fragment from pSM1316 and a PvuII-gapped vector, pSM217 (2μ URA3) (Chen et al., 1997), into SM1188 and by selecting for Ura+ transformants.
To express S. pombe mam4 in S. cerevisiae, we generated pSM1334 (CEN URA3 mam4) that contains the precise mam4 coding sequence flanked by STE14 5′- and 3′-noncoding sequences. The mam4 sequence was amplified by PCR from pST109-B1, generously provided by M. Yamamoto (University of Tokyo, Japan). The PCR product, containing 48 bp at each end homologous to STE14 5′- and 3′-untranslated sequences, was cotransformed with AflII and EcoRI-gapped pSM1237 (CEN URA3 STE14) into SM1188(Δste14-3), and Ura+ transformants were selected. The mam4 sequence and junctions were confirmed by DNA sequencing.
pSM1085 (CEN URA3 STE6::HA) was constructed by subcloning the SalI and NotI fragment containing STE6::HA from pSM500 (Paddon et al., 1996) into the same sites of pRS316 (Sikorski and Hieter, 1989).
Epitope Tagging of Ste14p
Ste14p was epitope tagged at one of three locations with the HA epitope. This tag and linker regions contain 43 amino acids. The HA epitope was inserted either at the N terminus after amino acid Q3 (pSM1248), at the C terminus after amino acid I239 (pSM1250), or internally after amino acid I226 (pSM1260). For each construct, a BglII site (AGATCT) was first inserted into STE14, pSM187 (CEN URA3 STE14), using site-directed mutagenesis (Kunkel et al., 1987), into which the triply iterated HA tag from pSM492 (Berkower et al., 1994) was subcloned using standard subcloning procedures. Two micron versions of these plasmids (pSM1335, pSM1337, and pSM1336, respectively) were constructed by recombinational cloning as described above using PvuI donor plasmids (pSM1248, pSM1260, and pSM1250, respectively) and a PvuII-gapped recipient plasmid, pSM218 (2μ LEU2) (Berkower and Michaelis, 1991). All epitope tag insertions were confirmed by DNA sequencing.
Mating Assays
Patch mating tests were performed essentially as described previously (Michaelis and Herskowitz, 1988). Briefly, patches of MATa cells grown on selective media were replica plated onto an SD plate spread with 0.3 ml of YEPD and a lawn of the MATα mating tester SM1068. Plates were incubated at 30°C for 2 d. Growth of prototrophic diploids is indicative of mating.
Production of Anti-Ste14p Antiserum
To generate polyclonal antibodies against Ste14p, rabbits were immunized with a GST fusion protein containing the C-terminal hydrophilic segment of Ste14p. The 3′-end of STE14, encoding I197-I239, was PCR amplified from pSM186 (Sapperstein et al., 1994) with oligonucleotides containing BamHI overhangs. The PCR fragment was subcloned into the BamHI site of pGEX-2T (Pharmacia, Piscataway, NJ). The junction between GST and STE14 was sequenced in the resulting plasmid pSM1353. Induction of the E. coli strain CAG456 (Baker et al., 1984) with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3 h produced a major 28-kDa product that was excised from an SDS-PAGE gel and used as an immunogen (Covance, Denver, PA).
Other Antibodies
The mouse anti-HA (12CA5) monoclonal antibody was purchased from BAbCo (Richmond, CA). The rabbit polyclonal anti-Pma1p, anti-hexokinase, anti-Sec23p, and anti-Kar2p antibodies were gifts from Dr. C. Slayman (Yale University, New Haven, CT), Dr. R. Jensen (Johns Hopkins University School of Medicine, Baltimore, MD), Dr. R. Schekman (University of California, Berkeley, Berkeley, CA), and Dr. M. Rose (Princeton University, Princeton, NJ), respectively. Horseradish peroxidase (HRP)-conjugated secondary antibodies (donkey anti-rabbit Ig and sheep anti-mouse Ig) used for immunoblotting were purchased from Amersham (Arlington Heights, IL). The Cy3-conjugated secondary antibodies (goat anti-mouse Ig and goat anti-rabbit Ig) and the FITC-conjugated goat anti-rabbit secondary antibody used for immunofluorescence were purchased from Jackson ImmunoResearch (West Grove, PA). The anti-mouse secondary antibodies were used to visualize Ste14p-HA and Och1p-HA; the anti-rabbit secondary antibodies were used to visualize Ste14p, Pma1p, hexokinase, Sec23p, and Kar2p.
Preparation of Cell Extracts
Cell extracts used to characterize the anti-Ste14p antiserum and to detect Ste14p-HA were prepared for immunoblots as described previously except that 5 OD600 units of cells were grown logarithmically in synthetic dropout media (Fujimura-Kamada et al., 1997).
To investigate the association of Ste14p with membranes, we used a variation on a previously published procedure (Feldheim and Schekman, 1994). Briefly, 50 0D600 units of midlog cells were harvested and lysed by agitation with zirconium beads in 250 OD600 units per ml of buffer G (0.1 M sorbitol, 50 mM KOAc, 2 mM EDTA, 20 mM HEPES, pH 7.4, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 2 μg/ml chymotrypsin, 1 μg/ml pepstatin). The lysate was cleared of intact cells and debris at 500 × g for 5 min at 4°C in a Beckman MicroCentrifuge 5415C (Fullerton, CA). The supernatant was diluted with an equal volume of either buffer G, 1.2 M NaCl, 0.2 M Na2CO3 (pH 11), 5 M urea, or 1% Triton X-100. Samples were incubated on ice for 30 min, and one-half of the sample was reserved as a total lysate fraction (T). The remaining one-half of each sample was centrifuged at 200,000 × g for 30 min at 4°C in a TLA 100.2 rotor (Beckman). The supernatant (S) fraction was removed, and the pellet fraction (P) was washed with buffer G and centrifuged as described above. After solubilizing all the fractions with Laemmli sample buffer to equivalent final volumes, the samples were subjected to SDS-PAGE, transferred to nitrocellulose, and processed for immunodetection as described below.
Immunoblotting Analysis
Samples in Laemmli buffer were heated to 65°C for 15 min and were resolved by 12.5% SDS-PAGE. Proteins were transferred to nitrocellulose for 2000 mA·hr in 10 mM Na2B4O7. The membrane was blocked with 5% milk in Tris-buffered saline with Tween (TBST; 10 mM Tris, pH 7.4, 150 mM NaCl, 0.2% Tween 20) for 2 h at room temperature and incubated with primary antibody (anti-Ste14 antiserum [depleted of nonspecific antibodies as described below] or anti-Sec23p antiserum diluted 1:500, anti-HA antibody or anti-hexokinase antiserum diluted 1:10,000, or anti-Pma1p antiserum diluted 1:2000 in 5% milk in TBST or 1% BSA in TBST) for 2 h. After three to five washes with TBST, the membrane was incubated with secondary antibody (anti-rabbit–HRP or anti-mouse–HRP diluted 1:10,000 or 1:5000 in 5% milk in TBST) for 45 min at room temperature. After three washes with TBST and one to two washes with TBS (10 mM Tris, pH 7.4, 150 mM NaCl), the membrane was developed by chemiluminescence (Boehringer Mannheim, Indianapolis, IN).
Before use for immunoblotting, the anti-Ste14p antiserum was depleted of nonspecific antibodies by incubation with a Δste14 extract immobilized on nitrocellulose. Cell extracts were prepared essentially as described above for determining the membrane association of Ste14p. The Δste14 lysate was incubated with a strip of nitrocellulose overnight, blocked as described above, washed with 1× TBS, and incubated with a 1:500 dilution of anti-Ste14p antiserum in 1% BSA in TBST, containing 0.01% NaN3, for 2 d at 4°C. The depleted antiserum was stored at 4°C.
Immunofluorescence
Preparation of cells for immunofluorescence was performed essentially as described previously except that the cell wall was removed by treating cells with 71 μM β-mercaptoethanol and 25 μg/ml zymolyase 100T (ICN Biochemicals, Costa Mesa, CA) at 30°C for 20 min (Berkower et al., 1994). Cell washes and antibody dilutions were into phosphate-buffered saline with Tween (40 mM K2HPO4, 10 mM KH2PO4, 150 mM NaCl, 0.1% Tween 20, 10 mg/ml BSA, 0.1% NaN3) unless otherwise stated. Fixed and permeabilized cells were processed according to previously described procedures (Berkower et al., 1994) except that the primary antibodies used were either anti-Ste14p antibody diluted 1:2000 (for Ste14p) or 1:500 (for Ste14p-HA [I226]) and depleted previously of nonspecific antibodies as described below, anti-HA antibody diluted 1:2000 (for Ste14p-HA) or 1:1000 (for Och1p-HA), anti-Pma1p antibody diluted 1:200, or anti-Kar2p antibody diluted 1:1000. The secondary antibodies used were either Cy3-conjugated anti-rabbit, Cy3-conjugated anti-mouse, or FITC-conjugated anti-rabbit diluted 1:2000 (Cy3-conjugated antibodies) or 1:500 (FITC-conjugated antibodies). The slides were viewed at 100× magnification with either a Zeiss Axiophot, a Zeiss Axioskop, or a Zeiss Axiovert microscope equipped with fluorescence optics (Zeiss, Thornwood, NY). Images were captured with either an AT200 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) and MetaMorph software (Universal Imaging Corporation, West Chester, PA), a MicroMAX CCD camera (Princeton Instruments, Trenton, NJ) and IP Lab Spectrum Software (Signal Analytics, Vienna, VA), or a Photometrics PXL CCD camera (Photometrics, Tucson, AZ) and IP Lab Spectrum Software.
Before use for immunofluorescence, nonspecific antibodies were depleted from the anti-Ste14 antiserum by incubation with a Δste14 strain, SM2926, using a modification of a published procedure (Payne and Schekman, 1985). Strain SM2926 (Δste14-3) was grown to midlog phase in SC-Ura media, and cells were prepared as described above for immunofluorescence. Fixed and permeabilized cells were incubated with a 1:2 dilution of anti-Ste14 antiserum in phosphate-buffered saline with Tween for 2 d at 4°C with gentle agitation. The cells were removed by centrifugation (500 × g for 5 min at 4°C), and the depleted antisera was stored at 4°C.
Sucrose Gradient Fractionation
The fractionation of subcellular organelles was based on sedimentation through a sucrose step gradient (Antebi and Fink, 1992). Briefly, 500 OD600 units of midlog cells were harvested by centrifugation (1000 × g for 5 min in a Beckman AccuSpin FR tabletop centrifuge), washed once with 10 mM sodium azide, and then resuspended to 100 OD600 units per ml in cold 10 mM NaN3 and 250 mM β-mercaptoethanol (see Figure 4) or cold 100 mM Tris and 10 mM dithiothreitol, pH 9.6 (see Figure 6). After a 10-min incubation on ice, the cells were either diluted with an equal volume of 2× buffer A (100 mM potassium phosphate, 2.8 M sorbitol, 20 mM NaN3, pH 7.6; see Figure 4) or recovered by centrifugation and resuspended to 50 OD600 units per ml in 1× buffer A (see Figure 6). Oxalyticase (Enzogenetics, Corvallis, Oregon) was then added to 1 μg/OD600 units, and the cell suspension was incubated at 30°C for 35–40 min. EDTA was added to a final concentration of 1 mM (see Figure 4 only), and spheroplasts were chilled on ice for 5–10 min and then harvested through a 2 M cushion of sorbitol (see Figure 4) or isolated directly by centrifugation (see Figure 6). The spheroplasts were resuspended to 250 OD600 units per ml of buffer B (0.3 M sorbitol, 10 mM triethanolamine, 1 mM EDTA, pH 7.2; see Figure 4) containing protease inhibitors (1 μg/ml leupeptin, 2 μg/ml pepstatin, 1 μg/ml chymostatin, 1 μg/ml aprotinin, and 5 μM phenylmethylsulfonyl fluoride) or to 100 OD600 units per ml of buffer C (50 mM Tris, pH 7.5, 0.2 M sorbitol, 1 mM EDTA; see Figure 6) containing protease inhibitors (same as previous list) and homogenized (25–30 strokes) with a 7-ml glass Dounce homogenizer (Wheaton Science Products, Millville, NJ). The homogenates were cleared of intact cells and debris by centrifugation for 5–10 min (1000 × g); this step was repeated to ensure the complete removal of cellular debris. The cleared homogenate (∼2–3 ml) was loaded on either an 11-step (see Figure 4) or 9-step (see Figure 6) sucrose gradient poured into a thin-walled SW28 ultracentrifuge tube (Beckman). The gradient was composed of 3.4-ml layers (18–54% w/w in 4% increments; see Figure 4) or 4-ml layers (20–55% w/w in 5% increments; see Figure 6) of sucrose layered over a 65% (w/w) sucrose pad (1.7 or 2 ml, see Figures 4 and 6, respectively) with each step prepared in 10 mM HEPES, pH 7.5, and 2 or 1 mM MgCl2 (respectively). The gradients were centrifuged at 100,000 × g (23,500 rpm) for 2–2.5 h at 4°C in a SW28 rotor (Beckman). Equivalent fractions (3.4 or 4.1 ml, see Figures 4 and 6A, respectively) were collected from the gradient after the bottom of the ultracentrifuge tube was punctured with a 20-gauge needle. All fractions were assayed for marker enzyme activities (see below), for protein concentration with the Bio-Rad Protein Assay reagent (Richmond, CA), and for the relevant distribution of marker proteins by immunoblotting. When not in use, fractions were stored at −80°C.
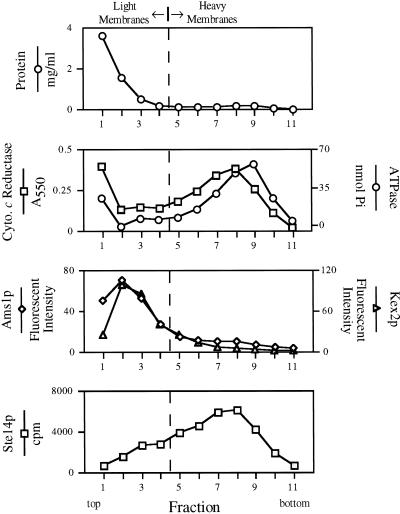
Figure 4.
Ste14p methyltransferase activity cofractionates with ER membranes by subcellular fractionation. A total yeast lysate derived from SM2915 that expresses a chromosomal copy of STE14 was subjected to fractionation on a sucrose step gradient. Equivalent volumes from each fraction were assayed for methyltransferase activity, for various organellar enzyme activities, and for protein concentration. Two populations of membranes can be identified. Light membranes marked by vacuolar α-d-mannosidase (Ams1p) and trans-Golgi network Kex2p activities are distributed at the top of the gradient (with peaks at fractions 2–3); heavier membranes marked by NADPH cytochrome c reductase (ER) and vanadate sensitive ATPase activities (plasma membrane) are distributed near the bottom of the gradient (fractions 6–10) with the peak distribution of these two compartments offset by one fraction. Methyltransferase activity cofractionates with the heavy ER membrane population. The dashed vertical line represents an arbitrary division between light and heavy membranes.
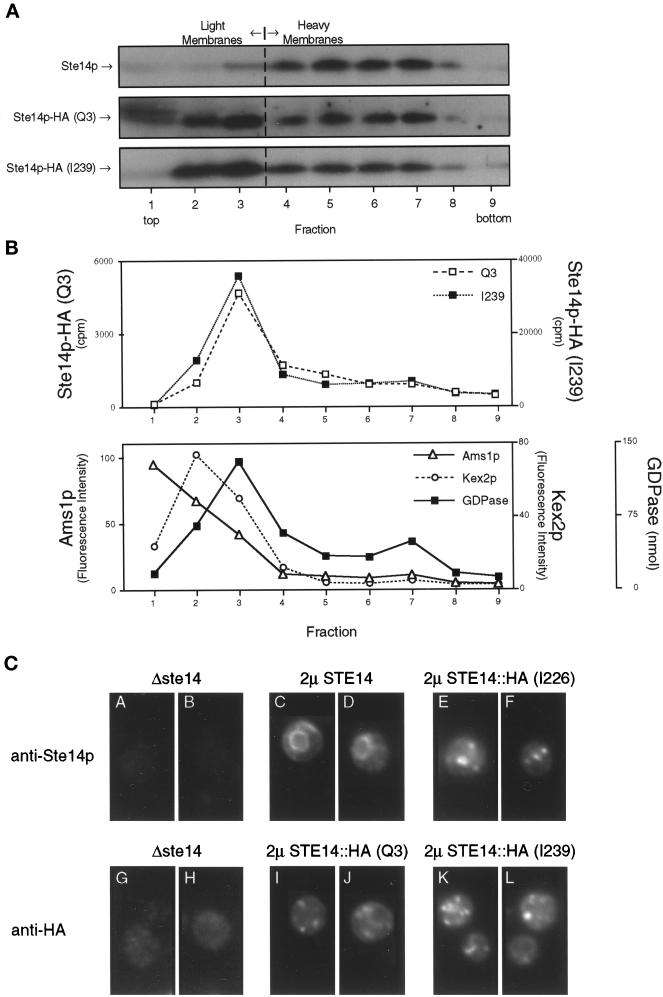
Figure 6.
The HA epitope tag mislocalizes Ste14p, as shown by subcellular fractionation and immunofluorescence. (A) Total yeast lysates derived from strains expressing CEN levels of wild-type and epitope-tagged (Q3 and I239) Ste14p (SM3185, SM3187, and SM3189, respectively) were subjected to fractionation on a sucrose step gradient similar to that described for Figure 4. Equivalent volumes from all gradient fractions were subjected to SDS-PAGE, transferred to nitrocellulose, and probed for Ste14p and epitope-tagged Ste14p. Fractions from wild-type and Q3 were probed with anti-Ste14p antiserum, whereas I239 fractions were probed with the anti-HA antibody. As expected, wild-type Ste14p fractionates with the heavy membranes (fractions 5–7). For fractions containing either N- or C-terminal epitope-tagged Ste14p, the majority of the methyltransferase cofractionates with light membranes (fractions 2–3). (B) Equivalent volumes from the Q3- and I239-derived fractions were assayed for methyltransferase activity and three light membrane activities. The Q3 and I239 methyltransferase activities cofractionate with Golgi GDPase activity and not with vacuolar α-d-mannosidase (Ams1p) or trans-Golgi network Kex2p activities. The three marker activities are averaged from the Q3 and I239 gradients. (C) Indirect immunofluorescence of Ste14p and epitope-tagged Ste14p is shown. Fixed and permeabilized cells were stained with either a 1:2000 dilution of rabbit anti-Ste14p antiserum that had been depleted previously of nonspecific antibodies (panels A-F) or a 1:2000 dilution of mouse anti-HA antiserum (12CA5) (panels G-L), followed by secondary decoration with either Cy3-conjugated goat anti-rabbit antiserum (panels A-F) or Cy3-conjugated goat anti-mouse antiserum (panels G-L).
Assays
NADPH cytochrome c reductase (Kubota et al., 1977; Feldman et al., 1987), Golgi guanosine diphosphatase (GDPase) (Abeijon et al., 1989), trans-Golgi network Kex2p (Cunningham and Wickner, 1989), vacuolar α-d-mannosidase (Opheim, 1978), and Ste14p carboxyl methyltransferase activities (see below) were determined using a fixed volume from each fraction (typically 10–25 μl). Plasma membrane ATPase activity (vanadate-sensitive pool) was determined after measuring total ATPase activity in the presence or absence of 100 μM orthovanadate (Rao and Slayman, 1993). The Kex2p substrate (Boc-gln-arg-arg-7-amino-4-methylcoumarin) was purchased from Peninsula Labs (Belmont, CA). All other reagents were purchased from Sigma (St. Louis, MO).
Methyltransferase Assay
Methyltransferase assays were performed on sucrose gradient fractions essentially as described previously (Philips and Pillinger, 1995; Volker et al., 1995). Briefly, an aliquot of each fraction (25 μl) was diluted with an equal volume of 2× buffer C (100 mM Tris, pH 7.4, 2 mM EDTA, 112 μCi/ml [1.4 μM] S-adenosyl-l-[methyl-3H]methionine [AdoMet], 200 μM N-acetyl-S-farnesyl-l-cysteine [AFC]) and incubated for 60 min at 30°C. Each reaction was processed according to the method of Philips (Philips and Pillinger, 1995) to determine the amount of methylated AFC, which is reported as cpm (see Figures 4 and 6B). AdoMet (84 Ci/mmol) was purchased from Dupont New England Nuclear (Boston, MA), and AFC was purchased from Biomol Research Laboratories (Plymouth Meeting, PA) and prepared as a 10–50 mM stock solution in dimethyl sulfoxide.
The method above was used to determine the methyltransferase activity for WT and epitope-tagged Ste14p and for S. pombe mam4p expressed in S. cerevisiae, except that the reactions (25 μl) contained 5 μg of membrane protein in 1× buffer C. Membranes were prepared as described for fractionation (see Figure 4) except that homogenates were centrifuged at 200,000 × g for 20 min in a TLA 100.2 rotor at 4°C. Membranes were washed once in buffer B and centrifuged again as described above. Membranes were resuspended in buffer B, and protein concentrations were determined with the Bio-Rad Protein Assay reagent.
For HA-tagged and untagged Ste14p, methyltransferase reactions were allowed to proceed for 0–1.5 min at an AFC concentration of 150 μM (4% v/v final dimethyl sulfoxide) and at different AdoMet concentrations (2.5–100 μM). For S. pombe mam4p, the reactions were performed as described above except that the reactions were allowed to proceed for 30 min. The activity of the enzymes is expressed in terms of pmol of [3H-methyl]AFC per mg of membrane protein as determined using a known concentration of [3H]AdoMet as a calibration standard. The Vmax values were determined using Lineweaver-Burke plots derived from triplicate experiments that were normalized to steady-state protein levels of Ste14p, Ste14p-HA (Q3), and Ste14p-HA (I239) as determined from quantitative immunoblots. Briefly, known concentrations of total membrane proteins were resolved by SDS-PAGE, transferred to nitrocellulose, probed with anti-Ste14p or anti-HA antibodies, and detected as described above. The ratios of steady-state protein levels between Ste14p and Ste14p-HA (Q3) or Ste14p-HA (I239) were used as correction factors for calculating the enzymatic activities of the tagged proteins.
RESULTS
Detection of Ste14p in Yeast Cell Extracts
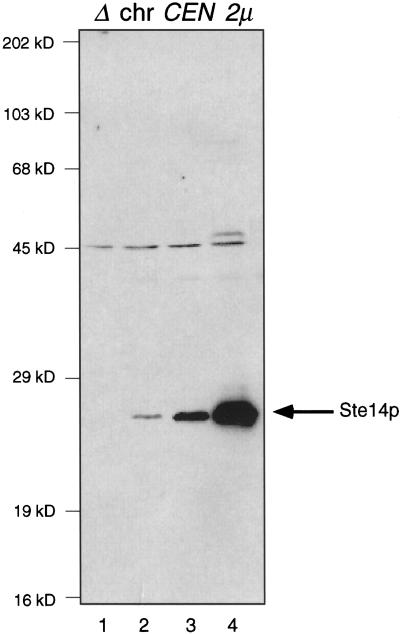
Analysis of the STE14 DNA sequence predicts a 239-amino-acid multiple membrane-spanning polypeptide of 27,887 Da (Sapperstein et al., 1994). To identify the Ste14p polypeptide in S. cerevisiae and examine its localization, we generated rabbit polyclonal antiserum directed against a GST fusion protein containing the C-terminal 42 residues of Ste14p. This antiserum was used to detect Ste14p in crude yeast cell extracts by immunoblotting, after first depleting the antiserum of nonspecific antibodies by incubating the antiserum with an extract from a Δste14 strain (Figure 1). A 24-kDa species is present in a wild-type cell extract (Figure 1, lane 2) and is absent in a Δste14 cell extract (Figure 1, lane 1). This 24-kDa species was present in increasing quantities in strains expressing CEN and 2μ levels of Ste14p (Figure 1, lanes 3 and 4, respectively). We conclude that this band represents Ste14p although this protein migrates at an apparent molecular weight of 24 kDa, which is less than the predicted molecular weight of 27.9 kDa. Aberrant migration is frequently observed for membrane proteins (Ehring et al., 1980). The aberrant migration of Ste14p observed in Figure 1 may be because Ste14p is a predicted transmembrane protein containing five to six transmembrane spans (Sapperstein et al., 1994) (see Figure 7B).
Figure 1.
Detection of Ste14p in whole cell lysates. Crude yeast cell extracts (0.4 OD600 cell equivalents) were resolved by 12.5% SDS-PAGE and transferred to nitrocellulose. Ste14p was detected using rabbit polyclonal anti-Ste14p antibodies that were depleted previously of nonspecific antibodies (described in MATERIALS AND METHODS). Lane 1, SM2926 (Δste14-3); lane 2, SM3041 (chromosomal STE14); lane 3, SM3185 (CEN STE14); and lane 4, SM3495 (2μ STE14).
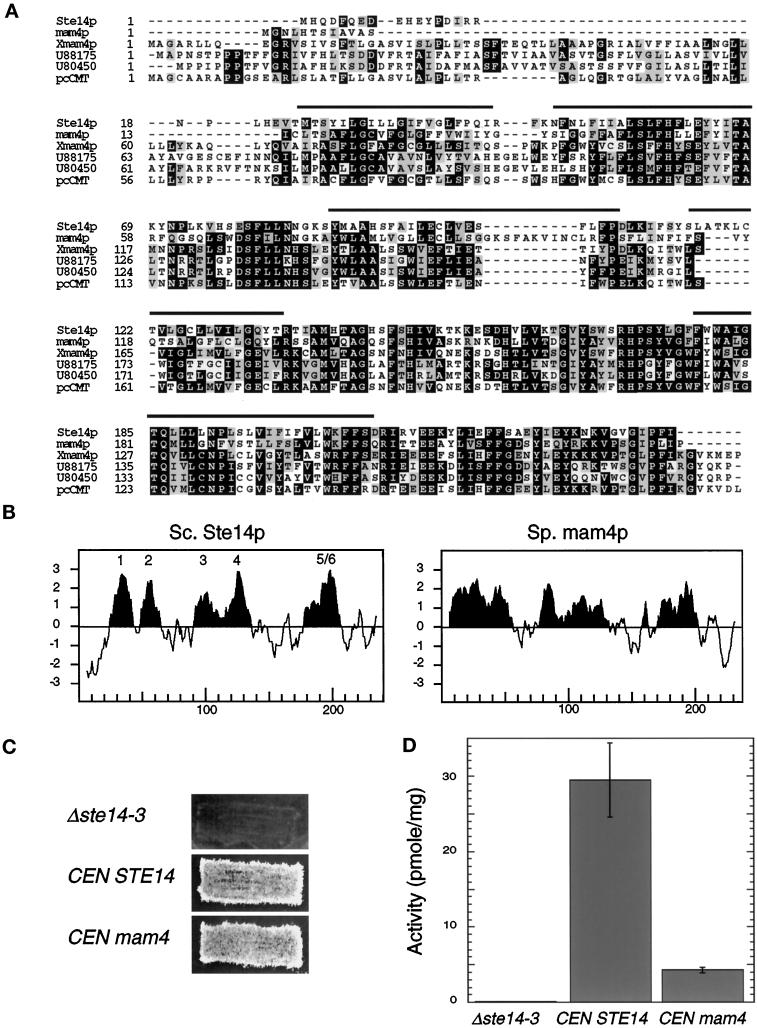
Figure 7.
A S. pombe homologue complements a Δste14 defect. (A) Alignments of S. cerevisiae Ste14p, S. pombe mam4p, X. laevis Xmam4p, two C. elegans open reading frames (accession numbers U88175 and U80450), and human pcCMT are shown. Protein sequences were aligned using ClustalW software (Thompson et al., 1994). Black boxes denote amino acid identity, and gray boxes denote amino acid similarity as determined with the Boxshade server. The bars above the sequence denote the putative membrane spans of Ste14p. (B) Comparison of the hydropathy plots of Ste14p and mam4p is shown. Hydropathy plots were generated according to the algorithm of Kyte and Doolittle (1982) with a window of 11 amino acids. Hydrophobic regions are shown in black, and the potential membrane spans of Ste14p are indicated. The hydropathy profile of S. cerevisiae Ste14p and of S. pombe mam4p was calculated using the data of Sapperstein et al. (1994) and of Imai et al. (1997), respectively. (C) The coding sequence of S. pombe mam4 was expressed from the S. cerevisiae STE14 promoter in SM1188 (Δste14-3) and assayed for complementing activity by the plate mating assay (as in Figure 5C). Strains tested were SM2926 (Δste14-3), SM3185 (Δste14-3,CEN STE14), and SM3583 (Δste14-3,CEN mam4). (D) Methyltransferase activity of Δste14, Ste14p, and mam4p is shown. Each reaction performed in triplicate contained 5 μg of membrane proteins, 100 μM AFC, and 0.7 μM [3H]AdoMet and was incubated at 30°C for 30 min. The mean activity ± SD are shown.
The antiserum recognized an additional species of 45 kDa. This species is present in the Δste14 extract and thus represents a nonspecific band. A faint band of ∼50 kDa is detected in the strain overexpressing Ste14p (lane 4) and is of the correct size for a Ste14p dimer, although we have no additional evidence of Ste14p dimerization.
Ste14p Is an Integral Membrane Protein
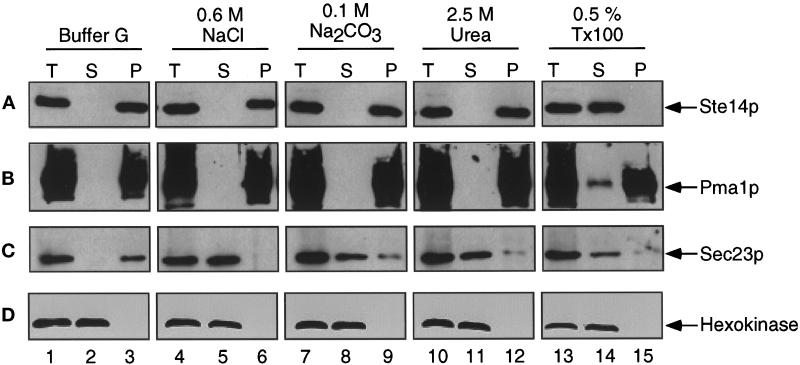
To determine directly whether Ste14p is associated with membranes and to assess the nature of the association, we prepared crude membrane fractions and treated them under conditions that solubilize either peripheral or integral membrane proteins. Consistent with predictions, Ste14p fractionated with membranes when yeast cell extracts were subjected to centrifugation at 200,000 × g (Figure 2A, lanes 1–3). In comparison, the integral membrane protein Pma1p and the peripheral membrane protein Sec23p also fractionated with membranes (Figure 2, B and C, lanes 1–3). In contrast, the soluble cytosolic protein marker hexokinase fractionated in the supernatant fraction (Figure 2D, lanes 1–3).
Figure 2.
Ste14p is an integral membrane protein. A crude yeast cell extract from SM3495 (2μ STE14) was prepared and treated with either buffer G, 0.6 M NaCl, 0.1 M Na2CO3 (pH 11), 2.5 M urea, or 0.5% Triton X-100. After incubation on ice for 20 min, one-half of each sample was reserved as total lysate (T). The remaining volume was separated into supernatant (S) and pellet (P) fractions by centrifugation at 200,000 × g. After being resuspended to equivalent volumes with Laemmli sample buffer, equivalent amounts of T, S, and P fractions were subjected to SDS-PAGE and transferred to nitrocellulose. The immunoblots were probed with (A) anti-Ste14p antiserum that had been depleted previously of nonspecific antibodies, (B) anti-Pma1p antiserum, (C) anti-Sec23p antiserum, and (D) anti-hexokinase antiserum.
Under conditions that released the peripheral membrane protein Sec23p from membranes (0.6 M NaCl, 0.1 M Na2CO3, pH 11, and 2.5 M urea), both Ste14p and Pma1p remained membrane associated (Figure 2, compare A, B, and C, lanes 4–12). Ste14p was only released into the supernatant fraction by treatment with 0.5% Triton X-100 (Figure 2A, lanes 13–15). The integral membrane protein marker Pma1p behaved essentially the same as Ste14p, although Pma1p was not as efficiently solubilized by Triton X-100 (Figure 2B, lanes 4–15). These results are consistent with the prediction that Ste14p is an integral membrane protein.
Ste14p Is Located in the ER Membrane of Yeast by Immunofluorescence
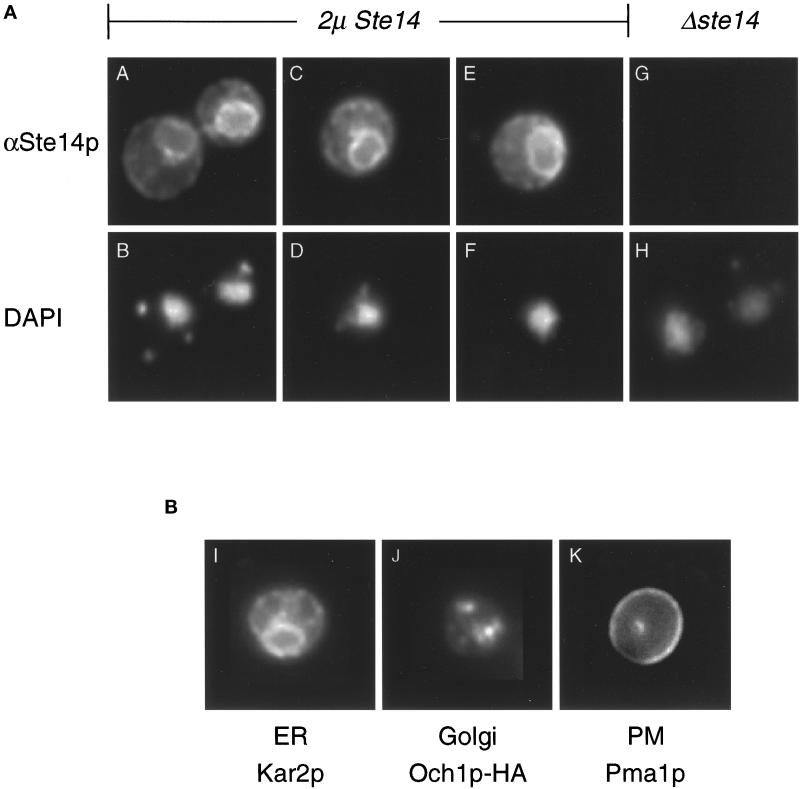
The Ste14p sequence does not contain any known localization signals. To determine the intracellular localization of Ste14p in yeast, we localized the protein by indirect immunofluorescence using anti-Ste14p antiserum (Figure 3A). In cells expressing high levels of Ste14p, we observed a ring-like staining pattern that mainly surrounds the nucleus (Figure 3A, panels A, C, and E) as marked by DAPI fluorescence (Figure 3A, panels B, D, and F); we were unable to detect Ste14p unless it was overexpressed. The perinuclear staining is similar to the immunofluorescence staining pattern of Kar2p, a well-characterized ER protein (Figure 3B, panel I), and is distinct from the patterns for the Golgi- or plasma membrane-localized proteins Och1p-HA or Pma1p, respectively (Figure 3B, panels J and K). No Ste14p staining was detected for a Δste14 strain (Figure 3A, panel G; panel H is DAPI stained). These indirect immunofluorescence data show that the majority of Ste14p is localized to the ER under steady-state conditions.
Figure 3.
Ste14p is localized to the ER membrane by immunofluorescence. (A) Immunofluorescence pattern of Ste14p. Indirect immunofluorescence of Ste14p was performed in Δste14-3 strains bearing either pSM1317 (2μ URA3 STE14) (panels A-F) or pRS316 (CEN URA3) (panels G and H). Cells were fixed with formaldehyde, spheroplasted, and probed with a 1:2000 dilution of anti-Ste14p antiserum (αSte14p) that had been depleted previously of nonspecific antibodies, followed by secondary decoration with Cy3-conjugated goat anti-rabbit antiserum (panels A, C, E, and G) and staining with DAPI (panels B, D, F, and H). The images in panels A, C, E, and G were exposed for equivalent amounts of time. (B) Immunofluorescence patterns for a group of marker proteins. The typical immunofluorescence staining pattern of an ER, Golgi, and plasma membrane marker is shown. Fixed and permeabilized cells from an SM1058 background expressing CEN URA3 plasmids or CEN URA3 OCH1::HA (SM3495) were stained with a 1:1000 dilution of anti-Kar2p antiserum (panel I), a 1:1000 dilution of anti-HA antiserum (panel J), or a 1:200 dilution of anti-Pma1p antiserum (panel K). The localization of Kar2p (ER), Och1p-HA (Golgi), and Pma1p (PM) shown here is consistent with previously observed localization data (Rose et al., 1989; Harris et al., 1994; Harris and Waters, 1996).
Ste14p Methyltransferase Activity Cofractionates with ER Membranes by Subcellular Fractionation
On the basis of the membrane association and immunofluorescence data, the Ste14 protein is localized to the ER membrane; however, it is not known whether Ste14p is active at this membrane. We used subcellular fractionation to determine whether the distribution of Ste14p methyltransferase activity coincided with the steady-state distribution of Ste14p. One advantage of this approach is that it allowed us to detect chromosomal levels of Ste14p; in contrast, only overexpressed levels of Ste14p could be detected by immunofluorescence.
A total yeast homogenate was subjected to fractionation on a sucrose step gradient (Antebi and Fink, 1992). The strain used in this experiment carries a chromosomal copy of the STE14 gene. A ste14 deletion strain was also examined by subcellular fractionation and found to lack detectable methyltransferase activity (Schmidt and Michaelis, unpublished observations). The gradient fractions were assayed for protein concentration and several enzymatic activities that serve to characterize various subcellular organelles. As shown in Figure 4, two major membrane populations can be defined by the distribution of marker enzyme activities. The upper part of the gradient (fractions 1–4) contains light membranes defined by α-mannosidase (Ams1p; vacuole) and Kex2p (trans-Golgi network) activities. Heavy membranes are found near the bottom of the gradient in fractions 6–10. NADPH cytochrome c reductase activity that marks the ER was recovered predominantly in fractions 6–9, whereas plasma membrane ATPase activity was recovered in fractions 7–10; note that the peak activities for these two marker enzymes are offset by one fraction (with peaks at fractions 8 and 9, respectively). Cytosolic proteins do not enter the gradient, as is evident by the high concentration of protein at the top of the gradient. The relative distribution of the marker enzymatic activities on this gradient is consistent with that reported previously (Antebi and Fink, 1992).
We determined the methyltransferase activity in each fraction using an in vitro assay that has been used previously to monitor methyltransferase activity (Philips and Pillinger, 1995; Volker et al., 1995). The fractionation profile of methyltransferase activity was most similar to the distribution of NADPH cytochrome c reductase activity (fractions 6–9). These results suggest that Ste14p activity is localized to the ER. The partially overlapping distribution of ER and plasma membranes does not allow us to exclude the possibility that some Ste14p activity is partly localized to the plasma membrane. As detailed above, however, Ste14p was not observed to be plasma membrane localized by immunofluorescence (Figure 3). In addition, there appears to be a slight shoulder of Ste14p activity (Figure 4, fraction 3) that cofractionates with light membranes. This shoulder may represent a small fraction of Ste14p that is in the Golgi, or it may simply reflect the normal fractionation of ER membranes, because the ER marker NADPH cytochrome c reductase also has a shoulder in the light membranes.
Insertion of an Epitope Tag into Ste14p
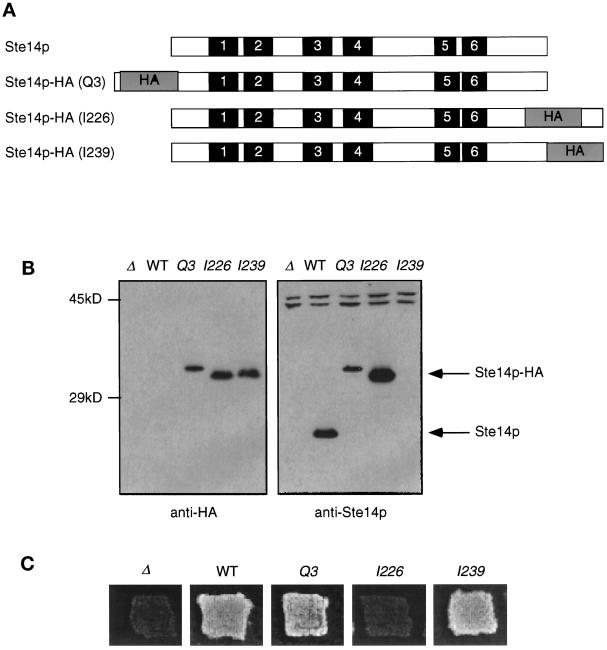
To aid in the analysis of the topology and localization of Ste14p, we inserted the triply iterated hemagglutinin epitope HA into Ste14p at the N terminus (Q3), at the C terminus (I239), or internally (I226) (Figure 5A). Epitope tags inserted at other internal sites interfered with Ste14 protein production (Romano and Michaelis, unpublished observations). The anti-HA antibodies were able to detect all of the epitope-tagged Ste14 proteins but not wild-type untagged Ste14p (Figure 5B). A slight mobility shift is detected for Ste14p-HA (Q3) compared with the other two tagged constructs (Figure 5B); the reason for this shift is unclear, but it is not because of multiple inserts. For comparison, the same samples were probed with anti-Ste14p antiserum (Figure 5B). The difference in mobility between Ste14p and Ste14p-HA (I226) is consistent with the insertion of the triply iterated HA epitope, which contains ∼40 amino acids. All Ste14p species could be detected with anti-Ste14p antiserum, except for Ste14p-HA (I239). Both the I226 and I239 constructs contain the HA tag in the C-terminal domain used to generate the anti-Ste14p antiserum (Figure 5B); however, I226 can be detected with anti-Ste14p antiserum, whereas I239 cannot. This suggests that the anti-Ste14p antibodies may recognize an epitope at the extreme C terminus of Ste14p very near I239. Using more quantitative immunoblots, we found that the protein levels of Ste14p-HA (Q3) and Ste14p-HA (I239) were approximately eightfold and fivefold lower, respectively, than the levels of wild-type Ste14p (Romano and Michaelis, unpublished observations). These results may indicate that the epitope tag can affect the metabolic stability of Ste14p.
Figure 5.
Introduction of epitope tags into Ste14p. (A) Diagram showing the placement of the triple HA epitope tag in Ste14p. Putative membrane spans (solid bars) and the triple HA epitope (gray bars) are indicated and shown in approximate proportions. The epitope tag (HA) was placed at the N terminus (Q3), internally (I226), or at the C terminus of Ste14p (I239). (B) Detection of tagged and untagged Ste14p and Ste14p-HA by anti-HA and anti-Ste14p antisera. Crude cell extracts were subjected to 12.5% SDS-PAGE and transferred to nitrocellulose, and immunoblots were probed with either anti-HA antiserum or anti-Ste14p antiserum depleted previously of nonspecific antibodies as indicated. Lanes contain SM2926 (Δ), SM3185 (WT), SM3187 (Q3), SM3431 (I226), and SM3189 (I239). (C) Patch mating assay. Patches of the indicated MATa strains were replica plated onto a lawn of the MATα mating tester (SM1068) spread on an SD plate and incubated at 30°C for 2 d. Mating is detected by the growth of prototrophic diploids as indicated for WT, Q3, and I239. No mating is observed for Δste14 or I226.
Epitope-Tagged Ste14p (Q3) and (I239) Are Functional
To determine whether insertion of the epitope tag affected protein function, we tested the ability of strains expressing epitope-tagged Ste14p to produce active a-factor using a plate mating assay. The activity of Ste14p is essential for the production of the mating pheromone a-factor; hence, the mating assay is an indirect measure of Ste14p activity. As shown in Figure 5C, a Δste14 strain is unable to mate. Both Ste14p-HA (Q3) and (I239) were able to support mating in a plate mating assay, suggesting that the a-factor pheromone is active and therefore methylated (Figure 5C). In a quantitative filter mating assay, nearly wild-type levels of mating were observed (80% relative to wild-type) for strains expressing either Ste14p-HA (Q3) or Ste14p (I239) (Romano and Michaelis, unpublished observations). The lower protein levels of Ste14p-HA (Q3) and (I239) noted above therefore do not appear to affect mating significantly, presumably because methyltransferase activity and/or a-factor itself are normally present in excess of that required for efficient mating. Mating is not detected for internally tagged Ste14p (I226), suggesting that a-factor is not methylated and that perhaps the insertion of the epitope at this location interferes with Ste14p methyltransferase activity (Figure 5C).
Because mating assays measure the activity of Ste14p only indirectly, we measured the enzymatic activity of epitope-tagged Ste14p using an in vitro methyltransferase assay (Philips and Pillinger, 1995; Volker et al., 1995). No Ste14p activity was detected for Δste14 or a strain expressing STE14::HA (I226), in agreement with the mating assay results. The Vmax values for wild-type Ste14p, Ste14p-HA (Q3), and Ste14p-HA (I239) were normalized for Ste14p expression levels (see MATERIALS AND METHODS) and determined to be 24.6 ± 1.7, 21.3 ± 3.2, and 24.3 ± 0.2 pmole per mg of membrane protein per min, respectively. Thus, the N- and C-terminal tags do not significantly impair Ste14p methyltransferase activity.
The ER Localization of Ste14p Is Sensitive to the Addition of an Epitope Tag
Many localization signals are contained in the N or C terminus of proteins (e.g., signal sequence, ER localization sequences). To determine whether insertion of the epitope tag at either the N or C terminus of Ste14p influenced the ER localization of the methyltransferase, we examined the sucrose gradient fractionation profile and indirect immunofluorescence pattern of the HA-tagged Ste14p constructs.
For subcellular fractionation, lysates from strains expressing wild-type Ste14p, Ste14p-HA (Q3), and Ste14p-HA (I239) were subjected to sucrose gradient centrifugation. Gradient fractions were analyzed by immunoblotting with anti-Ste14p or anti-HA antiserum. Consistent with our previous results in Figure 4, wild-type untagged Ste14p fractionated with heavy membranes (Figure 6A, fractions 5–7). In contrast, the peak distribution of both HA epitope-tagged Ste14p species was in fractions 2–3 of the gradient that contain light membranes. The light membrane compartments that fractionate in the upper part of this gradient include vacuolar, trans-Golgi network, and Golgi membranes (Figure 6B). Epitope-tagged Ste14p activity cofractionates mainly with the activity of the Golgi marker GDPase and not with the activities derived from vacuolar and trans-Golgi network compartments. These results suggest that the modification of either end of Ste14p adversely affects its localization, resulting in the mislocalization of Ste14p-HA (Q3) and (I239). It should be noted that the gradient used in this experiment contains fewer sucrose layers than did the gradient described for Figure 4. This change was empirically determined to improve the separation of lighter membranes.
To confirm that epitope-tagged Ste14p is mislocalized in the cell, we analyzed the intracellular localization of the Ste14p-HA constructs by indirect immunofluorescence. In contrast to the ER-staining pattern observed for wild-type Ste14p, strains overexpressing Ste14p-HA (I226) or Ste14p-HA (I239) display a punctate-staining pattern (Figure 6C, compare panels C and D with panels E, F, K and L). A strain expressing Ste14p-HA (Q3) shows mainly a punctate-staining pattern (55.1% of cells; n = 334), whereas a minor population of cells displays both ER- and punctate-staining patterns (36.8%); examples of each are shown in Figure 6C, panels I and J, respectively. A small population of cells shows only an ER-staining pattern (8.1%). For comparison, the immunofluorescence patterns of known ER, Golgi, and plasma membrane proteins are shown in Figure 3B, panels I-K (Rose et al., 1989; Harris et al., 1994; Harris and Waters, 1996). The punctate stainings observed for N- and C-terminal–tagged Ste14p are consistent with these proteins being Golgi localized instead of ER or plasma membrane localized. These immunofluorescence data taken together with the fractionation data suggest that the addition of an epitope tag to Ste14p results in mislocalization to the Golgi. Although we originally intended to use the Ste14p-HA constructs for topology studies, their aberrant localization precluded their use as reliable sensors of Ste14p topology.
A S. pombe Homologue Complements a Δste14 Defect
Recently a number of proteins that show significant amino acid conservation to Ste14p has been cloned (Figure 7A). One such homologue from S. pombe, mam4p, has been described and is 44% identical to Ste14p at the amino acid level (Imai et al., 1997). The size and hydropathy profile of the mam4 protein is similar to that of Ste14p (Figure 7B), and a mam4p mutant shows a mating defect that is likely caused by the absence of a methyl group on the S. pombe-mating pheromone M-factor (Imai et al., 1997).
To determine whether mam4p can functionally replace Ste14p, we analyzed the ability of mam4 to complement a Δste14 defect using a plate mating assay. In addition, we determined the activity of mam4p using an in vitro methyltransferase assay. As shown in Figure 7C, mam4p is able to complement a Δste14 mating defect in a plate mating assay, even though the in vitro methyltransferase activity of mam4p is sevenfold lower than that for Ste14p (Figure 7D). This lower activity may be attributable to a lower level of expression of mam4p in S. cerevisiae compared with that of Ste14p. Our results indicate that mam4p can replace Ste14p and is therefore a functional homologue. In addition, we determined that mam4p cofractionates with Ste14p and hence localizes to the ER in S. cerevisiae (Schmidt and Michaelis, unpublished observations).
DISCUSSION
In S. cerevisiae, the gene products that mediate CAAX processing have all recently been identified. Although the enzymology of these components is beginning to be elucidated, the intracellular site(s) where these processing reactions occur has not yet been determined. We show here by immunofluorescence and subcellular fractionation that the yeast Ste14p methyltransferase, which catalyzes the last step in this processing pathway, is localized to the ER membrane. In addition, we have evidence that the yeast CAAX proteases, Rce1p and Ste24p, that mediate the previous proteolysis step are also ER membrane-localized (Schmidt and Michaelis, unpublished observations). Thus, although prenylation appears to be performed in the cytoplasm (He et al., 1991), the final two steps of CAAX processing occur at the ER membrane in yeast. These latter two steps of CAAX processing are mediated by multispanning-membrane proteins that presumably contain their catalytic sites on the cytosolic face of the ER membrane. Consistent with our results is the microsomal association of prenylcysteine carboxyl methyltransferase activity from rat liver cells (Stephenson and Clarke, 1992) and the recent finding that the human homologue of Ste14p pcCMT is localized to the ER by immunofluorescence in tissue culture cells (Dai et al., 1998).
Proteins of the ER membrane are known to perform numerous cellular functions. These range from protein translocation and its associated activities, such as glycosylation, to a variety of metabolic and catabolic functions, such as certain steps of sterol biosynthesis and the detoxification of compounds by cytochrome p450s. Our finding that Ste14p is an ER membrane protein suggests a new role for the ER membrane, namely as the site of CAAX processing. The ER represents a way station, because this compartment is not the ultimate destination for most CAAX proteins. For instance, Ras1p, Ras2p, and the G-protein γ subunit (Ste18p) are localized to the plasma membrane, Ykt6p is found on ER to Golgi transit vesicles (Sogaard et al., 1994), Ydj1p is partly ER associated where it functions in facilitating protein translocation (Caplan et al., 1992a,b), and Pex19p may be associated with peroxisomes (Gotte et al., 1998). In addition, the mating pheromone a-factor is exported from the cell, presumably across the plasma membrane, and its intracellular precursors are associated with as yet uncharacterized membranes (Chen et al., 1997). How these lipid-modified proteins are trafficked from the ER membrane to their final destinations remains an unanswered question. Three possibilities can be considered: 1) via carrier-mediated transport, 2) by movement along the outer surface of the vesicular secretory pathway, or 3) by diffusion.
The present study raises the question of how Ste14p is retained in the ER. Signals have been identified that are involved in the ER localization of yeast and mammalian proteins. Two signals present at the C terminus of ER proteins, the KDEL sequence (HDEL in yeast) of lumenal ER proteins and the dilysine motif of type I transmembrane ER proteins, are necessary and sufficient for the retrieval of ER proteins that have escaped to the Golgi (Pelham et al., 1988; Jackson et al., 1990, 1993; Gaynor et al., 1994). An additional ER localization signal, the N-terminal diarginine motif, has been identified for the localization of mammalian type II transmembrane proteins (Schutze et al., 1994). ER localization signals in yeast have also been found in transmembrane domains. For instance, the transmembrane domain for Sec12p was found to contain an ER localization signal that is dependent on the Rer1 protein for ER localization (Sato et al., 1996). Two other membrane proteins, Sec71p and Sec63p, were also found to depend on Rer1p for their ER localization (Sato et al., 1997). Ste14p, a multispanning-membrane protein, contains neither an HDEL nor a dilysine motif at its C terminus but does contain two adjacent arginines at its N terminus. These arginines, however, are located at amino acid positions 16 and 17, and Schutze et al. (1994) has reported that ER localization only occurs when the arginines are located within four residues of the initiator methionine. To date, the role of the diarginine motif in the ER localization of yeast proteins has not been examined. It will be of interest to determine whether Ste14p contains a novel retention or retrieval signal, whether Ste14p depends on Rer1p for its localization suggesting a possible transmembrane localization signal, or whether it is ER-retained via an interaction with other ER membrane proteins. The ER-localized CAAX proteases, Ste24p and Rce1p, are attractive candidates in regard to this latter possibility.
As a first step in examining Ste14p localization signals, we show here that the insertion of a triply iterated HA epitope tag either at the N terminus (Q3), at the C terminus (I239), or internally but near the C terminus (I226) results in the mislocalization of Ste14p. Our immunofluorescence and subcellular fractionation data suggest that Q3, I226, and I239 are mainly localized to the Golgi instead of the ER. The insertion of an HA epitope tag in Ste14p may disrupt an unidentified ER retrieval or retention signal or interfere with the interaction of Ste14p with other components required for its ER localization. These results suggest that both the N- and C-terminal ends of Ste14p play a role in its proper localization. Even though the use of epitope tags is routine and often does not affect the function of proteins being examined, our data indicate that caution should be taken when interpreting results based on tagged membrane proteins, as evidenced by the mislocalization of HA-tagged versions of Ste14p.
Despite their mislocalization, Ste14p-HA (Q3) and (I239) are enzymatically active. The normalized Vmax values of Ste14p-HA (Q3) and Ste14p-HA (I239) as determined by in vitro methyltransferase assays are essentially the same as that of wild-type Ste14p. Both Ste14p-HA (Q3) and Ste14p-HA (I239) are also able to support a wild-type level of mating, providing additional evidence that these tagged molecules are functional in terms of a-factor production. Even though the tagged proteins are mislocalized, there is a measurable population of Ste14p that remains at the ER as detected by subcellular fractionation. This population of Ste14p-HA may provide sufficient methyltransferase activity for full mating. We cannot rule out the possibility, however, that HA-tagged versions of Ste14p could function to modify a-factor at the Golgi membrane. In contrast, Ste14p-HA (I226) is inactive, as measured by both plate mating and in vitro methyltransferase assays. The complete lack of enzymatic activity as well as the mislocalization of Ste14p-HA (I226) suggests that the HA epitope tag in this construct may disrupt a site important for substrate binding or catalysis in Ste14p, in addition to affecting localization.
Methyltransferases catalyze the transfer of a methyl group from methyl donors such as S-adenosylmethionine (AdoMet) to various methyl acceptor substrates, including DNA, RNA, lipid, small molecules, and proteins (Clarke, 1993). It has been shown that most methyltransferases share a tripartite consensus sequence that is thought to function in AdoMet binding (Kagan and Clarke, 1994). Interestingly, Ste14p does not have any significant sequence similarity to other families of methyltransferases and lacks the tripartite AdoMet-binding sequences present in most methyltransferases (Cheng et al., 1993; Kagan and Clarke, 1994). Thus, the site(s) of AdoMet binding in Ste14p remains to be determined. We are presently screening ste14 mutant alleles for potential defects in AdoMet binding to help clarify this issue.
Ste14p was the first member of the prenylcysteine protein carboxyl methyltransferase family to be cloned and sequenced (Blair, 1979; Wilson, 1985; Sapperstein et al., 1994). Recently, several homologues of Ste14p have been identified (shown in Figure 7A). These include S. pombe mam4p, X. laevis Xmam4p, and the human homologue pcCMT (Imai et al., 1997; Dai et al., 1998). In addition, database searches identified a mouse EST (accession number AA022288) (Imai et al., 1997) and two C. elegans open reading frames (accession numbers U88175 and U80450) that have significant amino acid sequence homology to Ste14p. There is strong conservation between these family members, with mam4p having the highest (44%) amino acid sequence identity to Ste14p (Imai et al., 1997). The members of this family have similar hydropathy profiles and comprise one of the few subgroups of methyltransferases that contain multiple membrane spans. Most of the amino acid conservation between these family members occurs at their C-termini, suggesting that residues important for CAAX methyltransferase activity are located at the C-terminus. Consistent with this idea is the fact that the insertion of the HA epitope tag near the C terminus (I226) of Ste14p results in the disruption of methyltransferase activity. Like S. cerevisiae Ste14p, the homologues S. pombe mam4p, Xenopus mam4p, and human pcCMT have been shown to possess in vitro CAAX methyltransferase activity (Imai et al., 1997; Dai et al., 1998). These CAAX methyltransferases are able to transcomplement other CAAX methyltransferase family members in vivo. We show here by complementation of a Δste14 mutant that S. pombe mam4p is functional in S. cerevisiae, providing additional transcomplementational evidence. Elsewhere we demonstrate that human pcCMT is also functional in S. cerevisiae (Dai et al., 1998). In addition, Imai. et al. (1997) have shown that Xmam4p is functional in S. pombe. Together these proteins form a novel family of protein prenylcysteine carboxyl methyltransferases.
Much remains to be learned about the structure, mechanism, localization signals, and topology of the Ste14p family of prenylcysteine carboxyl methyltransferases. Ste14p contains no known protein motifs, and the residues involved in its function and localization have not been identified. Given the hydrophobic nature of their prenylated substrates, it is likely that all the members of this family are ER membrane proteins and that their active sites face the cytosol and/or are partially buried in the membrane. S. cerevisiae is a genetically tractable model system for investigating this family of methyltransferases. Studies on Ste14p are expected to elucidate the general mechanism and structure of CAAX methyltransferases and to add to our knowledge of the processing of CAAX proteins. Furthermore, the ability to functionally express other methyltransferases in this system will speed the isolation and characterization of members in this methyltransferase family.
ACKNOWLEDGMENTS
The authors thank B. Eipper, C. Machamer, L. Roman, D. Kuehn, G. Nijbroek, and A. Tam for critical reading of the manuscript. The authors thank M. Philips for providing the pcCMT sequence before its publication. The authors also thank R. Jensen, D. Murphy, and W. Guggino for access to their microscopes. This work was supported by a grant (GM-41223) and fellowship (GM-18641) from the National Institutes of Health to S.M. and W.K.S., respectively.
Abbreviations used:
- AdoMet
S-adenosylmethionine
- AFC
N-acetyl-S-farnesyl-l-cysteine
- BSA
bovine serum albumin
- CCD
charge-coupled device
- DAPI
4′,6-diamidino-2-phenylindole
- ER
endoplasmic reticulum
- GDPase
guanosine diphosphatase
- GST
glutathione S-transferase
- HA
triple hemagglutinin tag
- HRP
horseradish peroxidase
- Ig
immunoglobulin
- pcCMT
prenylcysteine carboxyl methyltransferase
- PCR
polymerase chain reaction
- TBST
Tris-buffered saline with Tween
- WT
wild type
REFERENCES
- Abeijon C, Orlean P, Robbins PW, Hirschberg CB. Topography of glycosylation in yeast: characterization of GDP mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund PS. Post-translational processing of RhoA. J Biol Chem. 1997;272:33175–33180. doi: 10.1074/jbc.272.52.33175. [DOI] [PubMed] [Google Scholar]
- Baker TA, Grossman AD, Gross CA. A gene regulating the heat shock response in Escherichia coli also affects proteolysis. Proc Natl Acad Sci USA. 1984;81:6779–6783. doi: 10.1073/pnas.81.21.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C, Michaelis S. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 1991;10:3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LC. Genetic analysis of mating type switching in yeast. Ph.D. Thesis. Eugene, OR: University of Oregon; 1979. [Google Scholar]
- Boyartchuk V, Ashby M, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. Ydj1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992a;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Tsai J, Casey PJ, Douglas MG. Farnesylation of Ydj1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem. 1992b;267:18890–18895. [PubMed] [Google Scholar]
- Chen P, Sapperstein SK, Choi JC, Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Kumar S, Posfai J, Pflugrath JW, Roberts RJ. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein methylation. Curr Opin Cell Biol. 1993;5:977–983. doi: 10.1016/0955-0674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Wickner WT. Yeast KEX2 protease and mannosyltransferase I are localized to distinct compartments of the secretory pathway. Yeast. 1989;5:25–33. doi: 10.1002/yea.320050105. [DOI] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Human prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- Ehring R, Beyreuther K, Wright JK, Overath P. In vitro and in vivo products of E. coli lactose permease gene are identical. Nature. 1980;283:537–540. doi: 10.1038/283537a0. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Feldheim D, Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol. 1994;126:935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RI, Bernstein M, Schekman R. Product of SEC53 is required for folding and glycosylation of secretory proteins in the lumen of the yeast endoplasmic reticulum. J Biol Chem. 1987;262:9332–9339. [PubMed] [Google Scholar]
- Finegold AA, Johnson DI, Farnsworth CC, Gelb MH, Judd SR, Glomset JA, Tamanoi F. Protein geranylgeranyltransferase of Saccharomyces cerevisiae is specific for Cys-Xaa-Xaa-Leu motif proteins and requires the CDC43 gene product but not the DPR1 gene product. Proc Natl Acad Sci USA. 1991;88:4448–4452. doi: 10.1073/pnas.88.10.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau WH, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Na S, Zhu X, Seto-Young D, Perlin DS, Teem JH, Haber JE. Dominant lethal mutations in the plasma membrane H(+)-ATPase gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:10531–10535. doi: 10.1073/pnas.91.22.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Chen P, Chen SY, Vancura KL, Michaelis S, Powers S. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc Natl Acad Sci USA. 1991;88:11373–11377. doi: 10.1073/pnas.88.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna CA, Clarke S. Farnesyl cysteine C-terminal methyltransferase activity is dependent upon the STE14 gene product in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5071–5076. doi: 10.1128/mcb.10.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Davey J, Kawagishi-Kobayashi M, Yamamoto M. Genes encoding farnesyl cysteine carboxyl methyltransferase in Schizosaccharomyces pombe and Xenopus laevis. Mol Cell Biol. 1997;17:1543–1551. doi: 10.1128/mcb.17.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for the retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kubota S, Yoshida Y, Kumaoka H, Furumichi A. Studies on the microsomal electron-transport system of anaerobically grown yeast. V. Purification and characterization of NADPH-cytochrome c reductase. J Biochem (Tokyo) 1977;81:197–205. doi: 10.1093/oxfordjournals.jbchem.a131436. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Marcus S, Caldwell GA, Miller D, Xue C-B, Naider F, Becker JM. Significance of C-terminal cysteine modifications to the biological activity of the Saccharomyces cerevisiae a-factor mating pheromone. Mol Cell Biol. 1991;11:3603–3612. doi: 10.1128/mcb.11.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opheim DJ. Alpha-d-mannosidase of Saccharomyces cerevisiae. Characterization and modulation of activity. Biochim Biophys Acta. 1978;524:121–130. doi: 10.1016/0005-2744(78)90110-9. [DOI] [PubMed] [Google Scholar]
- Paddon C, Loayza D, Vangelista L, Solari R, Michaelis S. Analysis of the localization of STE6/CFTR chimeras in a Saccharomyces cerevisiae model for the cystic fibrosis defect CFTRΔF508. Mol Microbiol. 1996;19:1007–1017. doi: 10.1046/j.1365-2958.1996.444973.x. [DOI] [PubMed] [Google Scholar]
- Payne GS, Schekman R. A test of clathrin function in protein secretion and cell growth. Science. 1985;230:1009–1014. doi: 10.1126/science.2865811. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Sala D, Tan EW, Canada FJ, Rando RR. Methylation and demethylation reactions of guanine nucleotide-binding proteins of retinal rod outer segments. Proc Natl Acad Sci USA. 1991;88:3043–3046. doi: 10.1073/pnas.88.8.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips MR, Pillinger MH. Prenylcysteine-directed carboxyl methyltransferase activity in human neutrophil membranes. Methods Enzymol. 1995;256:49–63. doi: 10.1016/0076-6879(95)56009-2. [DOI] [PubMed] [Google Scholar]
- Pillinger MH, Volker C, Stock JB, Weissmann G, Philips MR. Characterization of a plasma membrane-associated prenylcysteine-directed alpha carboxyl methyltransferase in human neutrophils. J Biol Chem. 1994;269:1486–1492. [PubMed] [Google Scholar]
- Rao R, Slayman C. Mutagenesis of conserved residues in the phosphorylation domain of the yeast plasma membrane H(+)-ATPase. Effects on structure and function. J Biol Chem. 1993;268:6708–6713. [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol Cell Biol. 1994;14:1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc Natl Acad Sci USA. 1997;94:9693–9698. doi: 10.1073/pnas.94.18.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K, Nakano A. Endoplasmic reticulum localization of Sec12p is achieved by two mechanisms: Rer1p-dependent retrieval that requires the transmembrane domain and Rer1p-independent retention that involves the cytoplasmic domain. J Cell Biol. 1996;134:279–293. doi: 10.1083/jcb.134.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze MP, Peterson PA, Jackson MR. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Sollner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Stephenson RC, Clarke S. Identification of a C-terminal protein carboxyl methyltransferase in rat liver membranes utilizing a synthetic farnesyl cysteine-containing peptide substrate. J Biol Chem. 1990;265:16248–16254. [PubMed] [Google Scholar]
- Stephenson RC, Clarke S. Characterization of a rat liver protein carboxyl methyltransferase involved in the maturation of proteins with the -CXXX C-terminal sequence motif. J Biol Chem. 1992;267:13314–13319. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker C, Pillinger MH, Philips MR, Stock JB. Prenylcysteine analogs to study function of carboxylmethylation in signal transduction. Methods Enzymol. 1995;250:216–225. doi: 10.1016/0076-6879(95)50074-x. [DOI] [PubMed] [Google Scholar]
- Wilson KW. Identification and regulation of cell-type-specific genes required for mating in Saccharomyces cerevisiae. Ph.D. Thesis. San Francisco, CA: University of California, San Francisco; 1985. [Google Scholar]
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]