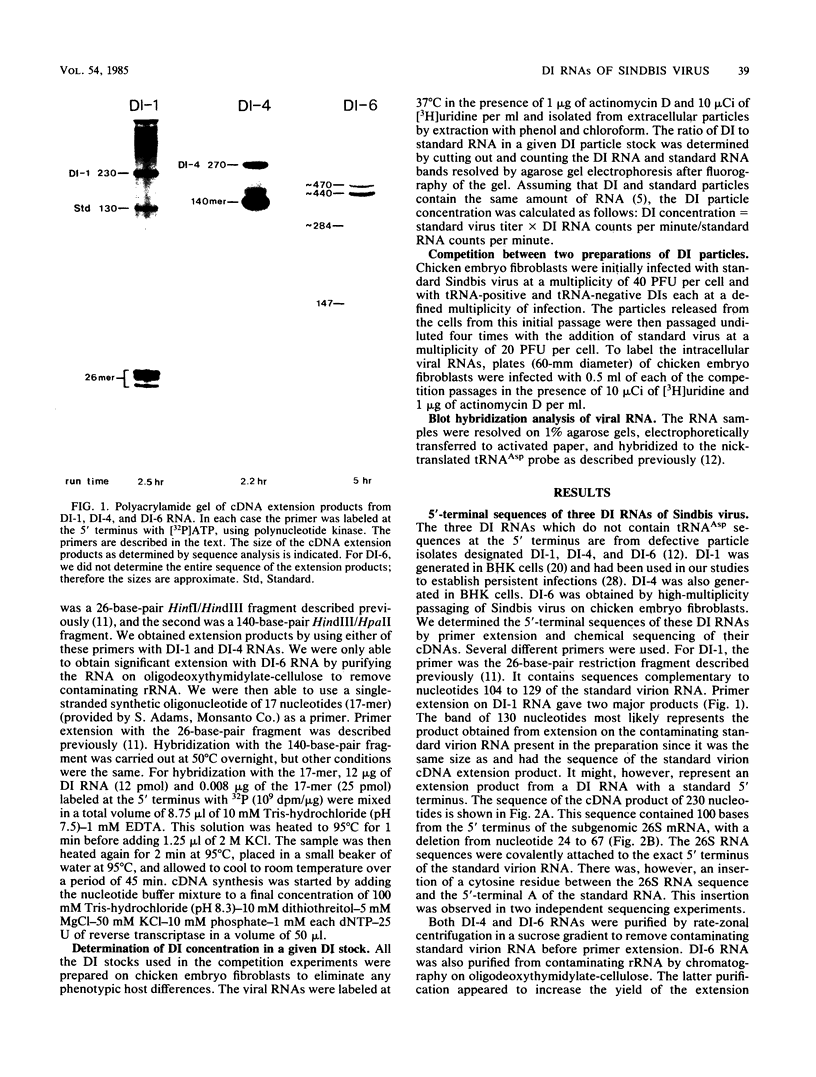
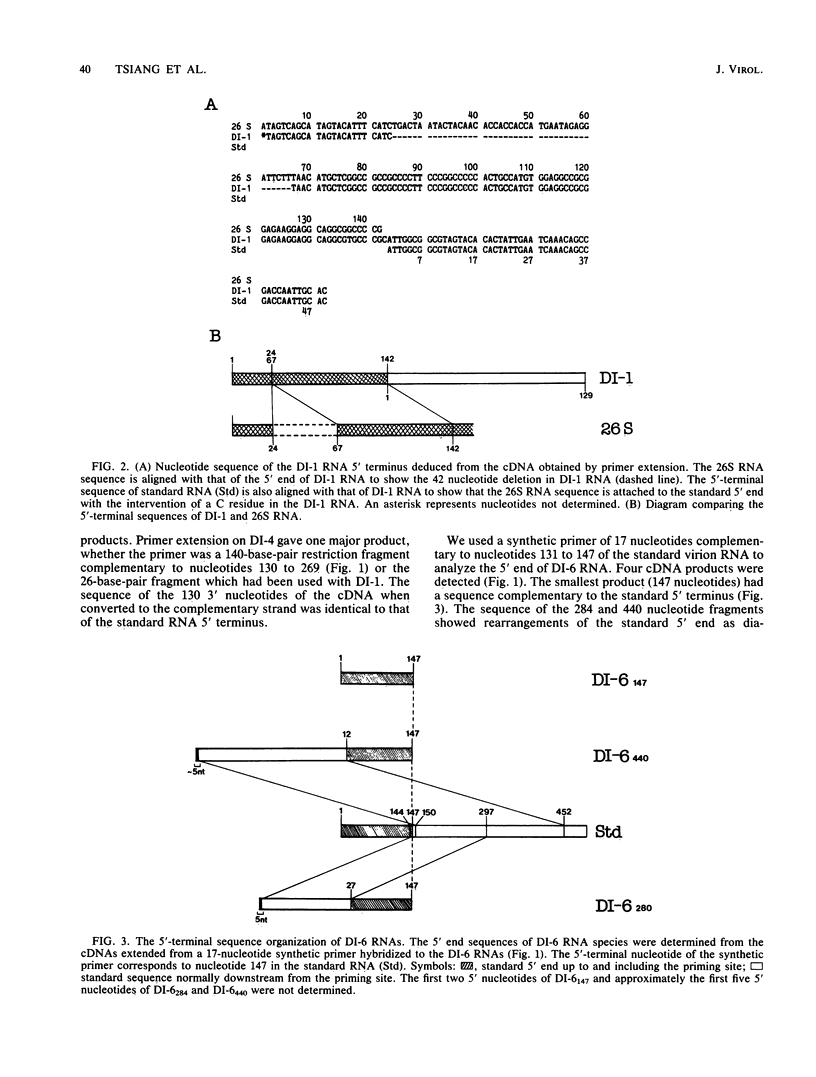
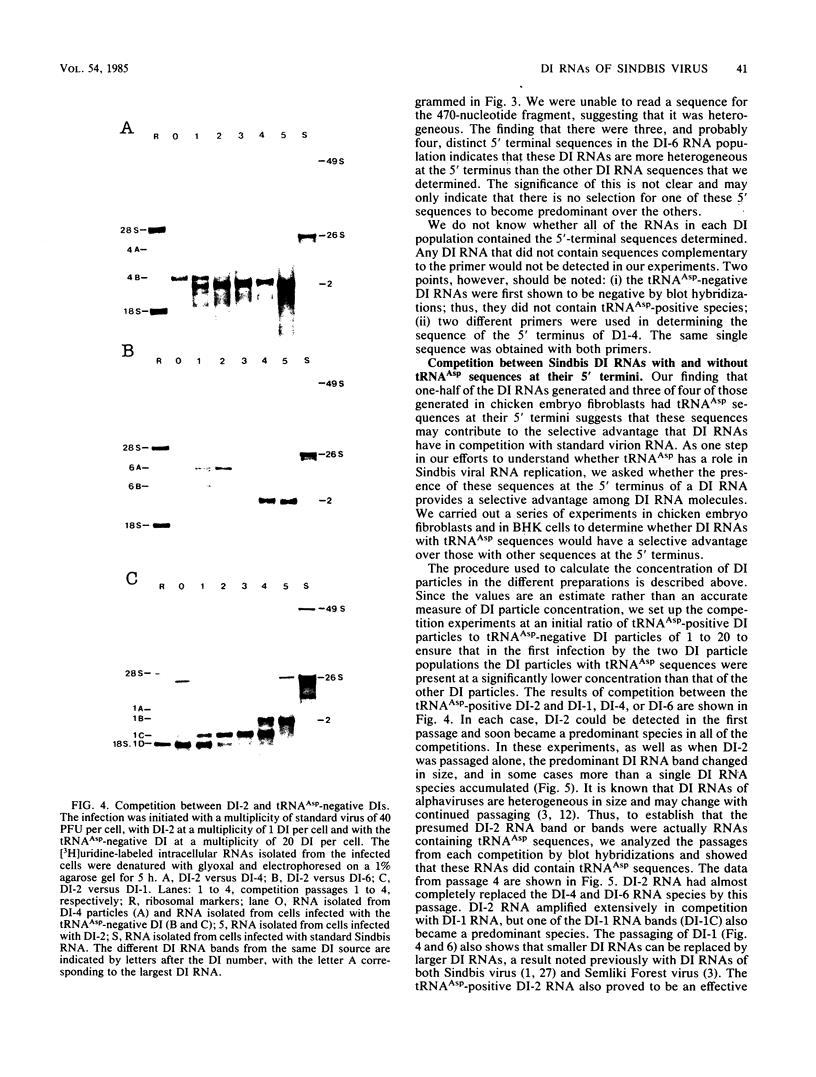
Abstract
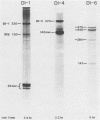
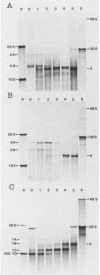
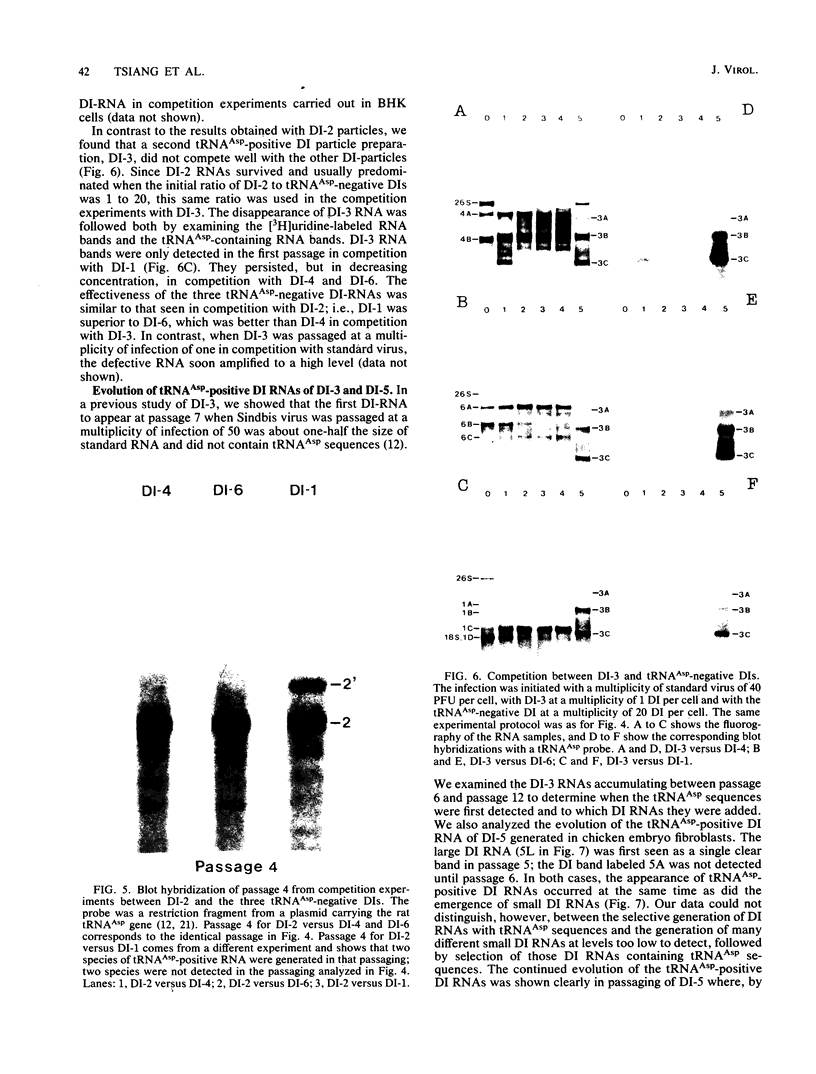
Three of six independently derived defective interfering (DI) particles of Sindbis virus generated by high-multiplicity passaging in cultured cells have tRNAAsp sequences at the 5' terminus of their RNAs (Monroe and Schlesinger, J. Virol. 49:865-872, 1984). In the present work, we found that the 5'-terminal sequences of the three tRNAAsp-negative DI RNAs were all derived from viral genomic RNA. One DI RNA sample had the same 5'-terminal sequence as the standard genome. The DI RNAs from another DI particle preparation were heterogeneous at the 5' terminus, with the sequence being either that of the standard 5' end or rearrangements of regions near the 5' end. The sequence of the 5' terminus of the third DI RNA sample consisted of the 5' terminus of the subgenomic 26S mRNA with a deletion from nucleotides 24 to 67 of the 26S RNA sequence. These data showed that the 5'-terminal nucleotides can undergo extensive variations and that the RNA is still replicated by virus-specific enzymes. DI RNAs of Sindbis virus evolve from larger to smaller species. In the two cases in which we followed the evolution of DI RNAs, the appearance of tRNAAsp-positive molecules occurred at the same time as did the emergence of the smaller species of DI RNAs. In pairwise competition experiments, one of the tRNAAsp-positive DI RNAs proved to be the most effective DI RNA, but under identical conditions, a second tRNAAsp-positive DI RNA was unable to compete with the tRNAAsp-negative DIs. Therefore, the tRNAAsp sequence at the 5' terminus of a Sindbis DI RNA is not the primary factor in determining which DI RNA becomes the predominant species in a population of DI RNA molecules.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Guild G. M., Stollar V. Defective interfering particles of Sindbis virus. V. Sequence relationships between SVSTD 42 S RNA and intracellular defective viral RNAs. Virology. 1977 Mar;77(1):175–188. doi: 10.1016/0042-6822(77)90416-0. [DOI] [PubMed] [Google Scholar]
- Jennings P. A., Finch J. T., Winter G., Robertson J. S. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983 Sep;34(2):619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- Kennedy S. I., Bruton C. J., Weiss B., Schlesinger S. Defective interfering passages of Sindbis virus: nature of the defective virion RNA. J Virol. 1976 Sep;19(3):1034–1043. doi: 10.1128/jvi.19.3.1034-1043.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S. I. Sequence relationships between the genome and the intracellular RNA species of standard and defective-interfering Semliki Forest virus. J Mol Biol. 1976 Dec;108(2):491–511. doi: 10.1016/s0022-2836(76)80132-5. [DOI] [PubMed] [Google Scholar]
- Käriäinen L., Pettersson R. F., Keränen S., Lehtovaara P., Söderlund H., Ukkonen P. Multiple structurally related defective-interfering RNAs formed during undiluted passages of Semliki forest virus. Virology. 1981 Sep;113(2):686–697. doi: 10.1016/0042-6822(81)90197-5. [DOI] [PubMed] [Google Scholar]
- Lazzarini R. A., Keene J. D., Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981 Oct;26(2 Pt 2):145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- Lehtovaara P., Söderlund H., Keränen S., Pettersson R. F., Käriäinen L. 18S defective interfering RNA of Semliki Forest virus contains a triplicated linear repeat. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5353–5357. doi: 10.1073/pnas.78.9.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovaara P., Söderlund H., Keränen S., Pettersson R. F., Käriäinen L. Extreme ends of the genome are conserved and rearranged in the defective interfering RNAs of Semliki Forest virus. J Mol Biol. 1982 Apr 25;156(4):731–748. doi: 10.1016/0022-2836(82)90139-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Monroe S. S., Ou J. H., Rice C. M., Schlesinger S., Strauss E. G., Strauss J. H. Sequence analysis of cDNA's derived from the RNA of Sindbis virions and of defective interfering particles. J Virol. 1982 Jan;41(1):153–162. doi: 10.1128/jvi.41.1.153-162.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe S. S., Schlesinger S. Common and distinct regions of defective-interfering RNAs of Sindbis virus. J Virol. 1984 Mar;49(3):865–872. doi: 10.1128/jvi.49.3.865-872.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe S. S., Schlesinger S. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5' ends. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3279–3283. doi: 10.1073/pnas.80.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P. J., Nichol S. T., Horodyski F. M., Holland J. J. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell. 1984 Apr;36(4):915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- Ou J. H., Rice C. M., Dalgarno L., Strauss E. G., Strauss J. H. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Strauss E. G., Strauss J. H. The 5'-terminal sequences of the genomic RNAs of several alphaviruses. J Mol Biol. 1983 Jul 25;168(1):1–15. doi: 10.1016/s0022-2836(83)80319-2. [DOI] [PubMed] [Google Scholar]
- Perrault J. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., Rüegg K. J., Weissmann C. Nanovariant RNAs: nucleotide sequence and interaction with bacteriophage Qbeta replicase. J Mol Biol. 1977 Dec 25;117(4):877–907. doi: 10.1016/s0022-2836(77)80004-1. [DOI] [PubMed] [Google Scholar]
- Schlesinger S., Schlesinger M., Burge B. W. Defective virus particles from Sindbis virus. Virology. 1972 May;48(2):615–617. doi: 10.1016/0042-6822(72)90076-1. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Kuchino Y., Nishimura S. Mammalian tRNA genes: nucleotide sequence of rat genes for tRNAAsp, tRNAGly and tRNAGlu. Nucleic Acids Res. 1981 May 25;9(10):2239–2250. doi: 10.1093/nar/9.10.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Kennedy S. I. The generation and propagation of defective-interfering particles of Semliki Forest virus in different cell types. Virology. 1978 Aug;89(1):285–299. doi: 10.1016/0042-6822(78)90060-0. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Strauss J. H. Replication strategies of the single stranded RNA viruses of eukaryotes. Curr Top Microbiol Immunol. 1983;105:1–98. doi: 10.1007/978-3-642-69159-1_1. [DOI] [PubMed] [Google Scholar]
- Söderlund H., Keränen S., Lehtovaara P., Palva I., Pettersson R. F., Käriäinen L. Structural complexity of defective-interfering RNAs of Semliki Forest virus as revealed by analysis of complementary DNA. Nucleic Acids Res. 1981 Jul 24;9(14):3403–3417. doi: 10.1093/nar/9.14.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Levis R., Schlesinger S. Evolution of virus and defective-interfering RNAs in BHK cells persistently infected with Sindbis virus. J Virol. 1983 Dec;48(3):676–684. doi: 10.1128/jvi.48.3.676-684.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Rosenthal R., Schlesinger S. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J Virol. 1980 Jan;33(1):463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]