Abstract
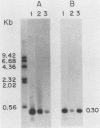
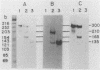
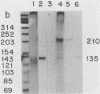
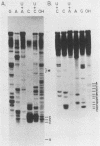
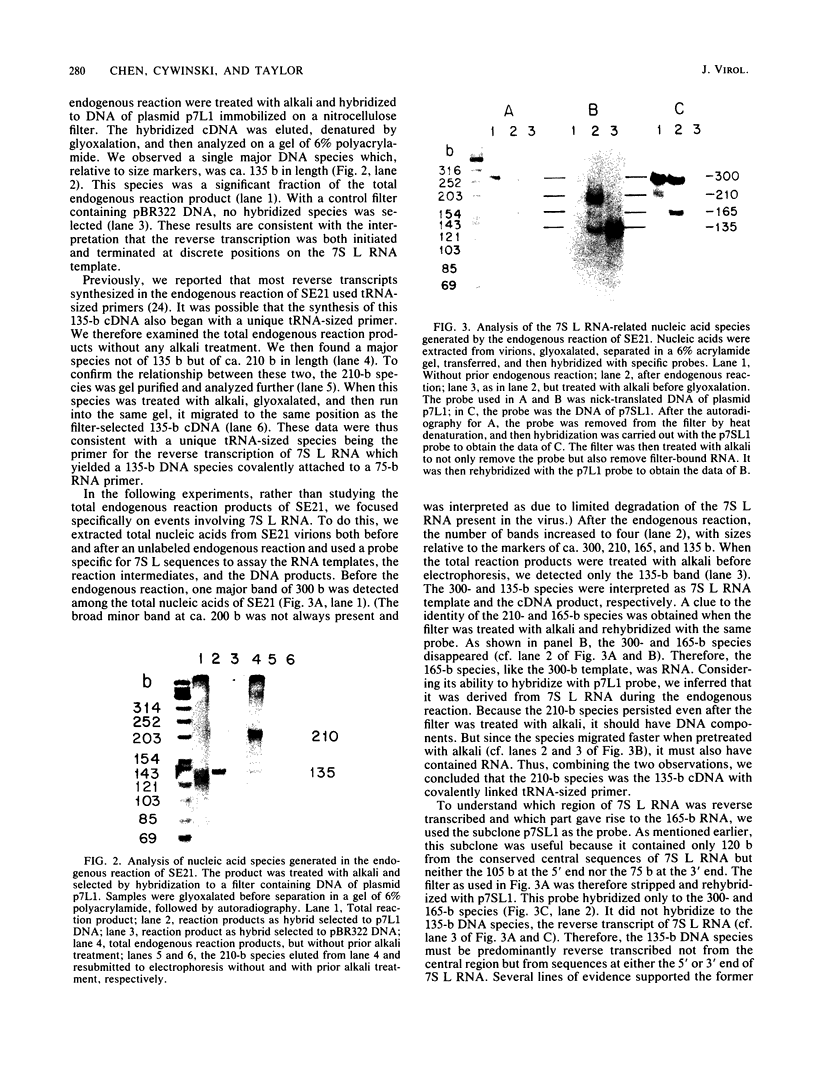
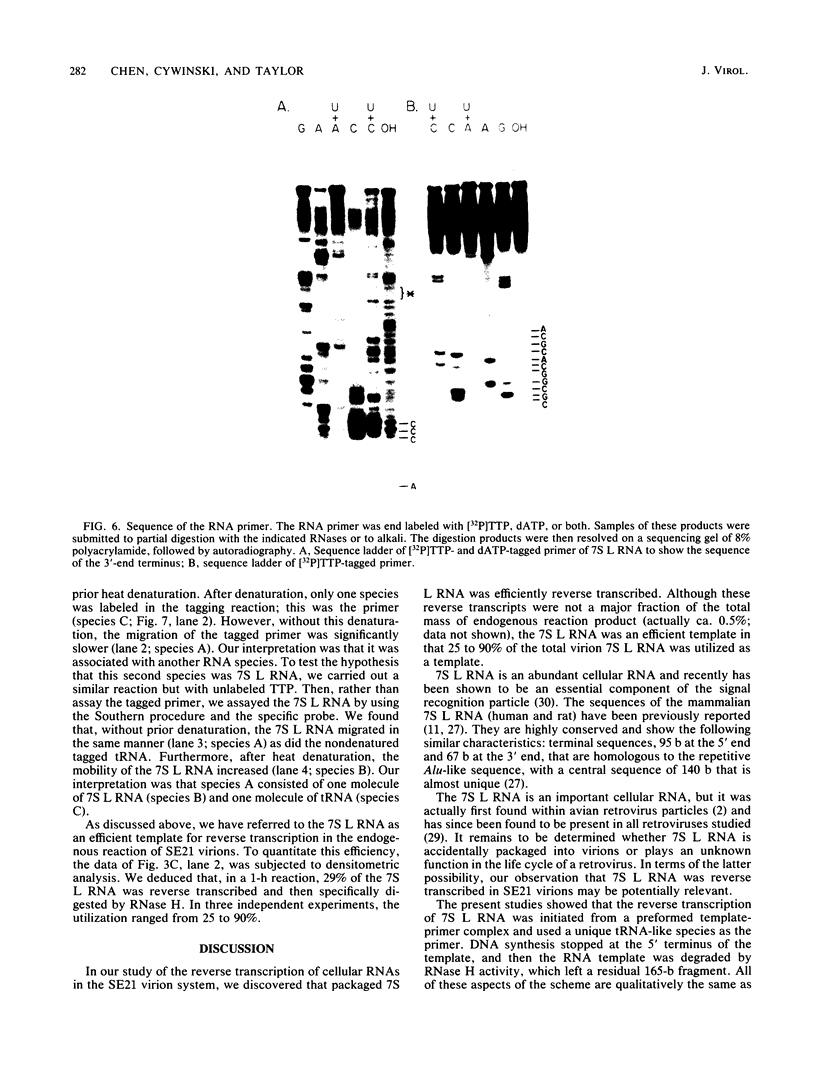
Linial et al. isolated a quail cell line, SE21Q1b, that is transformed by a single integrated provirus of Rous sarcoma virus. Virus particles are released from these cells, but because of a provirus defect, cellular rather than viral RNA is packaged. When these virus particles are disrupted with melittin in the presence of an appropriate reaction mixture containing actinomycin D, there is significant reverse transcription of packaged cellular RNA species. We have shown that (i) cellular 7S L RNA is an efficient template; (ii) initiation is on a unique tRNA-like primer; (iii) synthesis produces a 135-base strong-stop DNA product; and (iv) after synthesis, RNase H acts to remove the 135 bases of the 7S L RNA which acted as the template. A possible facilitator of such specific transcription may be that, in the virus particles but not in the cell, the majority of the 7S L RNA species exist complexed with the tRNA, even before the disruption of the virus. From the size and sequence features of the reverse transcript of 7S L RNA, we speculate that such events may have participated in the process by which animal cell genomes have, in the course of evolution, accumulated multiple copies of Alu-like elements.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Bishop J. M., Levinson W. E., Sullivan D., Fanshier L., Quintrell N., Jackson J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology. 1970 Dec;42(4):927–937. doi: 10.1016/0042-6822(70)90341-7. [DOI] [PubMed] [Google Scholar]
- Cheung K. S., Smith R. E., Stone M. P., Joklik W. K. Comparison of immature (rapid harvest) and mature Rous sarcoma virus particles. Virology. 1972 Dec;50(3):851–864. doi: 10.1016/0042-6822(72)90439-4. [DOI] [PubMed] [Google Scholar]
- Dahlberg J. E., Sawyer R. C., Taylor J. M., Faras A. J., Levinson W. E., Goodman H. M., Bishop J. M. Transcription of DNA from the 70S RNA of Rous sarcoma virus. I. Identification of a specific 4S RNA which serves as primer. J Virol. 1974 May;13(5):1126–1133. doi: 10.1128/jvi.13.5.1126-1133.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A., Dudock B. Methods of RNA sequence analysis. Methods Enzymol. 1983;100:431–453. doi: 10.1016/0076-6879(83)00072-5. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Levy C. C., Karpetsky T. P. The purification and properties of chicken liver RNase: An enzyme which is useful in distinguishing between cytidylic and uridylic acid residues. J Biol Chem. 1980 Mar 10;255(5):2153–2159. [PubMed] [Google Scholar]
- Li W. Y., Reddy R., Henning D., Epstein P., Busch H. Nucleotide sequence of 7 S RNA. Homology to Alu DNA and La 4.5 S RNA. J Biol Chem. 1982 May 10;257(9):5136–5142. [PubMed] [Google Scholar]
- Linial M., Medeiros E., Hayward W. S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell. 1978 Dec;15(4):1371–1381. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerath K., Gupta R. C., Randerath E. 3H and 32P derivative methods for base composition and sequence analysis of RNA. Methods Enzymol. 1980;65(1):638–680. doi: 10.1016/s0076-6879(80)65065-4. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Shank P. R., Linial M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: characterization of a deletion in the provirus. J Virol. 1980 Nov;36(2):450–456. doi: 10.1128/jvi.36.2.450-456.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Conversion of RNA to DNA in mammals: Alu-like elements and pseudogenes. Nature. 1983 Feb 10;301(5900):471–472. doi: 10.1038/301471a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Cordell-Stewart B., Rohde W., Goodman H. M., Bishop J. M. Reassociation of 4 S and 5 S RNA's with the genome of avian sarcoma virus. Virology. 1975 May;65(1):248–259. doi: 10.1016/0042-6822(75)90025-2. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Cywinski A. A defective retrovirus particle (SE21Q1b) packages and reverse transcribes cellular RNA, utilizing tRNA-like primers. J Virol. 1984 Aug;51(2):267–271. doi: 10.1128/jvi.51.2.267-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Esposito V., Melli M. Evolutionary conservation of the human 7 S RNA sequences. J Mol Biol. 1982 Oct 15;161(1):195–201. doi: 10.1016/0022-2836(82)90286-8. [DOI] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]