Abstract
Virtual screening by molecular docking has become a widely used approach to lead discovery in the pharmaceutical industry when a high resolution structure of the biological target of interest is available. The performance of three widely-used docking programs (Glide, GOLD, and DOCK) for virtual database screening is studied when they are applied to the same protein target and ligand set. Comparisons of the docking programs and scoring functions using a large and diverse data set of pharmaceutically interesting targets and active compounds are carried out. We focus on the problem of docking and scoring flexible compounds which are sterically capable of docking into a rigid conformation of the receptor. The Glide XP methodology is shown to consistently yield enrichments superior to the two alternative methods, while GOLD outperforms DOCK on average. The study also shows that docking into multiple receptor structures can decrease the docking error in screening a diverse set of active compounds.
I. Introduction
Virtual screening has become a widely used approach to lead discovery in the pharmaceutical industry. When a high resolution structure of the biological target of interest is available, the most common methodology for performing virtual screening involves the use of a flexible docking algorithm, in which conformational sampling methods are used to position the ligand in the receptor, and some sort of scoring function is applied to obtain a predicted free energy of binding. A number of powerful software programs, e.g. GOLD1–4, FlexX5, DOCK6,7, Glide8,9, Surflex10,11, LigandFit12, have been developed over the past several decades to carry out docking calculations, and good success in both binding mode and binding affinity prediction has often been achieved in selected test cases. A more challenging goal has been to improve the robustness of the methods with regard to both structural and energetic prediction; all of the above programs on occasion manifest both false negatives (active compounds which are not appropriately docked or scored by the methodology) and false positives (weakly binding compounds whose binding affinity is seriously overpredicted). A number of comparative evaluation of docking programs, conducted over the past several years, confirm this general picture13–17.
One of us has recently described a new approach9 that has been implemented in the Glide program (the extra precision, or XP, Glide methodology) which incorporates novel terms into the binding free energy scoring function (as compared to standard scoring approaches18–23, which are similar with regard to functional form), and appears, in preliminary tests, to substantially enhance the ability of Glide to pick out known active compounds from a random ligand database. Relative to the standard precision (SP) Glide scoring function8, improvements in enrichment of roughly an order of magnitude are reported for XP Glide9, for a large and diverse set of ligands and receptors. However, this comparison is entirely internal to the Glide program, and does not provide any calibration as to how well, or poorly, alternative approaches would fare with the particular data set under study. The present paper is aimed at addressing this question, employing in addition to Glide, the DOCK and GOLD programs which are both widely used in academia and in the pharmaceutical and biotechnology industries. We experiment with a number of docking and scoring approaches available in each program in order to obtain as much comparative data as possible.
In addition to evaluating the relative abilities of the various docking methods to identify known active compounds embedded in a random database, another objective of this work is the validation of a new approach to enrichment studies described in reference 9. In the great majority of enrichment studies in the literature, no attempt is made to separate misdockings due to induced fit effects from scoring errors; ligands are simply docked into a single rigid version of the receptor, and their scores are compared with those of database ligands. However, if the goal is to fairly calibrate the accuracy of the scoring function (or to improve the parametrization by fitting theory to experimental data), it makes no sense to include ligands that do not “fit” into a particular receptor structure, due to significant steric clashes, in the enrichment test set. These ligands presumably dock in grossly incorrect poses, and would require a substantial modification of the receptor active site conformation in order to dock correctly. The score associated with such a grossly incorrect pose cannot be expected to correspond to an experimentally measured value, and inclusion of such data in an “enrichment factor” does not provide a reasonable measure of the accuracy of the scoring function when induced fit effects are not an issue.
Computation of induced fit effects, in cases where the ligand does not fit into a specified receptor conformation, is quite feasible, as we have shown in a recent publication24. However, such an approach requires the use of a flexible receptor as well as ligand, and hence is in general much more computationally intensive than rigid receptor docking. There are also questions about how to compare the scores of ligands docked to different receptor conformations; incorporation of receptor strain energy into binding affinity prediction is an area at present in its infancy. For these reasons, we believe there is considerable value in separating the docking/scoring problem into two components: (1) calculation of induced fit effects (including strain energy estimation) when these are necessary to achieve a reasonable prediction of the binding mode; (2) docking and scoring of compounds which are sterically capable of docking more or less correctly into a specified rigid conformation of the receptor. We consider only component (2) in the present paper (as was also done in reference 9).
An approach to implement the separation suggested above is to simply eliminate the ligands from the training and test sets which do not dock in a “reasonable” pose (“non-fitting”) into the receptor conformation selected for the study. This is the approach adopted in reference 9. The assessment of docking accuracy was performed using RMSDs when a crystal structure was available, and otherwise employing visual inspection, with the correct pose inferred by analogy with known complexes in the PDB with ligands of a similar structure. The detailed criteria were documented in reference 9. A possible objection to this approach is that the ligand was misdocked specifically by Glide, but would be docked successfully with alternative programs. In the present paper, we examine “misdocked” ligands from the data sets of reference 9, docking these ligands with the GOLD and DOCK programs. This investigation provides an important cross-check on our previous protocols and assumptions with regard to sorting ligands into the categories of properly or improperly docked, reducing the bias inherent in the use of a single program. While misdocking with multiple programs does not prove that the compound cannot be docked into a rigid receptor, it certainly strongly suggests that this is the case, and, additionally, provides a check on whether our assessment of misdocking was unduly influenced by the score assigned to the ligand by Glide.
The paper is organized as follows. Section II discusses the data sets that we have assembled, including the initial division of ligands into “fitting” and “non-fitting” into the receptor conformation used in the study. In section III, we briefly discuss the methodologies employed in GOLD, DOCK, and Glide, with regard to both sampling and enrichment. Section IV presents results comparing enrichment factors, using fitting compounds only, for the various test cases. Results obtained with all three programs for the non-fitting compounds are then separately analyzed. Section V, the conclusion, provides a brief summary and discusses future directions.
II. Data set
We have used the same set of receptors and ligands as were employed to evaluate the Glide XP scoring function in reference 9. This data set is divided into two components; a training set, which was used to parametrize Glide XP, and an independent test set. Fourteen targets which are of pharmaceutical interest are contained in the training set, as shown in Table 1a. One of them (p38 MAP kinase) is represented by two alternative cocrystallized receptor sites. The crystallographic resolution of all these 15 proteins is less than 3.0 Å (9 of them are less than 2.2 Å). The receptors for these screens cover a wide variety of receptor types and therefore provide a proper test of the docking methods. All protein structures were prepared using the procedure as stated in the previous Glide methodology paper8.
Table 1.
Data set used to compare virtual database screening. All active compounds have experimental activities less than 10 μM except those for neuraminidase.
| (a) Glide training set | |||
|---|---|---|---|
| PDB code | description | num. well-docked actives | num. poor-docked actives |
| 1fjs | Factor Xa | 9 | 4 |
| 1bji | Neuraminidase | 9 | 0 |
| 1hpx | HIV-1 Protease | 9 | 5 |
| 1cx2 | Cyclooxygenase-2 | 13 | 0 |
| 1e66 | Acetylcholinesterase | 20 | 0 |
| 1ett | Thrombin | 15 | 1 |
| 1kim | Thymidine Kinase | 4 | 0 |
| 1aq1 | Human Cyclin Dep. Kinase | 6 | 4 |
| 1bl7 | p38 Map Kinase | 27 | 9 |
| 1kv2 | Human p38 Map Kinase | 10 | 0 |
| 1ml7 | EGRF Tyrosine Kinase | 107 | 10 |
| 1qpe | Lck Kinase | 87 | 34 |
| 1rt1 | HIV Reverse Transcriptase | 23 | 6 |
| 1tmn | Thermolysin | 5 | 1 |
| 3ert | Human Estrogen Receptor | 8 | 2 |
| (b) Glide test set | |||
| PDB code | description | num. well-docked actives | num. poor-docked actives |
| 1m4h | BACE | 34 | 43 |
| 1dan | Factor VIIa | 40 | 53 |
| 1fm6 | PPARγ (closed form) | 32 | 61 |
| 1fm9 | PPARγ (open form) | 25 | 68 |
| 1y6b | Vegfr2 (closed form) | 21 | 90 |
| 1ywn | Vegfr2 (open form) | 26 | 85 |
| 1aq1 | Human Cyclin Dep. Kinase | 143 | 110 |
| 1ett | Thrombin | 15 | 25 |
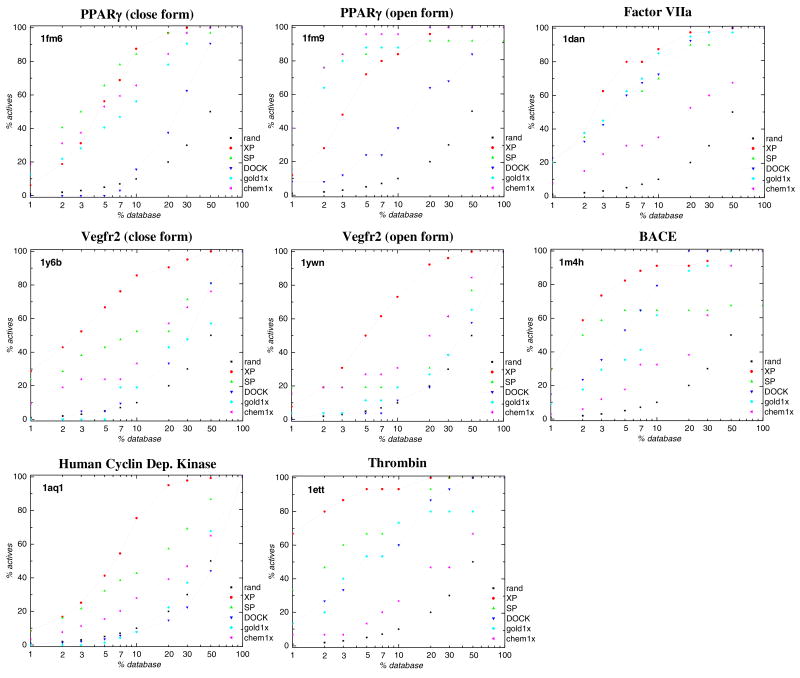
The test set, described in Table 1b, consists of four new receptors, with appropriate sets of cognate ligands, and two receptors (Human Cyclin Dependent Kinase 2, or CDK2, and Thrombin) studied previously, with new sets of ligands. Among the four new receptors, two of them (PPARγ and Vegfr2) are investigated using two different conformations of the receptor --- a closed form and an open form – which are appropriate for binding different classes of ligands. While a larger test set would be desirable (and will be employed in subsequent publications), development of suitable data sets is highly labor intensive; and the current test set is capable of providing an assessment as to whether there is large overfitting in the Glide XP results with the training set.
For PPARγ and Vegfr2 targets, only a small fraction (from 19% to 34%) of the active compounds can be correctly docked by Glide XP if only one form of the receptor (either the closed or open form) is used. However, if both forms of receptor are used, 61% and 42% of all active compounds can be correctly docked for PPARγ and Vegfr2 receptors, respectively. Therefore, for these cases, docking into multiple receptor structures instead of a single structure is an effective way to decrease the misdocking due to steric clashes, which is a major error in screening a large, diverse set of active compounds.
Comparing Table 1a and 1b, there are many more poorly-docked active compounds in the test set than in the training set. When the training set was constructed, in many cases a relatively small number of active compounds were included, and even in cases where a larger number of compounds are employed, they were typically derived from a small number of literature sources. However, for the test set, we have collected a significant number of compounds from the literature, using a number of different literature sources. Consequently, the diversity of the test set is significantly larger than that of the training set, leading to a larger number of compounds being misdocked into a given structure of the receptor. In a realistic laboratory application of virtual screening, where the data set to be screened is typically a highly diverse pharmaceutical compound collection, we would thus expect the fraction of misdocked compounds for all but the most rigid receptors to be substantial. The consequences of this observation are discussed further below.
In the present study, the same set of active ligands, including both well-docked and poorly-docked ones, are being docked with each program. The classification of the well-docked or poorly-docked are based on Glide XP results. As database ligands, we employed “druglike” decoys that averaged 400 in molecular weight (the “d1-400” dataset) in all cases except thymidine kinase (1kim). For 1kim, which has a very small active site, we used a similar (but in this case more competitive) set with an average molecular weight of 360 (the “d1-360” dataset). The detailed approach to creating these test databases and their property distributions were described in a previous publication8. We believe these compounds are representative of the chemical sample collections of pharmaceutical and biotechnology companies. As such, they should provide a fair and stringent test of the efficacy of the docking method. Each screen used 1000 decoy database ligands and between 4 and 253 known active binders as shown in Table 1. All selected known binders have experimental activities less than 10 μM except those for neuraminidase. The references for their structures and biological activities can be found in a previous publication9. Like the database ligands, the known binders were also MMFF94s-optimized. In these cases, we used input geometries obtained via a MacroModel25 conformational search.
While the training set described above has been used as such for parametrization of Glide XP (and, to a small extent, Glide SP as well), it is worth pointing out that neither GOLD nor DOCK have been trained using this data set. Thus, while comparison of Glide XP to GOLD and DOCK is one (major) objective of the paper (and is best addressed by examining the test set comparisons), the “training” set provides additional data with which to evaluate the absolute level of performance of GOLD and DOCK, their performance relative to each other, and the relative performance of the various options within each program that are evaluated below. To our knowledge, an evaluation of GOLD or DOCK using exclusively “fitting” compounds has not been reported in the literature; the present study provides this information using an extensive database of such compounds, comprising all the data summarized in Table 1a and 1b.
III. Docking Methodologies
Glide (Schrodinger, Inc.)
The Glide (Grid-Based Ligand Docking With Energetics, version 4.0) algorithm approximates a systematic search of positions, orientations, and conformations of the ligand in the receptor binding site using a series of hierarchical filters8,9, 26. The shape and properties of the receptor are represented on a grid by several different sets of fields that provide progressively more accurate scoring of the ligand pose. The fields are computed prior to docking. The binding site is defined by a rectangular box confining the translations of the mass center of the ligand. A set of initial ligand conformations is generated through exhaustive search of the torsional minima, and the conformers are clustered in a combinatorial fashion. Each cluster, characterized by a common conformation of the “core” and an exhaustive set of “rotamer group” conformations, is docked as a single object in the first stage8. The search begins with a rough positioning and scoring phase that significantly narrows the search space and reduces the number of poses to be further considered to a few hundred. In the following stage, the selected poses are minimized on precomputed OPLS-AA van der Waals and electrostatic grids for the receptor. In the final stage, the 5–10 lowest-energy poses obtained in this fashion are subjected to a Monte Carlo procedure in which nearby torsional minima are examined, and the orientation of peripheral groups of the ligand is refined. The minimized poses are then rescored using the GlideScore function, which is an expanded version of ChemScore19 with force field–based components and additional terms accounting for solvation and repulsive interactions. The choice of the best pose is made using a model energy score (Emodel) that combines the energy grid score, GlideScore, and the internal strain of the ligand8. We investigated both Standard Precision mode (SP) and Extra Precision mode (XP) of Glide in this comparative study.
The Glide XP methodology has been described in detail in reference 9; we briefly summarize the important features here. The starting point for XP scoring is a modified version of ChemScore, as in the case of SP; however, novel terms are used to handle physical effects that are missing from ChemScore. Desolvation penalties are applied by docking explicit waters into the highest scoring docked complexes, and evaluating the solvation of polar and charged ligand and protein groups by counting the number of neighboring waters and comparing these values to statistics extracted from a database of correctly docked active ligands. Molecular recognition motifs based on the concept of hydrophobic enclosure of the ligand by the protein are defined, and incremental increases in binding affinity are added to the ligand score when the appropriate motifs are recognized. In order to properly evaluate these new terms, a considerable augmentation of the sampling algorithm, which is carried out at higher resolution, is required; the algorithm itself, based on growing side chains from core positions identified by SP docking, is discussed further in reference 9. Additional terms involving special treatment of salt bridges, pi-cation interactions, and various other specialized medicinal chemistry motifs, are described in reference 9. The XP scoring function has been parametrized using the training set of 15 receptor structures and cognate “fitting” ligands, as is discussed further below.
GOLD (Cambridge Crystallographic Data Center)
Version 2.2 of the GOLD (Genetic Optimization for Ligand Docking) docking program was evaluated in the present study. The GOLD program uses a genetic algorithm (GA) to explore the full range of ligand conformational flexibility and the rotational flexibility of selected receptor hydrogens1–4. The mechanism for ligand placement is based on fitting points. The program adds fitting points to hydrogen-bonding groups on the protein and ligand, and maps acceptor points in the ligand on donor points in the protein and vice versa. Additionally, GOLD generates hydrophobic fitting points in the protein cavity onto which ligand CH groups are mapped. The genetic algorithm optimizes flexible ligand dihedrals, ligand ring geometries, dihedrals of protein OH and NH3 groups, and the mappings of the fitting points. The docking poses are ranked based on a molecular mechanics–like scoring function. There are two different built-in scoring functions in the GOLD program — GoldScore and ChemScore. Note that the ChemScore function implemented in GOLD4 is an optimized version of the original chemscore function developed by Eldridge et al.19. In parallel, the performance of two combined docking protocols was also studied. In the first combined protocol, “GoldScore-reChemScore”, the dockings produced with the GoldScore function are rescored and reranked using the ChemScore function; in the second combined protocol, “ChemScore-reGoldScore”, the docking produced with the ChemScore function are rescored and reranked using the GoldScore function. In both protocol, the Simplex algorithm (local optimization) is used to relax each docking in the alternative scoring function. We also compared two different speed modes — default settings (1x) and 7–8 times speedup settings (8x). In the present work, the binding site was defined as a spherical region which encompasses all protein atoms within 5.0 Å of each crystallographic ligand atom. Protein and ligand input structures were prepared as described above. Default GA settings were used for all calculations.
DOCK(UCSF)
The version 5.2.0 of DOCK6,7 was used in these studies. DOCK characterizes concavities on a protein surface using sets of spheres positioned on top of a Connolly surface generated on the binding pocket. The centers of these spheres characterize positions where ligand atoms can be found in the binding pocket. DOCK uses a graph matching algorithm to position the atoms of a ligand onto the centers of the spheres. A minimization of the ligand poses is performed allowing DOCK to refine the ligand position in the binding pocket. Flexibility of the ligands is modelled by treating the ligand as a series of fragments, where a central fragment (the anchor) is docked first followed by sequentially docking the outer fragments around the anchor. As each fragment is docked, neighbor fragments are combined.
For each target, the Connolly molecular surface was calculated using a probe radius of 1.4 Å. Inside the binding pocket, spheres are generated therein with the DOCK program SPHGEN. SPHGEN outputs the spheres in clusters that overlap each other. Clusters were examined for each target, and the cluster covering the known binding site was chosen by selecting those spheres within 7.5 Å of the the co-crystalized ligand. Compounds were docked allowing for ligand flexibility, using the grid-based energy scoring option for minimization after initial placement in the site. The box for the scoring grid was defined such that all spheres were enclosed with an extra 5.0 Å added in each dimension. Scoring grids for contact and energy scores were calculated with a grid spacing of 0.3 Å. The bump check was set such that compounds with atoms closer than half the sum of the van der Waals radii of the respective atoms were rejected. A 6–12 Lennard-Jones van der Waals potential was used along with a Coulomb potential using a distance-dependent dielectric constant of 4r to simulate solvation effects. The energy cutoff was 10.0 Å. The radii used were those in the vdw_AMBER_parm99.defn set. Ligand atoms were matched to receptor spheres using the anchor first search with the anchor size set to 10 atoms. The automatic matching option was used, and conformations were generated on the fly with the torsion drive option.
IV. Results and Discussions
The objective of the present study is to compare how well three widely-used molecular docking programs perform during virtual database screening when applied to the same protein target and ligand set. In the context of virtual screening, the measure of performance is the ability of the program to prioritize seeded active compounds for a particular target relative to the decoy compounds in the database. We have obtained enrichment data for the training set and the test set as described above.
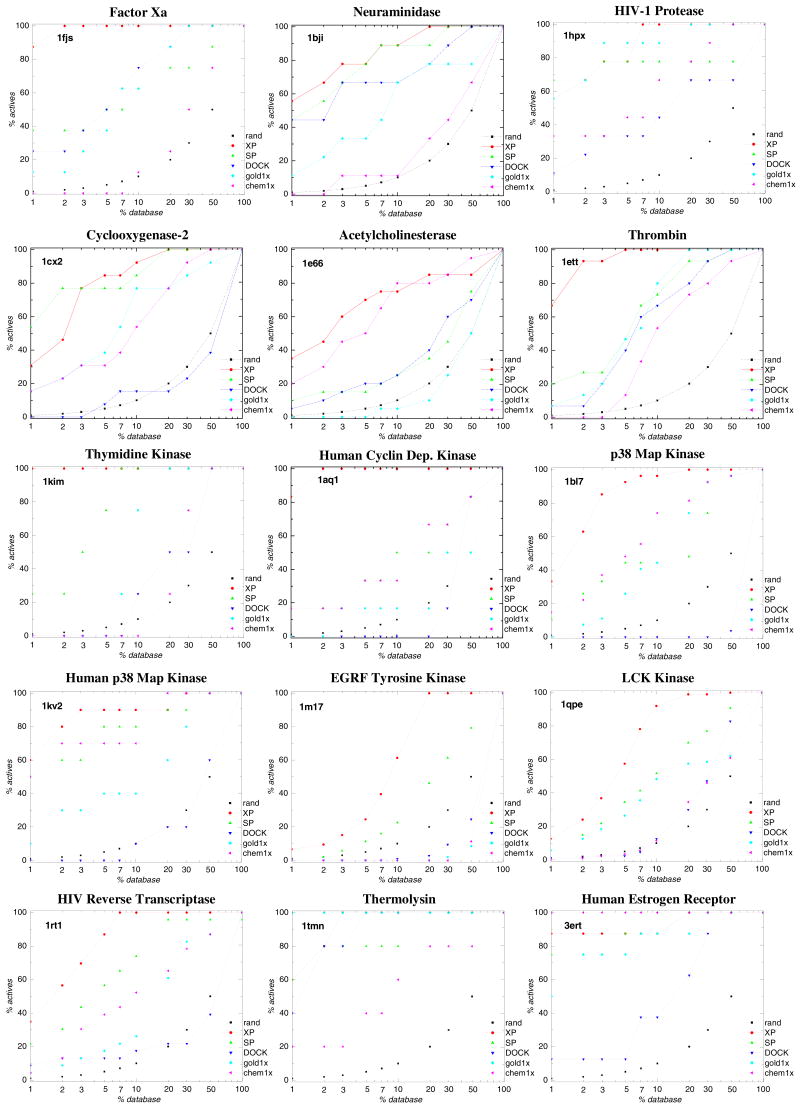
Figure 1 displays the percent of known actives recovered as a function of the percent of the ranked database sampled for Glide XP, Glide SP, DOCK, GOLD 1xGoldScore and GOLD 1xChemScore, for all test cases in the training set and test set. The enrichment curves show that Glide XP gives the best performance for six out of eight cases when evaluated on the test set. On the training set with 15 cases, Glide XP outperforms the other methods in database enrichment ability for 13 cases.
Figure 1.
Percent of known actives found (y axis) vs percent of the ranked database screened (x axis) for Glide XP(XP, red solid), Glide SP (SP, green dash), DOCK (DOCK, blue dash dot), GOLD GoldScore1x (gold1x, Cyan dash dot dot) and GOLD ChemScore1x (chem1x, Magenta short dash). Black dotted lines (rand) show results expected by chance. The listed PDB codes are defined in Table 1. (a) Glide training set; (b) Glide test set.
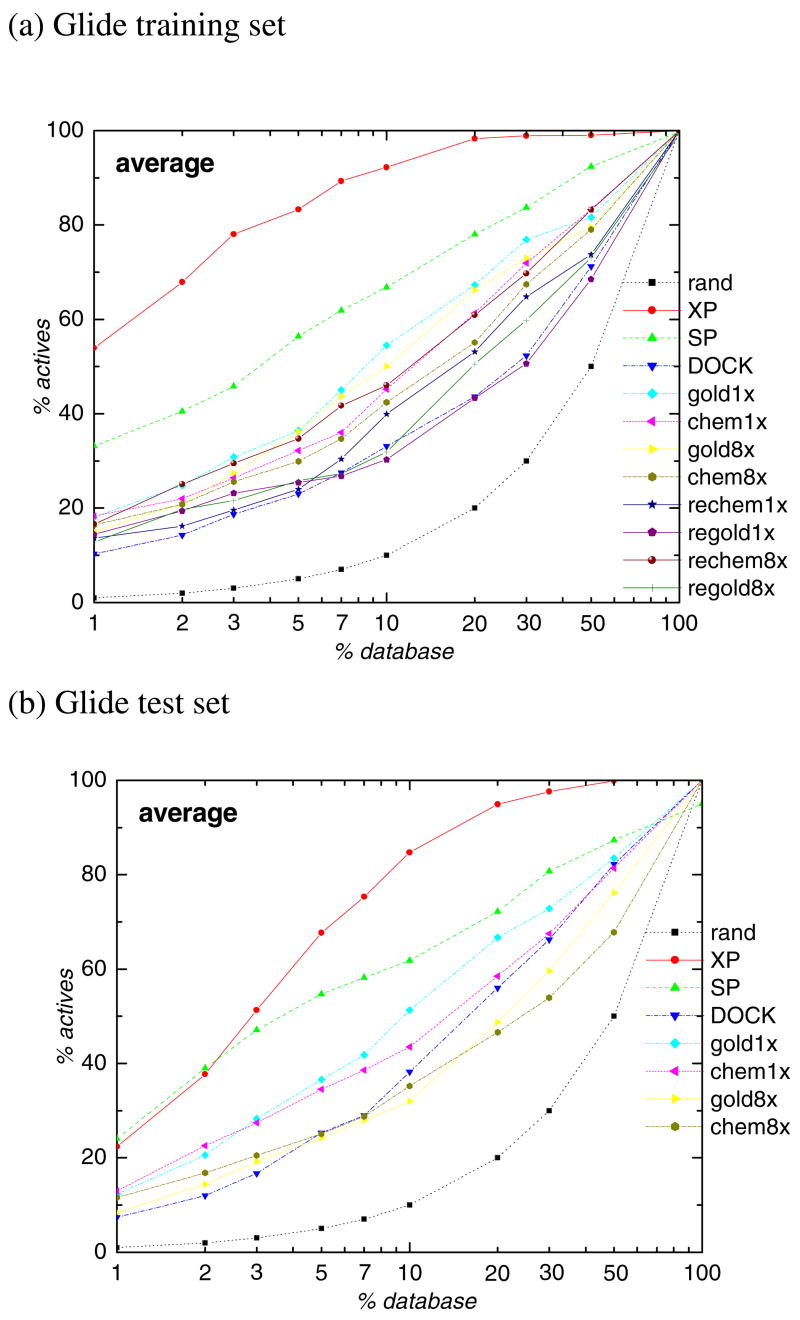
Figure 2a shows the average enrichment curves over all 15 training set targets for all 11 docking methods in the present study. All docking methods could identify active compounds from the ligand database since all enrichment curves are significantly better than the random selection curve. GOLD methods have similar performance as DOCK. They found between 30% and 55% of known actives in the top 10% of the ranked database on average. Glide SP found 67% of actives in the top 10% of the ranked database. Glide XP achieves better enrichment than the other methods, unsurprisingly since this training set was used to parametrize Glide XP. On average, Glide XP found 92% of the known actives in the top 10% of the ranked database. For the GOLD program, the performance of GoldScore is somewhat better than ChemScore. The known actives found in the top 10% of the ranked database are 55% and 45% for 1xGoldScore and 1xChemscore, respectively. However, the docking speed of GoldScore mode (8.5 minutes per ligand for 1x settings) is about 3 times slower than the Chemscore mode(2.8 minutes per ligand for 1x settings). The combined docking protocols which correspond to docking with one scoring function and rescoring with the other do not improve the performance relative to the single scoring functions. The average docking time for each method is listed in Table 2.
Figure 2.
Average percent of known actives found over training set or test set (y axis) vs percent of the ranked database screened (x axis) for Glide XP (XP, red solid), Glide SP (SP, green dash), DOCK (DOCK, blue dash dot), GOLD GoldScore1x (gold1x, Cyan dash dot dot), GOLD ChemScore1x (chem1x, Magenta short dash), GOLD GoldScore8x (gold8x, yellow short dot), GOLD ChemScore8x (chem8x, dark yellow short dash dot), GOLD GoldScore1x-reChemScore (rechem1x, Navy solid), GOLD ChemScore1x-reGoldScore (regold1x, purple solid), GOLD GoldScore8x-reChemScore (rechem8x, wine solid), GOLD ChemScore8x-reGoldScore (regold8x, Olive solid). Black dotted lines (rand) show results expected by chance, (a) Glide training set; (b) Glide test set.
Table 2.
Average docking time (minutes) per ligand on a 2.2 GHz AMD (Athlon MP 2800+) single processor. Times for combined “GoldScore-reChemScore” are identical to these for the GoldScore functions. Times for combined “ChemScore-reGoldScore” are identical to these for the ChemScore functions.
| method | description | minute per ligand |
|---|---|---|
| XP | Glide XP | 7.0 |
| SP | Glide SP | 0.42 |
| DOCK | DOCK | 4.0 |
| 1xgold | GOLD GoldScore 1x | 8.5 |
| 1xchem | GOLD ChemScore 1x | 2.8 |
| 8xgold | GOLD GoldScore 8x | 1.0 |
| 8xchem | GOLD ChemScore 8x | 0.35 |
| 1xrechem | GOLD GoldScore1x-reChemScore | 8.5 |
| 1xregold | GOLD ChemScore1x-reGoldScore | 2.8 |
| 8xrechem | GOLD GoldScore8x-reChemScore | 1.0 |
| 8xregold | GOLD ChemScore8x-reGoldScore | 0.35 |
Figure 2b presents the average enrichment curves for the independent test set (Table 1b) for Glide XP and SP, DOCK, and the four non-composite GOLD methods. DOCK achieves slightly better results for the test set than the training set (38% vs. 33% of the known actives in the top 10% of the ranked database. This difference likely corresponds to statistical variation based on sample size). On average, DOCK performs similarly to the GOLD results for the test set. They found between 32% and 51% of the known actives in the top 10% of the ranked database. Glide SP found 62% of the actives in the top 10% of the ranked database. Glide XP achieves the best enrichment. On average, for the test set Glide XP found 85% of the known actives in the top 10% of the ranked database. In summary, compared with training set results, there is some quantitative degradation in the enrichment for all methods except DOCK, but no significant overfilling is found at this percentage of active compound recovery. In fact, it is possible that the slight degradation in performance of XP (seen for GOLD as well) is not because of overfitting, but because one is dealing with a more challenging set of receptors and/or active compounds. A further discussion of XP performance on the test set will be presented below.
On average, in the top 2% of the ranked database, Glide XP found 68% and 38% of known actives for the training set and test set, respectively. There are several possible explanations for the significant decrease in performance seen at this level of accuracy, which is a highly demanding one (i.e. ranking active compounds ahead of nearly all of the decoy ligands). Firstly, at least some of the difference could be due to overfitting of Glide XP for the active ligands in the training set (thus enabling training set ligands to be ranked higher than they would be otherwise). Secondly, it is possible that there are novel molecular recognition motifs in one or more of the additional receptors in the test set, which have not been incorporated into the Glide XP scoring function as of yet. Glide XP is a combination of physics based approach (to determine functional forms of the scoring terms) with an expert system component (to identify particular chemistries/geometries that should be rewarded or penalized as compared to “normal” scoring); the expert system performance is dependent upon having relevant examples in the training set. The existence of novel examples missing from the training set is a problem distinct from “overfitting”, which in fact would represent a very different sort of difficulty (recognition of motifs identified from previous targets which should not be rewarded or penalized with the same weights in the current targets). Analysis of which problem described above is dominant awaits further investigation.
To characterize the overall performance of each docking method for database screening, Table 3 reports, for both training and test sets, a new measure of enrichment defined as the average number of database decoys outranking the active compounds in the database. Specifically, the number of database decoys with a score that is superior to each active is tabulated, these values are summed, and the result is then divided by the total number of active compounds in the data set. We believe that this metric is superior to standard definitions of enrichment, which punish active ligands when they are outranked by other active ligands; this is a particularly serious problem when the active test suite contains a large number of compounds. A “perfect” score based on this metric would thus be zero (no database ligands outranking any active compounds), and smaller numbers are better. Also, this metric can differentiate the following two circumstances with the same enrichment factor: suppose there are 10 actives in the top 50 database rankings, (a) in one situation, the ranks of 10 actives are from 1 to 10; (b) in the other, the ranks of 10 actives are from 41 to 50. Obviously, the former is better. This new metric (0 vs, 40) can clearly distinguish these two situations. The average metric for all 15 targets in Table 3a indicates the following order of performance Glide XP, Glide SP, GOLD, DOCK. Compared with the training set results in Table 3a, Glide XP results for the test set (Table 3b) are significantly degraded (20 for the training set vs. 39 for the test set), but it is still the best method in the test set by a significant margin. Surprisingly, the performance of DOCK improves on the test set (341 for the training set vs. 265 for the test set).
Table 3.
New enrichment metrics defined as the average number of outranking decoy ligands over well-docked actives for (a) Glide XP training set; (b) Glide XP test set. (see table 2 for the meaning of table headings)
| (a) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pdb ID | XP | SP | DOCK | 1x gold | 1x chem | 8x gold | 8x chem | 1xre chem | 1xre gold | 8xre chem | 8xre gold |
| 1fjs | 1 | 187 | 87 | 82 | 428 | 273 | 427 | 414 | 554 | 521 | 465 |
| 1bji | 25 | 37 | 92 | 248 | 417 | 224 | 340 | 263 | 224 | 132 | 222 |
| 1hpx | 16 | 167 | 290 | 18 | 98 | 155 | 362 | 131 | 138 | 327 | 261 |
| 1cx2 | 24 | 22 | 511 | 133 | 115 | 91 | 89 | 107 | 316 | 66 | 200 |
| 1e66 | 111 | 344 | 353 | 514 | 123 | 435 | 100 | 235 | 307 | 108 | 428 |
| 1ett | 2 | 70 | 98 | 58 | 180 | 159 | 313 | 289 | 406 | 299 | 377 |
| 1kim | 0 | 29 | 259 | 100 | 252 | 68 | 220 | 208 | 582 | 148 | 199 |
| 1aq1 | 3 | 216 | 456 | 380 | 272 | 291 | 256 | 352 | 376 | 270 | 320 |
| 1bl7 | 7 | 183 | 806 | 126 | 95 | 92 | 79 | 285 | 96 | 123 | 66 |
| 1kv2 | 26 | 57 | 451 | 157 | 42 | 119 | 51 | 211 | 139 | 46 | 181 |
| 1ml7 | 41 | 279 | 714 | 845 | 756 | 802 | 669 | 749 | 759 | 589 | 670 |
| 1qpe | 13 | 157 | 316 | 414 | 426 | 474 | 435 | 503 | 573 | 402 | 620 |
| 1rt1 | 11 | 83 | 512 | 233 | 212 | 230 | 264 | 238 | 387 | 220 | 287 |
| 1tmn | 9 | 32 | 10 | 2 | 157 | 50 | 345 | 356 | 414 | 207 | 274 |
| 3ert | 14 | 23 | 165 | 46 | 0 | 114 | 0 | 1 | 5 | 2 | 221 |
| ave | 20 | 126 | 341 | 224 | 238 | 238 | 263 | 289 | 352 | 231 | 319 |
| (b) | |||||||||||
| pdb ID | XP | SP | DOCK | 1x gold | 1x chem | 8x gold | 8x chem | ||||
| 1aq1 | 25 | 206 | 574 | 403 | 379 | 283 | 338 | ||||
| 1dan | 30 | 75 | 55 | 59 | 351 | 96 | 510 | ||||
| 1ett | 9 | 52 | 95 | 204 | 357 | 286 | 520 | ||||
| 1fm6 | 45 | 48 | 276 | 110 | 77 | 153 | 113 | ||||
| 1fm9 | 44 | 82 | 245 | 21 | 7 | 267 | 83 | ||||
| 1m4h | 35 | 315 | 48 | 99 | 246 | 634 | 754 | ||||
| 1y6b | 52 | 221 | 347 | 456 | 302 | 435 | 352 | ||||
| 1ywn | 69 | 310 | 477 | 433 | 285 | 396 | 273 | ||||
| ave | 39 | 164 | 265 | 223 | 250 | 319 | 368 | ||||
In order to check the consistency of our new measure of enrichment with the standard definition, the average standard enrichment factors at 10% of ranked database are shown in Table 4. The standard enrichment factor (EF) is defined as: EF=(HITSsampled/HITStotal)/(Nsampled/Ntotal)
Table 4.
The average standard enrichment factors at 10% of ranked database for different methods. The standard enrichment factor is defined in the text, (see table 2 for the meaning of table headings)
| set | XP | SP | DOCK | 1x gold | 1x chem | 8x gold | 8x chem | 1xre chem | 1xre gold | 8xre chem | 8xre gold |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Training set | 9.2 | 6.7 | 3.3 | 5.5 | 4.5 | 5.0 | 4.2 | 4.0 | 3.0 | 4.6 | 3.2 |
| Test set | 8.5 | 6.2 | 3.8 | 5.1 | 4.4 | 3.2 | 3.5 |
Here, Ntotal is the number of ligands in the docked database, Nsampled is the number of ligands in the docked database to be examined, HITStotal is the total number of the known active ligands, and HITSsampled is the number of known active ligands found in the top Nsampled ligands of the docked database. Compared to the last rows in Table 3a and Table 3b, the new enrichment metric and the standard enrichment factor give very similar trends with regard to the performance. The advantage of the new enrichment metric, however, is that it is not dependent on the total number of active ligands making a better comparison between different databases possible.
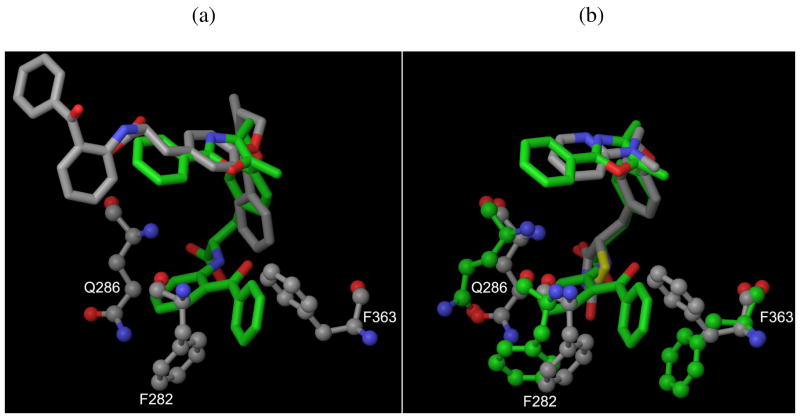
Besides the well-docked active set, we also compare the performance of all methods for the set that was poorly-docked by Glide XP. We visually inspect the binding modes of these known actives generated by docking programs and find they are quite different or missing some key interactions compared with their experimental binding modes or analogues. For example, as shown in Figure 3a, docking of ligand 1293-1 into the 1fm6 (PPARγ closed form) structure yields a ligand pose with an RMSD of 10.9 A when compared to the structure of 1293-1 in its cognate receptor 1fm9 (PPARγ open form). The primary reason is that 1fm6 was cocrystallized with a much smaller ligand 1241-2, in which some side-chain atoms of a number of residues protrude into the binding site, thus blocking binding of the larger ligand (1293-1) in the correct pose. The most significant differences between the two structures are Phe363, Phe282 and Gln286, which in 1fm6 is rotated to a conformation that in rigid docking would block the terminal phenyl groups of 1293-1 (Figure 3b). Table 5 reports the number of poorly-docked active compounds in the top 10% of the ranked database. For the training set, Glide XP prioritizes most poorly-docked actives for 7 targets of 10. From the total number of poorly-docked actives in the top 10% of the ranked database (as shown in the last row of Table 5a), Glide XP and SP appear to outperform other methods in ranking poorly-docked actives.
Figure 3.
An example of misdocked ligand due to steric clashes, (a) Misdocked pose (carbon atoms are colored in grey) generated by Glide XP for ligand 1293-1 docking into 1fm6 crystal structure (carbon atoms are colored in grey). The “correct pose” (carbon atoms are colored in green) is shown for comparison, (b) Crystal structure of the binding site of 1fm9 (PPARγ open form) with its native ligand 1293-1 (carbon atoms are colored in green) superimposed on the 1fm6 (PPARγ closed form) structure with its native ligand 1241-2 (carbon atoms are colored in grey). Only residues that involve steric clashes with ligand are shown. Hydrogen atoms are not shown. The molecular representations for ligands and proteins are “tube” and “ball and stick”, respectively. The color schemes for elements are Carbon (grey or green), Oxygen (red) and Nitrogen (blue).
Table 5.
Number of poorly-docked active compounds in the top 10% of the ranked database, (see table 2 for the meaning of table headings)
| (a) Glide training set | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pdb ID | # all misdock | XP | SP | DOCK | 1x gold | 1x chem | 8x gold | 8x chem | 1xre chem | 1xre gold | 8xre chem | 8xre gold |
| 1fjs | 5 | 5 | 4 | 1 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| 1hpx | 5 | 5 | 4 | 4 | 4 | 3 | 2 | 0 | 1 | 2 | 1 | 0 |
| 1ett | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| 1aq1 | 4 | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 1bl7 | 9 | 1 | 1 | 0 | 2 | 2 | 2 | 3 | 1 | 2 | 3 | 3 |
| 1ml7 | 11 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1qpe | 34 | 20 | 15 | 2 | 8 | 5 | 5 | 3 | 4 | 3 | 7 | 1 |
| 1rt1 | 6 | 3 | 2 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 2 | 1 |
| 1tmn | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| 3ert | 2 | 1 | 0 | 1 | 1 | 2 | 0 | 2 | 2 | 1 | 2 | 2 |
| Total | 78 | 41 | 32 | 11 | 19 | 15 | 13 | 11 | 9 | 9 | 17 | 8 |
| (b) Glide test set | ||||||||||||
| pdb ID | #all misdock | XP | SP | DOCK | 1x gold | 1x chem | 8x gold | 8x chem | ||||
| 1aq1 | 110 | 7 | 8 | 7 | 19 | 6 | 13 | 5 | ||||
| 1dan | 53 | 29 | 30 | 36 | 31 | 9 | 26 | 9 | ||||
| 1ett | 25 | 10 | 15 | 16 | 15 | 4 | 14 | 3 | ||||
| 1fm6 | 61 | 17 | 22 | 9 | 26 | 34 | 13 | 30 | ||||
| 1fm9 | 68 | 10 | 42 | 6 | 25 | 34 | 21 | 28 | ||||
| 1m4h | 43 | 15 | 19 | 34 | 24 | 8 | 6 | 1 | ||||
| 1y6b | 90 | 13 | 5 | 12 | 7 | 15 | 6 | 12 | ||||
| 1ywn | 85 | 22 | 10 | 12 | 5 | 13 | 5 | 9 | ||||
| Total | 535 | 123 | 151 | 132 | 152 | 123 | 104 | 97 | ||||
For the test set, the advantage displayed by Glide XP in ranking poorly docked actives disappears, and that of Glide SP is significantly diminished. This suggests that the somewhat better ability to recognize partially correct structures may be more dependent upon the fitting data set than the performance of the scoring function for well-docked actives. The principal conclusion is that none of the programs tested performed very well when assessing the ability to rank poorly docked compounds in the top 10% of the ranked database.
V. Conclusion
We have carried out extensive comparisons of several docking programs and scoring functions using a large data set of pharmaceutically interesting targets and active compounds. The Glide XP methodology was shown to consistently yield enrichments superior to the alternative methods, not only for the training set (Table 1a) used to develop XP, but also for the independent test set (Table 1b). Glide SP scoring shows improvement as compared to the scoring in GOLD and DOCK, presumably in part because it has some component of XP scoring mixed in with the more standard terms originally derived from ChemScore (the same starting point as was used to develop GOLD). Most versions of GOLD significantly outperform DOCK on average, although results vary for individual receptors; the various scoring options in GOLD do equally well, with the exception of “rescoring” with GOLD which appears to result in degradation of performance. These conclusions apply to well docked compounds; for misdocked compounds, based on the test set results, all methods perform roughly equally poorly.
From the point of view of computational efficiency, the CPU time required on average for Glide XP calculations (7.0 minutes per ligand) is larger than other methods except the most accurate version of Goldscore (8.5 minutes per ligand). This extra cost for Glide XP is the trade-off for the higher enrichment factors obtained. Glide SP delivers the second best overall enrichment performance while providing a considerable speedup (0.42 minute per ligand) as compared to all approaches with the exception of the fast version of GOLD Chemscore setting.
While the XP scoring function can be improved, the dominant error at this point in screening a large, diverse set of active compounds with a single receptor is clearly going to be misdocking due to steric clashes which arise because the receptors are modeled as rigid structures. If virtual screening is to deliver reliable results, covering a wide range of chemotypes, this problem has to be successfully attacked. There are various approaches that are promising, including docking into multiple receptor structures (illustrated here by the PPARγ and Vegfr2 cases - note that the fractions of compounds misdocked into both closed and open forms of PPARγ receptors and Vegfr2 receptors are quite small) and also using induced fit techniques24. The multiple receptor structures could be obtained from multiple X-ray crystal structures or structurally diverse high quality models generated using a torsion angle sampling tool27. An extensive investigation as to how well these alternative possible solutions work will be necessary in order to make significant progress.
Acknowledgments
This work was supported in part by a grant (GM-30580) from the National Institutes of Heath, and by NIH NRSA fellowships to Zhiyong Zhou (R90 DK071502). We thank Matt Repasky from Schrodinger Inc. for providing the data sets (both training set and test set) of receptors and active ligands.
Footnotes
Supporting Information Available
1K Drug-Like Ligand Decoys Set was created by selecting 1000 ligands from a one million compound library that were chosen to exhibit “drug-like” properties. This decoy set is available at https://www.schrodinger.com/ProductInfo.php?mID=6&sID=6&cID=18
References
- 1.Jones G, Willett P, Glen RC. Molecular Recognition of Receptor-Sites Using a Genetic Algorithm with a Description of Desolvation. Journal of Molecular Biology. 1995;245(1):43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 2.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 3.Nissink JWM, Murray C, Hartshorn M, Verdonk ML, Cole JC, Taylor R. A new test set for validating predictions of protein-ligand interaction. Proteins-Structure Function and Genetics. 2002;49(4):457–471. doi: 10.1002/prot.10232. [DOI] [PubMed] [Google Scholar]
- 4.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins-Structure Function and Genetics. 2003;52(4):609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 5.Kramer B, Rarey M, Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins-Structure Function and Genetics. 1999;37(2):228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 6.Ewing TJA, Kuntz ID. Critical evaluation of search algorithms for automated molecular docking and database screening. Journal of Computational Chemistry. 1997;18(9):1175–1189. [Google Scholar]
- 7.Ewing TJA, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases. Journal of Computer-Aided Molecular Design. 2001;15(5):411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 8.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 9.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 10.Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. Journal of Medicinal Chemistry. 2003;46(4):499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 11.Pham TA, Jain AN. Parameter estimation for scoring protein-ligand interactions using negative training data. Journal of Medicinal Chemistry. 2006;49(20):5856–5868. doi: 10.1021/jm050040j. [DOI] [PubMed] [Google Scholar]
- 12.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. Journal of Molecular Graphics & Modelling. 2003;21(4):289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 13.Kontoyianni M, McClellan LM, Sokol GS. Evaluation of docking performance: Comparative data on docking algorithms. Journal of Medicinal Chemistry. 2004;47(3):558–565. doi: 10.1021/jm0302997. [DOI] [PubMed] [Google Scholar]
- 14.Leach AR, Shoichet BK, Peishoff CE. Prediction of protein-ligand interactions. Docking and scoring: Successes and gaps. Journal of Medicinal Chemistry. 2006;49(20):5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 15.Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins-Structure Function and Bioinformatics. 2004;56(2):235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 16.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. Journal of Medicinal Chemistry. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 17.Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP. Comparison of automated docking programs as virtual screening tools. Journal of Medicinal Chemistry. 2005;48(4):962–976. doi: 10.1021/jm049798d. [DOI] [PubMed] [Google Scholar]
- 18.Bohm HJ. The Development of a Simple Empirical Scoring Function to Estimate the Binding Constant for a Protein Ligand Complex of Known 3-Dimensional Structure. Journal of Computer-Aided Molecular Design. 1994;8(3):243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 19.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions .1. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. Journal of Computer-Aided Molecular Design. 1997;11(5):425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 20.Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. Journal of Molecular Biology. 2000;295(2):337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 21.Muegge I, Martin YC. A general and fast scoring function for protein-ligand interactions: A simplified potential approach. Journal of Medicinal Chemistry. 1999;42(5):791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 22.Wang RX, Lai LH, Wang SM. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. Journal of Computer-Aided Molecular Design. 2002;16(1):11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 23.Wang RX, Lu YP, Wang SM. Comparative evaluation of 11 scoring functions for molecular docking. Journal of Medicinal Chemistry. 2003;46(12):2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 24.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. Journal of Medicinal Chemistry. 2006;49(2):534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 25.MacroModel, version 9.1. Schrodinger, L.L.C.; New York, NY: 2005. [Google Scholar]
- 26.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 27.Knight JL, Zhou ZY. Gallicchio, E., Himmel, D. M., Friesner, R. A., Arnold, E., Levy, R. M., Modeling maximal structural diversity in X-ray crystallographic refinement using Protein Local Optimization by torsion angle sampling. doi: 10.1107/S090744490800070X. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]