Abstract
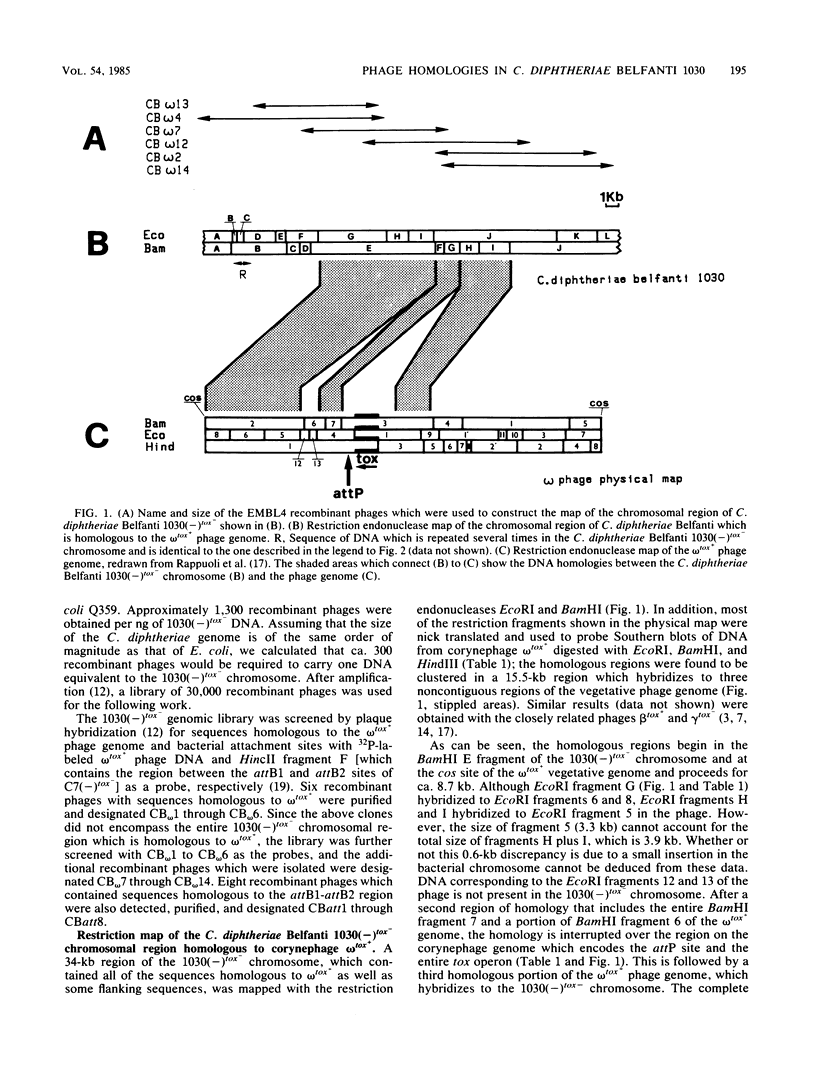
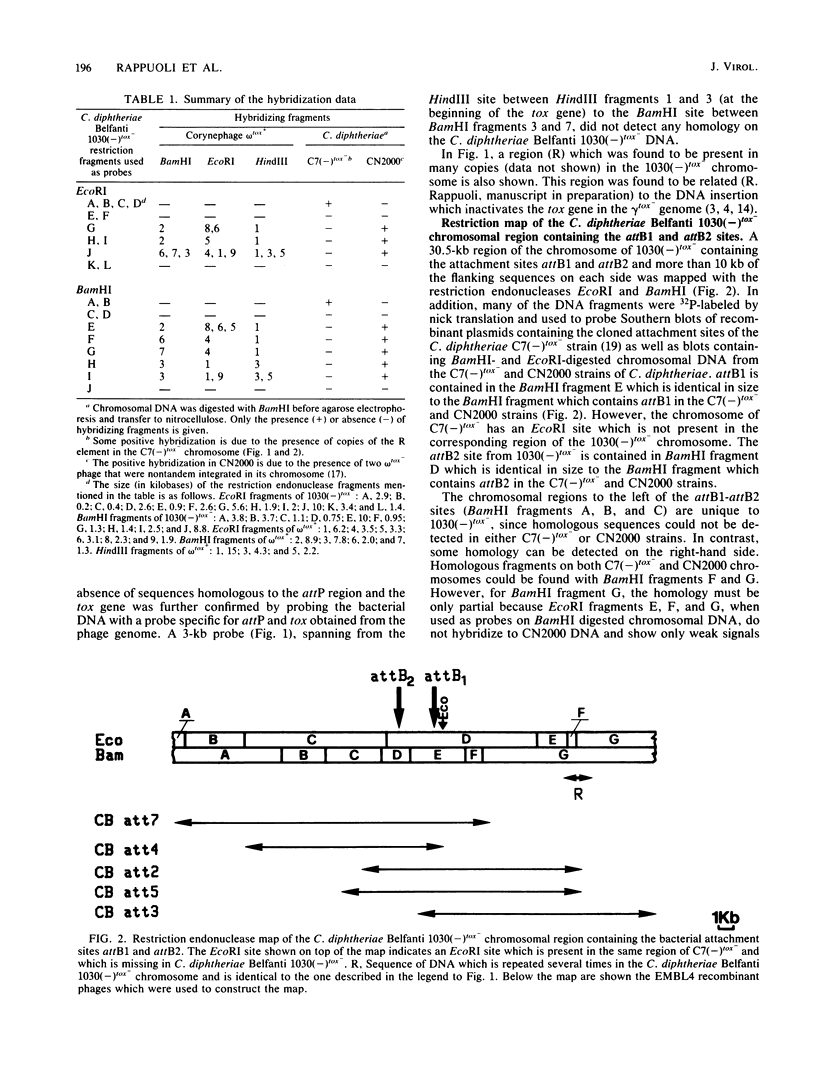
A library of chromosomal DNA from Corynebacterium diphtheriae Belfanti 1030(-)tox- was cloned in the lambda phage vector EMBL4 and screened for sequences homologous to corynephage omega tox+ and the attB1-attB2 region of the C7(-)tox- chromosome. Two portions of the 1030(-)tox- chromosome, 35 and 30.5 kilobases long which contain, respectively, the entire region homologous to corynephage omega tox+ and the attB1-attB2 sites, were mapped with the restriction endonucleases BamHI and EcoRI. Chromosomal DNA from 1030(-)tox- was shown to contain a 15.5-kilobase region that was homologous to ca. 42% of the corynephage omega tox+ genome. These sequences were found to hybridize to three regions of the phage genome and do not contain either the diphtheria tox operon or the attP site. These sequences are distant from the chromosomal region that contains the attB1-attB2 sites. Moreover, unlike other known defective prophages, the physical map of this prophage starts at the cos site and is colinear with the vegetative phage map. The 30.5-kilobase region of the 1030(-)tox- chromosome, which contains the attB1-attB2 sites, has a central core region that is almost identical to the corresponding region of the C7(-)tox- chromosome; however, the flanking sequences in these two strains of C. diphtheriae are different.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BARDSDALE W. L., PAPPENHEIMER A. M., Jr Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J Bacteriol. 1954 Feb;67(2):220–232. doi: 10.1128/jb.67.2.220-232.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G. A., Groman N. B. Genetic elements novel for Corynebacterium diphtheriae: specialized transducing elements and transposons. J Bacteriol. 1981 Oct;148(1):143–152. doi: 10.1128/jb.148.1.143-152.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G. A., Groman N. B. Identification of deoxyribonucleic acid restriction fragments of beta-converting corynebacteriophages that carry the gene for diphtheria toxin. J Bacteriol. 1981 Oct;148(1):153–162. doi: 10.1128/jb.148.1.153-162.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G. A., Groman N. B. Physical mapping of beta-converting and gamma-nonconverting corynebacteriophage genomes. J Bacteriol. 1981 Oct;148(1):131–142. doi: 10.1128/jb.148.1.131-142.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R. K. Characterization and genetic mapping of nontoxinogenic (tox) mutants of corynebacteriophage beta. J Virol. 1976 Jul;19(1):195–207. doi: 10.1128/jvi.19.1.195-207.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K., Murray N. E. On the nature of sbcA mutations in E. coli K 12. Mol Gen Genet. 1980;179(3):555–563. doi: 10.1007/BF00271745. [DOI] [PubMed] [Google Scholar]
- Kaiser K., Murray N. E. Physical characterisation of the "Rac prophage" in E. coli K12. Mol Gen Genet. 1979 Sep;175(2):159–174. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L. New bacteriophage lambda vectors with positive selection for cloned inserts. Methods Enzymol. 1983;101:3–19. doi: 10.1016/0076-6879(83)01004-6. [DOI] [PubMed] [Google Scholar]
- Maximescu P. New host-strains for the lysogenic Corynebacterium diphtheriae Park Williams No. 8 strain. J Gen Microbiol. 1968 Aug;53(1):125–133. doi: 10.1099/00221287-53-1-125. [DOI] [PubMed] [Google Scholar]
- Michel J. L., Rappuoli R., Murphy J. R., Pappenheimer A. M., Jr Restriction endonuclease map of the nontoxigenic corynephage gamma c and its relationship to the toxigenic corynephage beta c. J Virol. 1982 May;42(2):510–518. doi: 10.1128/jvi.42.2.510-518.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Michel J. L., Murphy J. R. Integration of corynebacteriophages beta tox+, omega tox+, and gamma tox- into two attachment sites on the Corynebacterium diphtheriae chromosome. J Bacteriol. 1983 Mar;153(3):1202–1210. doi: 10.1128/jb.153.3.1202-1210.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Michel J. L., Murphy J. R. Restriction endonuclease map of corynebacteriophage omega ctox+ isolated from the Park-Williams no. 8 strain of Corynebacterium diphtheriae. J Virol. 1983 Feb;45(2):524–530. doi: 10.1128/jvi.45.2.524-530.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Ratti G. Physical map of the chromosomal region of Corynebacterium diphtheriae containing corynephage attachment sites attB1 and attB2. J Bacteriol. 1984 Apr;158(1):325–330. doi: 10.1128/jb.158.1.325-330.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J., Groman N., Coyle M. Plasmids in Corynebacterium diphtheriae and diphtheroids mediating erythromycin resistance. Antimicrob Agents Chemother. 1980 Nov;18(5):814–821. doi: 10.1128/aac.18.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]



