Abstract
Induction of the fibroblast growth factor-2 (FGF-2) gene and the consequent accumulation of FGF-2 in the nucleus are operative events in mitotic activation and hypertrophy of human astrocytes. In the brain, these events are associated with cellular degeneration and may reflect release of the FGF-2 gene from cell contact inhibition. We used cultures of human astrocytes to examine whether expression of FGF-2 is also controlled by soluble growth factors. Treatment of subconfluent astrocytes with interleukin-1β, epidermal or platelet-derived growth factors, 18-kDa FGF-2, or serum or direct stimulation of protein kinase C (PKC) with phorbol 12-myristate 13-acetate or adenylate cyclase with forskolin increased the levels of 18-, 22-, and 24-kDa FGF-2 isoforms and FGF-2 mRNA. Transfection of FGF-2 promoter–luciferase constructs identified a unique −555/−513 bp growth factor-responsive element (GFRE) that confers high basal promoter activity and activation by growth factors to a downstream promoter region. It also identified a separate region (−624/−556 bp) essential for PKC and cAMP stimulation. DNA–protein binding assays indicated that novel cis-acting elements and trans-acting factors mediate activation of the FGF-2 gene. Southwestern analysis identified 40-, 50-, 60-, and 100-kDa GFRE-binding proteins and 165-, 112-, and 90-kDa proteins that interacted with the PKC/cAMP-responsive region. The GFRE and the element essential for PKC and cAMP stimulation overlap with the region that mediates cell contact inhibition of the FGF-2 promoter. The results show a two-stage regulation of the FGF-2 gene: 1) an initial induction by reduced cell contact, and 2) further activation by growth factors or the PKC-signaling pathway. The hierarchic regulation of the FGF-2 gene promoter by cell density and growth factors or PKC reflects a two-stage activation of protein binding to the GFRE and to the PKC/cAMP-responsive region, respectively.
INTRODUCTION
Central to our understanding of ontogeny and plasticity of adult tissues is the elucidation of molecular mechanisms that control genetic programs for cell proliferation, growth, and differentiation. During development these programs are directed by sequentially expressed growth factors and their receptors (Arenander and de Vellis, 1989; Adamson, 1993). In mature tissues, such as brain, cell growth and proliferation may also be reactivated by an induction of intrinsic growth factor activities. In support of this hypothesis, we found that fibroblast growth factor-2 (FGF-2), a mitogenic and growth-promoting protein (Wagner, 1991; Mason, 1994), is induced in the reactive astrocytes of the adult human brain (Joy et al., 1997). None of the FGF-2 isoforms initiated at different start codons contains a secretory sequence (Florkiewicz and Sommer, 1989). Consequently, FGF-21 is primarily found in both the cytoplasm and the nucleus of expressing cells (Florkiewicz et al., 1991; Powell and Klagsbrun, 1991; Woodward et al., 1992; Puchacz et al., 1993; Stachowiak et al., 1994, 1996a,b). Our studies show that the induction of the FGF-2 gene and the consequent accumulation of FGF-2 protein in the nucleus and the nuclear accumulation of FGF receptor are operative events in mitotic activation and hypertrophy of human glial cells (Moffett et al., 1996; Stachowiak et al., 1996a, 1997; Joy et al., 1997). Given the pleiotropic effects of FGF-2 on human (Joy et al., 1997; Stachowiak et al., 1997b) and nonhuman neural cells (Hatten et al., 1988; Kniss and Burry, 1988; Engele and Bohn, 1992; Mayer et al., 1993; Vescovi et al., 1993, Grothe and Meisinger, 1997), the elucidation of the mechanisms that direct FGF-2 expression in these cells should promote our understanding of the control of neural development and plasticity.
In rat (Finklestein et al., 1988; Frautschy et al., 1991; Gomez-Pinilla et al., 1992; Liu and Chen, 1994) and human brains (Joy et al., 1997), the induction of FGF-2 in reactive astrocytes is associated with cellular degeneration and appears to reflect a release of the FGF-2 gene from cell contact inhibition (Westermann and Unsicker 1993; Moffett et al., 1996; Joy et al., 1997). In the human FGF-2 gene, this induction is mediated partially by the −650- to −512-bp FGF-2 promoter region (Moffett et al., 1996). Growth factors and cytokines secreted by glia, neurons, blood-borne monocytes, and macrophages stimulate reactive transformation of astrocytes (Yeh et al., 1991; Eng et al., 1992; McMillian et al., 1994) and could also play a role in the induction of FGF-2. In cultured neonatal rat astrocytes, these factors increase FGF-2 immunoreactivity (Araujo and Cotman, 1992). However, studies of human astrocytes showed that in confluent cultures, the FGF-2 gene is inactive because of cell contact-induced inhibition. Furthermore, stimulation with serum or growth factors does not restore the expression of nuclear FGF-2 in those cells (Moffett et al., 1996, Stachowiak et al., 1997b) (our unpublished observations).
The present study demonstrates that growth factors and cAMP- and protein kinase C (PKC)-signaling pathways can increase expression of FGF-2 gene products in adult human astrocytes after transition from a confluent to a subconfluent state. We identified a growth factor-responsive element (GFRE) and a separate region essential for the cAMP/PKC stimulation. Both elements contain unique protein-binding sequences. They are located within the FGF-2 gene promoter region, deletion of which attenuates promoter induction by reduced cell contact (Joy et al., 1996; Moffett et al., 1996, Stachowiak et al., 1997). Inactivation of the FGF-2 promoter by cell contact is the primary regulatory mechanism that renders the FGF-2 gene promoter unresponsive to stimulation by growth factors or PKC. Only when released from cell contact inhibition can the FGF-2 promoter be further activated by soluble growth factors and PKC.
MATERIALS AND METHODS
Materials
Culture media were from Life Technologies (Grand Island, NY), epidermal growth factor (EGF) from Upstate Biotechnology (Lake Placid, NY), platelet-derived growth factor (PDGF) from Genzyme (Cambridge, MA), FGF-2 from Boehringer Mannheim (Indianapolis, IN), and interleukin-1β (IL-1β) from Bachem (Torrance, CA). The remaining reagents were from Boehringer Mannheim, Sigma (St. Louis, MO), Bio-Rad (Richmond, CA), Stratagene (La Jolla, CA), and Du Pont New England Nuclear (Boston, MA).
Astrocytic Cell Cultures
Normal human astrocytes were cultured from brain tissue from trauma patients as described by Moffett et al. (1996). The astrocytic strains used in this study were isolated from the frontal lobe (QG culture) and frontal cortex (HG culture) of different individuals. The experiments were also repeated using primary cultures of the embryonal normal human brain astrocyte (NHA) (embryonal astrocytes from Clonetics, San Diego, CA) and produced the same results. All cultured cells expressed glial acidic fibrillary protein but did not express galactocerebroside, an oligodendrocytic marker (Joy et al., 1996). Cultures were routinely grown in Waymouth 87/3 medium (MAB) supplemented with 20% FBS. Treatments with growth factors, IL-1β, 10% FBS, phorbol 12-myristate 13-acetate (PMA), or forskolin were initiated after cells were maintained in medium in which the serum was replaced with 0.25% BSA for at least 24 h. For stimulation with forskolin or PMA, passages lower than 18 were used, because stimulation was reduced at the higher passages.
Western Blot Analysis of FGF-2
To determine total cell content of FGF-2, cells from two 100-mm plates were washed thoroughly in PBS and lysed at 4°C with lysis buffer (1% NP-40, 0.5% deoxycholate, 20 mM Tris, pH 7.5, 5 mM EDTA, 2 mM EGTA, 150 mM NaCl, and protease inhibitors [0.01 mM PMSF, 10 ng/ml aprotinin, 10 ng/ml leupeptin, and 10 ng/ml pepstatin]). The lysates were clarified by centrifugation at 20,000 × g for 15 min. Protein concentrations were determined using the Bio-Rad protein assay. Extracts from different cell preparations were adjusted to contain the same amount of protein and were purified on heparin-Sepharose. Western analysis of FGF-2 was performed using an FGF-2 monoclonal antibody (Upstate Biotechnology) followed by rabbit anti-mouse immunoglobulin G and 125I-labeled protein A as previously described (Puchacz et al., 1993; Stachowiak et al., 1994). Autoradiograms were exposed for different lengths of time to ensure that the signals were in the linear range. The levels of individual FGF-2 isoforms were estimated by densitometric scanning of autoradiograms using a Bio-Rad GS-670 imaging densitometer, and data were analyzed with a Molecular Analyst program (Bio-Rad). In some experiments media combined from two 100-mm dishes were processed and analyzed in a way similar to the cell extracts. No FGF-2 was detected in the media conditioned by astrocytes.
Immunocytochemical Staining for FGF-2
Astrocytes were immunostained for FGF-2 using the same primary monoclonal FGF-2 antibody as for the Western analysis, horseradish peroxidase-conjugated secondary antibody and diaminobenzidine-hydrogen peroxide reaction as previously described (Stachowiak et al., 1994, 1996a; Joy et al., 1997). The specificity of FGF-2 immunostaining was determined by several observations: 1) staining was not observed when primary FGF-2 antibody was omitted or substituted with preimmune serum; 2) similar nuclear and cytoplasmic staining was observed using different polyclonal or monoclonal antibodies and peroxidase or immunofluorescent staining; 3) neutralization of FGF-2 antibody with an excess of 18-kDa FGF-2 reduced FGF-2 staining; 4) FGF-2 immunorecativity was absent in glioma cells that do not express FGF-2 and was induced by transfection of the FGF-2-expressing plasmid; and 5) changes in FGF-2 immunoreactvity were confirmed by Western analysis (Puchacz et al., 1993; Stachowiak et al., 1994, 1996a; Joy et al., 1997).
Treatment of Astrocytes with FGF-2 Antisense and Sense FGF-2 Oligonucleotides
This experiment was performed as described by Stachowiak et al., (1994). Modified (phosphoro-thio-DNA backbone) sense (s-FGF-2) and antisense (as-FGF-2) oligonucleotides were custom synthesized by Bio-Synthesis (Lewisville, TX). Their sequence was identical (s-FGF-2; 5′-GGG ACC AUG GCA GCC-3′) or complementary (as-FGF-2; 5′-GGC TGC CAT GGT CCC-3′) to the 5′ region of human FGF-2 mRNA (Abraham et al., 1986), including the first methionine codon (underlined). Oligomers were purified by gel filtration, ethanol precipitated, lyophilized to dryness, and dissolved in culture medium. Cells were incubated with 2.5 μM oligonucleotides for 48 h.
Quantitative RT-PCR
PCR of reverse-transcribed total RNA (5 μg) was performed using 25-nucleotide (nt) FGF-2 primers as previously described (Moffett et al., 1996). A pair of primers complementary to the mRNA of constitutively expressed human histone gene H3.3 (Pieper et al., 1990; Moffett et al., 1996) was used as a control. The PCR products were electrophoresed on 2% agarose gels, stained with ethidium bromide, and photographed. The length of amplification products for human FGF-2 mRNA was consistent with the predicted length of 179 nt. The identity of RT-PCR–generated FGF-2 cDNA was confirmed by hybridization to a cloned 32P-labeled human FGF-2 cDNA (Abraham et al., 1986; Moffett et al., 1996). The relative abundance of FGF-2 mRNA was determined using two methods that yielded similar results. Photographs of the ethidium bromide-stained FGF-2 and H3.3 cDNAs were scanned using a Bio-Rad imaging densitometer. In addition, PCR-generated cDNA hybridized to 32P-labeled FGF-2 was excised from the nylon membrane, and its radioactivity was determined using a Beckman Instruments (Palo Alto, CA) scintillation counter. In both methods the FGF-2 mRNA content was expressed as the ratio of FGF-2 to H3.3 cDNA. The amount of input RNA in the reverse transcription reaction and the number of PCR cycles used to quantify the cDNA products were in a linear range of PCR (see also Moffett et al., 1996).
Construction of FGF-2Luc Reporter Plasmids and Transfection Assays
We constructed plasmid (−1800/+314)FGF-2Luc (numbers depict nucleotides relative to the transcriptional start site) (Stachowiak et al., 1994) using a −1800/+314-bp fragment of the human FGF-2 gene promoter (Shibata et al., 1991) and promoterless pGL2Basic (Promega, Madison, WI). Deletions of the FGF-2 promoter were produced using Bal31 nuclease and were identified by DNA sequencing (Stachowiak et al., 1994; Moffett et al., 1996). To construct the minimal FGF-2 promoter plasmids (-650/-453)(-103/+314)FGF-2Luc, (-555/-453)(-103/+314)FGF-2Luc, and (-512/-453)(-103/+314)FGF-2Luc, we used the AccI65I–AccI fragments -650/453, -555/-453, and -512/-453 isolated from the (-650/+314)FGF-2Luc, (-555/+314)FGF-2Luc, and (-512/+314)FGF-2Luc, respectively. The fragments were blunt ended with T4 polymerase and cloned into the Acc65I site of the (−103/+314)FGF-2Luc directly upstream from the −103/+314 promoter sequence. Proper orientation of the fragment was confirmed by DNA sequencing. RSVLuc, described by de Wet et al. (1987), expresses constitutively high levels of luciferase from the Rous sarcoma virus (RSV) promoter. The FGF-2Luc plasmids, RSVLuc, or pGL2Basic were transfected into human astrocytes by electroporation or calcium phosphate precipitation as described earlier (Stachowiak et al., 1994; Moffett et al., 1996). Both methods produced the same results, indicating that different transfection procedures did not influence regulation of the transfected FGF-2Luc genes. Transfected cells were plated into 12-well dishes. Twenty-four hours after transfection, cells were rinsed with PBS, and the serum-free medium containing 0.25% BSA was added. After another 24 h, drugs or growth factors were added or 10% FBS was added. Cells were lysed, and an aliquot of extract (10–50 μg of protein) was used to determine luciferase activity (Moffett et al., 1996). Differences in the efficiency of transfection of different plasmids and culture dishes were normalized by measuring the cell content of transfected plasmid DNA by dot blot hybridization in lysates used for the luciferase assay (Goc and Stachowiak, 1994; Stachowiak et al., 1994; Moffett et al., 1996). Luciferase activity was expressed in numbers of light units per picogram of transfected DNA per microgram of cellular protein. This method produces similar results but is more sensitive than cotransfection of the FGF-2Luc plasmid with RSV β-gal and measuring β-galactosidase and luciferase activities in the same extract (Stachowiak et al., 1994). The lack of luciferase stimulation in cells transfected with pGL2Basic or FGF-2 promoter mutants excluded posttranscriptional modification of luciferase activity.
Nuclear Extract Preparation, Electrophoretic Mobility Shift Assay (EMSA), and DNase I Footprinting
Nuclear extracts were prepared as described by Lee et al. (1988), except that buffer A contained 0.75 mM spermidine and 0.15 mM spermine. Double-stranded oligonucleotides, corresponding to different regions of the FGF-2 promoter, were synthesized. Consensus oligonucleotides used had the following sequences: cAMP-responsive element (CRE), 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′; AP2, 5′-GATCGAACTGACCGCCCGCGGCCCGT-3′; AP1, 5′-CGCTTGATGAGTCAGCCGGAA-3′; and NFκβ, 5′-AGTTGAGGGGACTTTCCCAGGC-3′. These oligonucleotides were purchased from Promega Biotech. Signal transducer and activator of transcription (STAT) consensus sequences: 5′-GATCCATTT(CTGG) AAATG-3′ (STAT1/2) and 5′-GATCCATTT(CCCGT)AAATC-3′ (STAT3/4), with different length spacing sequence (N) and STAT-binding specificity, were taken from Seidel et al. (1995).
The oligonucleotides were labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). EMSA was performed according to the method of Sawadogo et al. (1988) as described by Moffett et al. (1996). Nuclear extracts (1–5 μg of protein) were incubated for 10 min at room temperature in 20 μl of 5 mM HEPES (pH 7.8), 50 mM KCl, 1 mM EDTA, 5 mM MgCl2, 10% glycerol, 2 mM DTT, 2 mM PMSF, 2 μg of BSA, and 2 μg of poly(dI-dC). Labeled DNA probe (5 fmol, 2000–5000 cpm) was added, and the reaction was allowed to progress for another 20 min at room temperature. Products from the binding reactions were then resolved on 5% nondenaturing gels in an electrode buffer (pH 7.8) containing 10 mM Tris base, 0.0275 mM EDTA, and 9.24 mM sodium acetate/acetic acid. In several experiments, 0.25% NP-40 was added to the gel and to the binding buffer.
DNase I footprinting (Galas and Schmitz, 1978) was performed as described by Ohlsson and Edlund (1986). A 213-bp Acc65II–AccI fragment, corresponding to bp −650 to −453 of the FGF-2 promoter, was labeled on the coding strand using the Klenow fragment of DNA polymerase I and [α-32P] dNTP. Before the probe was added, 50-μl reactions containing 25 mM HEPES (pH 7.8), 50 mM KCl, 0.05 mM EDTA, 5 mM DTT, 2 μg of salmon sperm DNA, and 5% glycerol were allowed to incubate at room temperature for 10 min. The binding reaction was incubated at room temperature for 20 min before 0.1 μg of DNase I was added for an additional 1 min. One hundred microliters of stop buffer (100 mM Tris-HCl, pH 8.0, 100 mM NaCl, 15 mM EDTA, 300 mM sodium acetate, 200 μg/ml proteinase K, and 100 μg/ml tRNA) were then added, and the reactions were incubated at 37°C for 15 min. DNA was extracted with phenol-chloroform, ethanol precipitated, and resuspended in 80% formamide, 50 mM EDTA, and 0.2% bromophenol blue-xylene cyanol. DNA was heated at 80°C for 15 min and analyzed on sequencing gels with a sequencing ladder used as a marker. DNase I footprinting autoradiographs were scanned with a Beckman LU70 spectrophotometer at 10 readings/mm. The intensity of bands was quantified using peak-crest absorbance.
Southwestern Blot of FGF-2 Promoter-binding Proteins
The assay was performed according to the method of Miskimins et al. (1985). Nuclear extracts were electrophoresed through SDS-polyacrylamide gels in standard gel buffer. The samples were not boiled before loading onto the gels. After electrophoresis, the gels were blotted onto nitrocellulose membranes overnight at 4°C in Tris-glycine buffer (25 mM Tris base and 192 mM glycine). The membranes were then blocked for 1 h at room temperature in preincubation buffer (5% dry milk, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 100 μg of calf thymus DNA). DNA binding was preformed in the same buffer containing 5–6 × 106 cpm of the FGF-2 promoter probes. After a 1-h incubation at room temperature, the blots were washed in DNA-binding buffer containing 300 mM NaCl without calf thymus DNA. The blots were exposed to x-ray film for 24 h.
Statistical and Sequence Analyses
Analysis of variance was used to test the overall statistical significance of differences in luciferase expression. Groups were compared using the least significant difference posthoc test. Direction of effects was inferred from the relation of mean values. The FGF-2 promoter was analyzed for known transcription factor binding sequences using a BLAST program (IntelliGenetics, Mountain View, CA).
RESULTS
Growth Factors Increase Intracellular Content of High- and Low-Molecular-Mass Isoforms of FGF-2 in Subconfluent Human Astrocytes
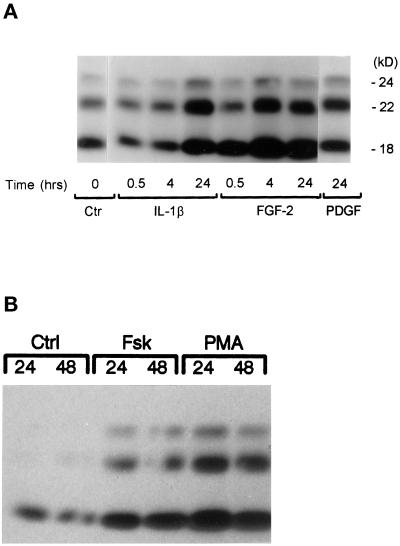
In confluent human astrocytes nuclear FGF-2 protein is depleted, and treatment with serum (Joy et al., 1996; Moffett et al., 1996; Stachowiak et al., 1997b) or growth factors (Stachowiak, Moffett, and Stachowiak, unpublished observations) does not induce FGF-2 expression. To determine whether cell density may affect FGF-2 regulation, cell lysates were prepared from the same astrocytic cultures as previously but maintained in a subconfluent state. Western blots with monoclonal FGF-2 antibodies of serum-free subconfluent astrocytic cultures detected proteins that migrated as three separate bands (Figure 1), consistent with our previous findings (Joy et al., 1997). Their molecular masses (18, 22, and 24 kDa) were similar to those of human FGF-2 proteins generated from alternate use of CUG or AUG translational codons (Florkiewicz and Sommer, 1989; Powell and Klagsbrun, 1991). The ratios of individual isoforms varied slightly between astrocytic cultures obtained from different subjects, but the 18- and 22-kDa isoforms were more abundant than the 24-kDa FGF-2 isoform.
Figure 1.
Effects of growth factors (A and B), forskolin, and PMA (C) on FGF-2 content in human astrocytes: Western analysis. Subconfluent astrocytic cultures were maintained in serum-free medium containing 0.25% BSA for 24 h before the incubation with growth factors, PMA, or forskolin. Total cell lysates were subjected to Western blot analysis as described in MATERIALS AND METHODS. (A and B) Growth factor concentrations used (PDGFAB, 1 × 10−10 M; FGF-2, 5 × 10−10 M; IL-1β, 2.8 × 10−10 M [this Figure]; and EGF, 5.0 × 10−9 M [Figure 2]) are similar to their respective Kd values (Binger et al., 1990; Sorkin et al., 1991; Ban et al., 1993; Stachowiak et al., 1997a) and maximally or near maximally increased or FGF-2 immunoreactivity in rat (Araujo and Cotman, 1992) and human astrocytes and astrocyte proliferation (Joy et al., 1997) (our unpublished observations). Similar results were obtained in two or three independent experiments with different astrocytic cultures. (C) Western analysis of PMA- and forskolin-induced changes in FGF-2 content in human astrocytes. Subconfluent astrocytes were maintained in serum-free medium for 24 h, after which the cells were treated with forskolin (10 μM) or PMA (100 nm) for the indicated times. Control cells were incubated with 0.007% DMSO used as a vehicle for forskolin and PMA. Similar results were obtained in three independent experiments.
Regulation of FGF-2 expression by the high-affinity growth factor receptors was examined by incubating serum-free subconfluent astrocytic cultures with concentrations of growth factors similar to the Kd values of their high-affinity receptors (Figures 1 and 2). Astroglial cells contain cytokine IL-1β receptors (Ban et al., 1993). Furthermore, in the human brain, expression of IL-1β correlates with astrocytosis and is thought to stimulate the formation of scar tissue after brain injury (Da Cunha et al., 1993). Incubation of subconfluent cultures with 2.5 nM IL-1β produced fourfold increases in the content of 24- and 22-kDa FGF-2 and twofold increases in the contents of the 18-kDa isoform (Figure 1A) as revealed by densitometric scanning. Shorter treatments (0.5 or 4 h) with IL-1β had no effect on the content of FGF-2. Similar results were observed in three separate experiments.
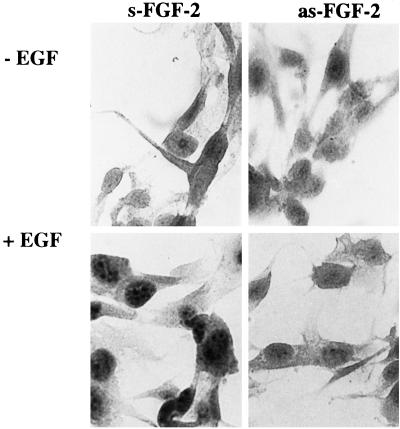
Figure 2.
Effect of antisense FGF-2 oligonucleotide on EGF-induced increase in FGF-2 protein content. Astrocytes were incubated for 48 h with 2.5 μM sense (s-FGF-2) or antisense (as-FGF-2). During the last 24 h, cells were incubated with or without (control) EGF (5.0 × 10−9 M). Cells were stained with monoclonal FGF-2 antibody, and the specificity of immunostaining was demonstrated as described in MATERIALS AND METHODS.
PDGF and EGF expressed in the brain are critical factors controlling the proliferation and differentiation of glial cells and activation of astrocytes during gliosis (Birecree et al., 1991; Yeh et al., 1991; Liu et al., 1994). Incubation of cultured astrocytes with PDGF (Figure 1A) increased the levels of FGF-2 protein. Like IL-1β, PDGF predominately increased the levels of 22- and 24-kDa FGF-2 isoforms. Likewise, the treatment of astrocytes with EGF increased the levels of 18-, 22-, and 24-kDa FGF-2 by 30, 60, and 130%, respectively (the effect of EGF on FGF-2 immunoreactivity in astrocytes is shown in Figure 2).
To determine whether FGF-2 can regulate its own expression in human astrocytes, we incubated subconfluent cultures with 0.5 nM 18-kDa FGF-2. At 30 min, the cellular content of the 18-kDa FGF-2 isoform increased, suggesting an uptake of extracellular FGF-2. Increases in the content of the 22- and 24-kDa FGF-2, however, were observed at 4 h and maintained for at least 24 h (Figure 1A). This delayed accumulation of the high-molecular-mass translational isoforms indicated an increased synthesis of endogenous FGF-2. IL-1β, recombinant 18-kDa FGF-2, or 10% FBS serum increased the levels of all FGF-2 isoforms also in HG astrocytic cultures (our unpublished data).
Expression of FGF-2 Is Increased by Direct Stimulation of cAMP and PKC Signaling Pathways
Growth factors regulate gene expression through multiple signaling molecules, including tyrosine kinases and calcium- and phospholipid-dependent PKC and cAMP-activated protein kinase A (PKA) (Fantl et al., 1993). To determine whether activation of cAMP and PKC pathways increases the levels of FGF-2, we treated astrocytes with adenylate cyclase-stimulating forskolin or with PKC-stimulating PMA. Forskolin at maximally effective 10 μM concentration (shown in Figure 1C) increased the content of all three FGF-2 isoforms. Treatment with 0.1 μM PMA produced larger maximal increases in FGF-2 content than treatment with forskolin.
Increases in FGF-2 mRNA Accompanied Increases in FGF-2 Protein Levels
The forskolin-induced increases in FGF-2 content and nuclear accumulation of FGF-2 were inhibited by cycloheximide or antisense FGF-2 oligonucleotide (Stachowiak et al., 1994). To determine whether delayed elevation of FGF-2 by growth factor also requires de novo FGF-2 synthesis, we specifically inhibited the synthesis of FGF-2 protein by pretreating astrocytes with 15-mer as-FGF-2 or s-FGF-2 (control) oligonucleotides. The as-FGF-2 inhibits induction of FGF-2 in glia (Morrison, 1991) and in adrenal medullary cells (Stachowiak et al., 1994). Two days of incubation with as-FGF-2 had little or no reducing effect on basal FGF-2 immunoreactivity in astrocytes, but it prevented an increase in nuclear and nucleolar FGF-2 content by EGF (Figure 2).
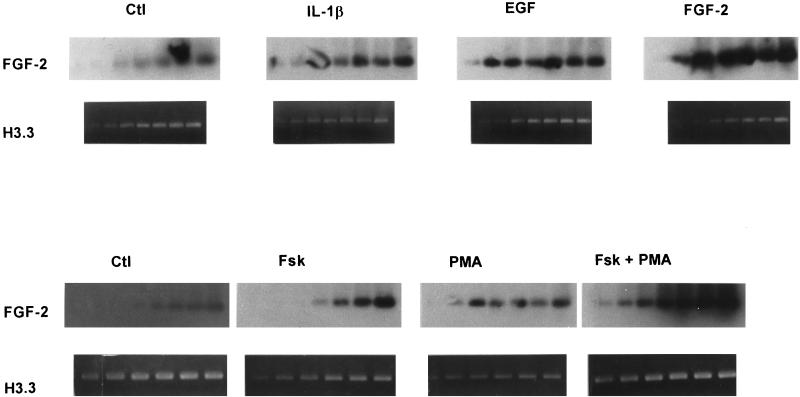
To determine whether the induction of FGF-2 proteins could reflect an induction of FGF-2 mRNA, we analyzed the abundance of FGF-2 mRNA using RT-PCR. Products of these reactions are shown in Figure 3. The amount of reaction products increased with the number cycles used (Figure 3) and the amount of input RNA. Radioactivity counting or densitometric scanning of the reaction products within the linear range of PCR showed that the FGF-2 mRNA increased in astrocytes treated with EGF (5.3-fold), 18-kDa FGF-2 (4.1-fold), forskolin (2.3-fold), or PMA (3.7-fold) when histone H3.3 mRNA levels were used to normalize the data. Treatment with 10% FBS increased FGF-2 mRNA content ∼3-fold. These results indicate that increases in FGF-2 mRNA levels underlie the increases in FGF-2 protein.
Figure 3.
Effects of growth factors, forskolin, or PMA on FGF-2 mRNA levels. Human astrocytic cultures at 40–60% confluence were maintained in serum-free medium for 24 h before they were treated for 24 h with EGF, FGF-2, or IL-1β (A) and with forskolin, PMA, or both (B) at the concentrations given in Figures 1 and 2. Total RNA was isolated and analyzed for FGF-2 and histone H3.3 mRNA levels using RT-PCR. Aliquots of FGF-2 cDNA were taken from cycles 24, 26, 28, 30, 32, 34, and 36 and of H3.3 cDNA from cycles 16, 18, 20, 22, 24, 26, and 28. DNA was electrophoresed through 2% agarose gels and stained with ethidium bromide (dark-field photograph). The FGF-2 cDNA products were also transferred to a nylon membrane and hybridized to 32P-FGF-2 cDNA (light-field autoradiograms) as described in MATERIALS AND METHODS.
Stimulation of Growth Factor Receptors, Adenylate Cyclase, or PKC Activates the FGF-2 Gene Promoter
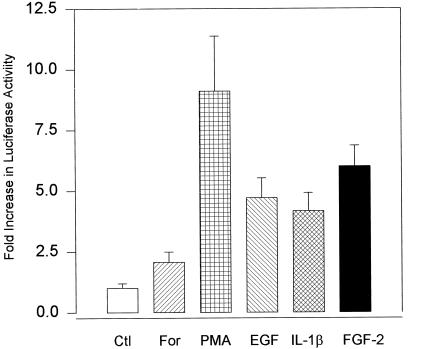
The −1800/+314-bp upstream region of the human FGF-2 gene contains the necessary cis elements to mimic the regulation of the endogenous FGF-2 gene by cell contact in human astrocytes (Moffett et al., 1996) and by second messenger pathways in bovine adrenal medullary cells (Stachowiak et al., 1994). To investigate the molecular mechanisms underlying the induction of FGF-2 mRNA, we initially used a (−1800/+314)FGF-2Luc reporter plasmid (Stachowiak et al., 1994). Luciferase activity (1104 ± 144 cpm · pg DNA−1 · μg protein−1) in frontal lobe QG astrocytes transiently transfected with (−1800/+314)FGF-2 Luc was similar to the activity (1076 ± 158 cpm · pg DNA−1 · μg protein−1) expressed by RSVLuc and >70-fold higher than the minimal activity (15.1 ± 6.1 cpm · pg DNA−1 · μg protein−1) detected in astrocytes transfected with promoterless pGL2Basic.
Treatment with EGF, FGF-2, IL-1β, 10% FBS, or PMA significantly increased the expression of luciferase in astrocytes transfected with (−1800/+314)FGF-2Luc (Figure 4). The magnitude of these increases was similar to the increases in FGF-2 mRNA. The inactive phorbol ester (4-α-phorbol didecanoate), which does not stimulate PKC, had no effect on expression of FGF-2Luc construct (our unpublished observations). The increases in luciferase expression were reproducibly observed in cells treated with forskolin. However, these effects were smaller than the effects of PMA and did not reach statistical significance. Activation of (−1800/+314)FGF-2Luc by growth factors or second messenger stimulators was also observed in the NHA (Figure 5C) and HG astrocytic cultures. These results demonstrate that the −1800/+314-bp fragment of the FGF-2 gene contains the cis elements sufficient for transcriptional regulation by growth factors, PKC, or cAMP in human astrocytes.
Figure 4.
Activation of the FGF-2 gene promoter by growth factors, forskolin, and PMA in astrocyte cultures. Human astrocytes (QG) were transfected with a luciferase reporter plasmid containing a −1800/+314-bp fragment (relative to the transcription start site) of the FGF-2 promoter. Treatment with growth factors, forskolin, or PMA was for 24 h in serum-free medium at the concentrations given in Figures 1 and 2 as described in MATERIALS AND METHODS. Bar graphs represent mean ± SEM from 6–15 samples obtained in two experiments. Analysis of variance showed an overall significant effect of treatments on luciferase expression (p < 0.000002). Luciferase expression was significantly increased in astrocytes treated with PMA (p < 0.00001), FGF-2 (p < 0.0005), IL-1β (p < 0.05), or EGF (p < 0.005).
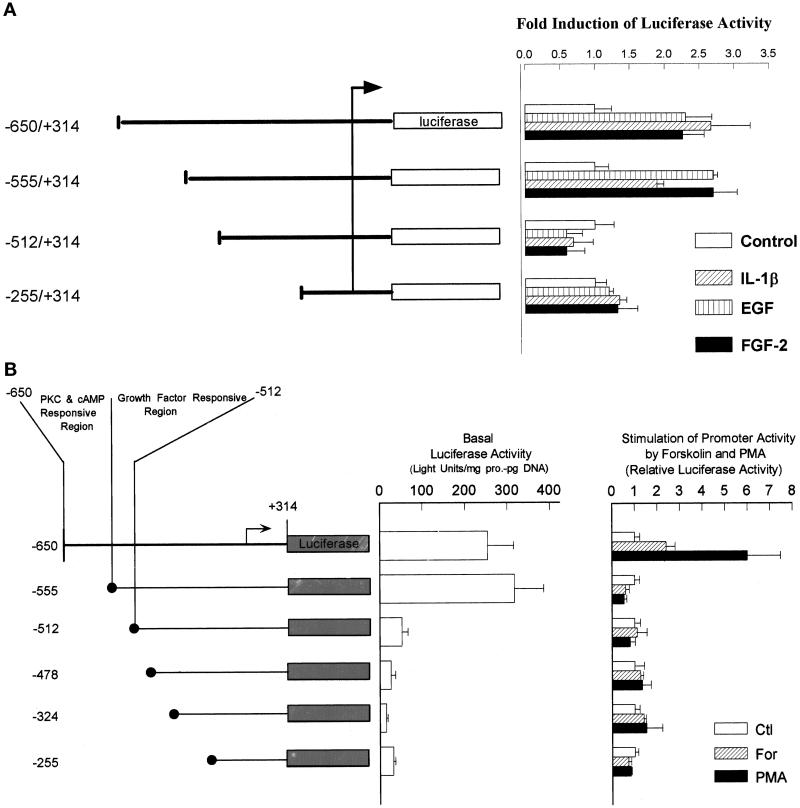
Figure 5.
Identification of the FGF-2 promoter regions responsible for the activation by growth factors, PMA, and forskolin and for the maintenance of basal promoter activity. Astrocytes were transfected with FGF-2 promoter–luciferase constructs given in the figure. The numbers refer to the start and end points of FGF-2 promoter relative to the transcription start site (marked by the arrow). The luciferase activity for each plasmid is expressed relative to the activity in nonstimulated cells. (A) QG astrocytes: activation by growth factors. Bars show mean ± SEM from six to eight samples obtained in two independent experiments except (−255/+314)FGF-2Luc. Each treatment had a statistically significant effect on the expression of (−650/+314)FGF-2Luc (p < 0.05) or (−555/+314)FGF-2Luc (p < 0.01) but not on the expression of (−512/+314)FGF-2Luc or (−255/+314)FGF-2Luc. (B) QG astrocytes: basal promoter activity and activation by forskolin and PMA. Cells were transfected with the FGF-2 promoter–luciferase constructs and incubated in control serum-free medium (basal luciferase expression) or treated with 100 nM PMA or 5 μM forskolin as described in MATERIALS AND METHODS. Basal luciferase activity was calculated as light units per microgram of cellular protein per picogram of transfected DNA. In cells stimulated with forskolin or PMA, the luciferase activity for each plasmid is expressed relative to the activity in nonstimulated cells. Deletion from −555 to −513 bp significantly reduced basal promoter activity (p < 0.005). PMA had a statistically significant effect on the expression of (−650/+314)FGF-2Luc (p < 0.005) but not on the expression of the remaining constructs. Stimulation of (−650/+314)FGF-2Luc by forskolin did not attain a statistically significant level. FGF-2 promoter sequence from −650 to −512 bp is shown. The crossed bar below the sequence indicates GFRE (−555/−512 bp), and the solid bar indicates (−625/−556 bp) essential for PMA and forskolin stimulation. (C) NHA astrocytes: activation of the minimal (−103/+314 bp) FGF-2 promoter by upstream growth factor- and PKC-responsive elements. Astrocytes were transfected with the FGF-2 promoter-luciferase constructs and treated as in A or B. Bars show mean ± SEM of four (PMA) or eight (EGF) samples. EGF had a statistically significant effect on the expression of (−650/+314)FGF-2Luc, (−550/+314)FGF-2Luc, (−650/−453)(−103/+314)FGF-2Luc, and (−555/−453)(−103/+314)FGF-2Luc (p < 0.005). PMA had a statistically significant effect on the expression of (−650/+314)FGF-2Luc and (−650/−453)(−103/+314)FGF-2Luc (p < 0.0001). (D) FGF-2 promoter sequence from −624 to −513 bp is shown. The open bar below the sequence indicates growth the factor-responsive element (−555/−513 bp), and the solid bar indicates the region essential for PKC/cAMP stimulation (−624/−556 bp).
The minimal luciferase activity in cells transfected with pGL2Basic was not affected by treatment of astrocytes with PMA, forskolin, or growth factors. This result and the lack of luciferase stimulation in cells transfected with FGF-2 promoter deletion mutants (see below) exclude posttranscriptional modification of the luciferase activity.
Identification of FGF-2 Promoter Regions Required for Stimulation with Growth Factors, cAMP, and PKC
Earlier experiments showed that basal expression of the FGF-2 gene and its activation by PMA or forskolin are mediated by sequences located upstream from its core promoter (Stachowiak et al., 1994). Consistent with these previous findings, the construct that contained the −255/+314-bp promoter fragment did not express luciferase above the levels of promoterless pGL2Basic (Figure 5B) and showed no induction after growth factor treatment (Figure 5A). Also, the expression of (−512/+314) was not significantly affected by growth factors. In contrast, the (−555/+314)FGF-2Luc showed both growth factor stimulation and an increase in basal expression. The further upstream region (−650/−556) had no additional effect on growth factor stimulation. Thus, the −555/−513-bp region conferred both stimulation by growth factors and high basal expression to the downstream FGF-2 promoter and was named a GFRE.
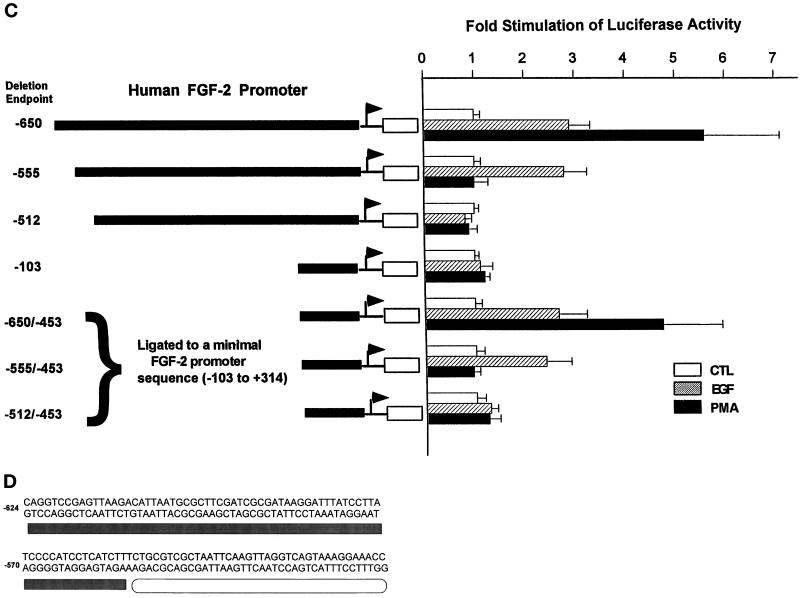
To determine whether cAMP/PKC and growth factors act through converging or independent pathways, we examined the effects of promoter deletions on forskolin and PMA stimulation (Figure 5B). Expression of the (−555/+314)FGF-2Luc construct was not stimulated by forskolin or PMA (Figure 5B), even though it retained the full response to stimulation with growth factors (Figure 5A). When additional −650/−556 bp were included in the promoter–reporter construct [(−650/+314)FGF-2Luc], PMA or forskolin stimulation was restored to a level similar to that in (−1800/+314)FGF-2 Luc (Figure 5B). We recently found that a deletion of the −650/−625-bp promoter fragment did not affect PMA or forskolin stimulation, narrowing the region essential for PKC/cAMP stimulation to −624/−556 bp. This outcome shows that cAMP/PKC and growth factors activate the FGF-2 gene through separate promoter regions.
Earlier studies indicated that the core FGF-2 promoter (−20/+50 bp) was sufficient to support basal expression and to confer p53 regulation to the chloramphenicol acetyltransferase reporter gene in the TE671 cell line (Shibata et al., 1991) and in glioma cells (Ueba et al., 1994), respectively. In the present study, however, a short FGF-2 promoter fragment (−103/+314 bp) was insufficient to respond to growth factor or PMA stimulation (Figure 5C). In contrast, when an upstream promoter region (−650/−453 bp) was ligated directly to the inactive −103/+314 bp minimal promoter [plasmid (−650/−453)(−103/+314)FGF2Luc; Figure 5C], both the EGF and PMA stimulations were restored to levels similar to that of (−650/+314)FGF-2Luc (Figure 5C). Growth factor stimulation was restored by ligation of −555/−453-bp sequence to the minimal promoter [plasmid (−555/−453)(−103/+314)FGF2Luc; Figure 5C], whereas the activation by PMA required a longer promoter fragment, including the sequence essential for PMA and forskolin stimulation [plasmid (−650/−453)(−103/+314)FGF2Luc]. The sequence directly downstream from the GFRE (−512/−453 bp) did not restore EGF or PMA stimulation when ligated to the minimal FGF-2 promoter. These experiments demonstrated that the upstream −650/−453-bp FGF-2 promoter region is both essential and sufficient to confer growth factor and PKC stimulation to the core FGF-2 promoter. They confirmed that the key sequence for the growth factor stimulation was located between −554 and −512 bp, whereas the sequence essential for the PMA activation was located upstream from the GFRE. The sequence between −452 and −103 bp was not required for promoter activation by growth factors or PMA.
Treatment with Growth Factors or PMA Does Not Overcome Inactivation of the FGF-2 Promoter by High Cell Density
In astrocytes the activity of the FGF-2 promoter decreases as the cell density increases from 30 to 70% confluence, reaching 95% inhibition in fully confluent cells (Moffett et al., 1996). Using a deletion analysis, we identified −650 to −513 bp as a region required for the activation of the FGF-2 promoter as astrocytes transit from the confluent to subconfluent state (Moffett et al., 1996). The GFRE (−555/−513 bp) and the sequence required for PMA and forskolin stimulation (−624/−556 bp) (Figure 5) lie within this region. Therefore, we examined whether cell density and growth factors or PMA control FGF-2 promoter activity in an interactive manner. Astrocytes transfected with (−1800/+314)FGF-2Luc or control RSVLuc plasmids were plated at subconfluent or confluent cell densities. The cells were then treated with FGF-2, EGF, or PMA for 24 h, after which luciferase activity was determined. As we have found previously (Moffett et al., 1996), FGF-2 promoter activity in confluent astrocytes was >20-fold lower than in subconfluent astrocytes, whereas the expression of the control RSV promoter was reduced only 2-fold (Table 1). EGF, 18-kDa FGF-2, or PMA did not activate the FGF-2 promoter in astrocytes in high-density cultures. Reducing cell density restored basal expression and the stimulation by growth factors or PMA. Thus the cell contact inhibition is the primary control of the FGF-2 gene promoter activity in human astrocytes. Only after the FGF-2 gene promoter is released from this inhibition does it respond to stimulation by growth factors and PKC. These results are consistent with our earlier findings that serum, growth factors, or PMA cannot induce FGF-2 protein or mRNA in confluent astrocytes (Joy et al., 1996; Moffett et al., 1997; Stachowiak et al., 1997b) (our unpublished observations) but do so in subconfluent astrocytes (present study).
Table 1.
Effect of cell density on stimulation of FGF-2 promoter activity by growth factors and PMA
| Treatment | FGF-2luciferase
|
RSVluciferase
|
||
|---|---|---|---|---|
| Low | High | Low | High | |
| Control | 1.00 ± 0.15 | 0.054 ± 0.01 | 1.0 ± 0.15 | 0.5 ± 0.13 |
| EGF | 2.79 ± 0.57a | 0.050 ± 0.006 | ||
| FGF-2 | 2.70 ± 0.34a | 0.03 ± 0.006 | ||
| PMA | 4.62 ± 1.20a | 0.08 ± 0.04 | 2.6 ± 0.04 | 1.1 ± 0.26 |
Astrocytes were transfected with (−650/+314)FGF-2luciferase or control RSVluciferase constructs as described in MATERIALS AND METHODS. After electroporation, cells were seeded into 12-well dishes at high (100% confluence) or low density (∼30% confluence). Attached cells were washed twice with PBS, and serum-free medium containing 0.25% BSA was added. Twenty-four hours later the cells were treated with EGF (5.0 × 10−9 M), 18-kDa FGF-2 (5 × 10−10 M), or PMA (100 nM) for an additional 24 h. The results are expressed relative to control luciferase activity in low-density cultures. Numbers for FGF-2luciferase represent mean ± SEM of 8–18 samples from two or three independent experiments. Three-way analysis of variance showed a significant interaction between growth factors or PMA and cell density (p < 0.002). Numbers for RSVluciferase from a single representative experiment and are shown for comparison.
Posthoc analysis: difference from control (p < 0.005).
Two-Stage Regulation of Protein Binding to FGF-2 Gene Promoter
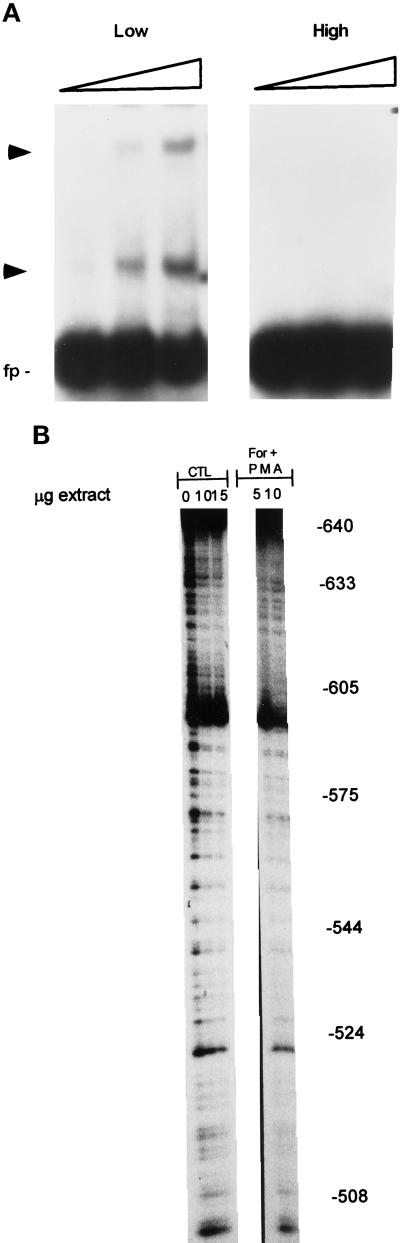
As previously found (Moffett et al., 1996), nuclear proteins isolated from confluent astrocytes did not bind to the −650- to −453-bp region of the FGF-2 promoter even when astrocytes were cultured with 10% FBS (Figure 6A). In contrast, nuclear proteins from subconfluent astrocytic cultures showed binding to the −650/−453 probe (Figure 6A).
Figure 6.
Protein binding to the upstream region of FGF-2 promoter. (A) EMSA of nuclear extracts from astrocytes grown at high and low density. Nuclear extracts were prepared from astrocytes grown at high (3.5 × 104 cells/cm2, 100% confluence) or low (1.2 × 103 cells/cm2, 40–50% confluence) density in 10% FBS, and binding reactions were performed as described in MATERIALS AND METHODS. One, 2.5, or 5 μg of nuclear proteins were used for each culture condition. The target DNA contained nucleotides corresponding to FGF-2 promoter sequences −650 to −512 bp and additional sequences downstream from GFRR (−511 to −453 bp) that do not bind nuclear proteins from subconfluent or confluent astrocytes (Moffett et al., 1996). (B) DNase I footprinting of FGF-2 promoter regions (coding strand) involved in growth factor and second messenger stimulation. Variable amounts of nuclear proteins (0–15 μg) from control astrocytes or astrocytes treated with second messenger stimulators were incubated with 32P-labeled FGF-2 promoter DNA followed by limited digestion with DNase I. The reaction products were then subjected to electrophoresis in 7% sequencing gels. The DNA fragments were visualized using autoradiography. The numbers represent location of bases within the FGF-2 gene promoter. Scanning showed that intensity of bands in protected subregions −624/−600, −575/−556, and −520/−510 nt was reduced by 48, 57, and 80, respectively, when 10 μg of PMA + forskolin extract was compared with control extract. In contrast, cell stimulation with PMA and forskolin had little (−10%) reducing effect on band intensity in the −595/−580-nt subregion and an increasing effect (+60%) in the −630/−625 subregion.
A computer search revealed no obvious homology between the GFRE (Figure 5D) and the target sequences for trans-acting factors published so far, including known growth factor-responsive sequences such as serum-responsive element, CRE, or AP1 binding sites (Boulikas, 1994). To identify target sequences for nuclear factors, we performed DNase I footprinting using reaction conditions established by EMSA for sequence-specific protein binding (see below). Extracts from subconfluent astrocytes caused a partial protection of almost the entire −625/−512-nt promoter sequence indicating that the whole region is engaged in protein binding (Figure 6B). Scanning of the autoradiographs has revealed that treatment with forskolin and PMA enhanced DNA protection by ∼50% within −624/−600- and −575/−556-nt subregions of the PKC/cAMP-responsive region (Figure 6B, legend).
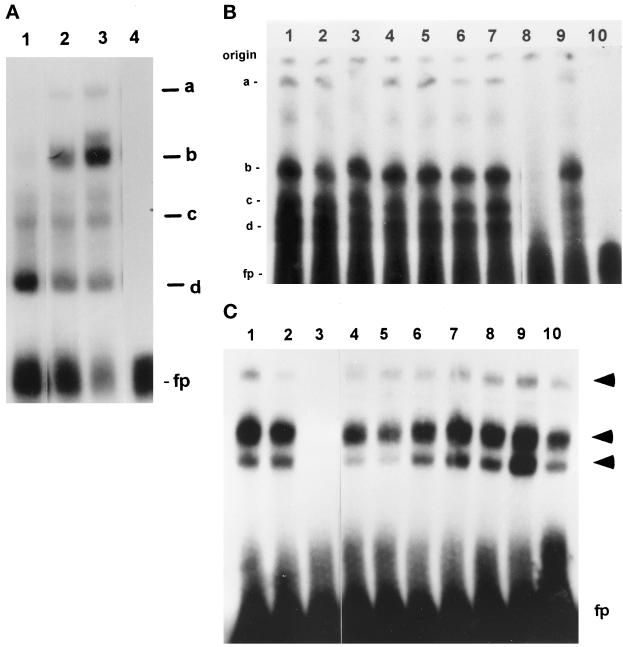
Protein binding to the FGF-2 promoter region essential for the activation by PMA and forskolin was further investigated by EMSA. Nuclear extracts from subconfluent astrocytes formed two complexes, “c” and “d,” with a −624/−555-bp promoter probe (Figure 7A). Treatment of astrocytes with PMA or forskolin reduced formation of complex d and induced two additional, lower-mobility complexes, “a” and “b.” Complexes formed also with shorter −641/−600- and −578/−553-bp probes, consistent with the results of footprinting in which DNase I protection was detected in the −624/−600- and −575/−556-nt subregions (Figure 6B). The −624/−555-bp sequence (Figure 5D) lacks homology to common cAMP- or PKC-responsive sequences: CRE, AP1, AP2-NFκβ, or serum-responsive element (Walton and Rehfuss, 1992; Boulikas, 1994). In addition, protein binding to the −624/−555-bp promoter fragment was competed out by an excess of target DNA but not by oligonucleotides containing target sequences of OCT1, CRE, AP1, AP2, NF[κ]B, or STAT (Figure 7B). Thus, the PKC and cAMP stimulation may not be mediated by common PKC or cAMP trans-activators acting through atypical sequences.
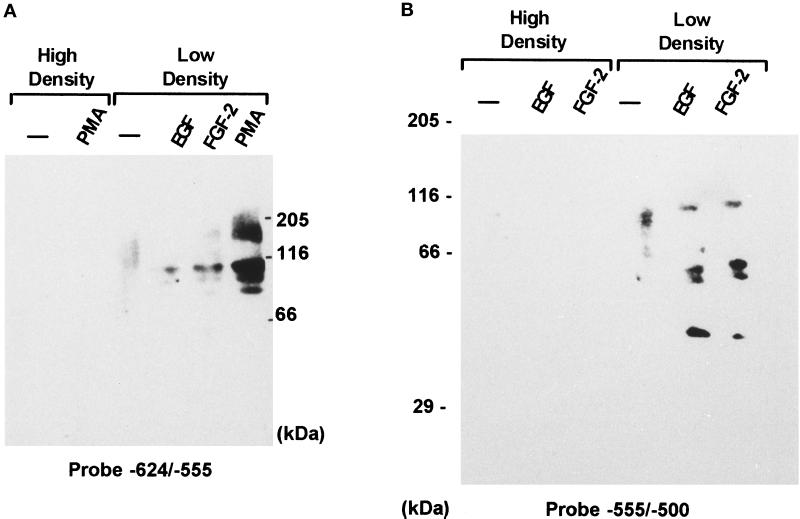
Figure 7.
EMSA of DNA–protein complexes formed with the PKC/cAMP- and growth factor-responsive FGF-2 gene promoter regions. (A) EMSA of DNA–protein complexes formed with the cAMP/PKC-responsive region of the FGF-2 gene promoter. Nuclear proteins (5 μg) were used from subconfluent serum-free astrocytes that were untreated (lane 1) or treated with forskolin (10 μM, 24 h; lane 2) or PMA (100 nM, 24 h; lane 3). A double-stranded oligonucleotide corresponding to −624/−555-bp FGF-2 promoter sequences was used as a target DNA. (a–d) DNA–protein complexes; fp, free probe. (B) Binding to PKC/cAMP-responsive promoter region (−624/−555 bp): competition with unlabeled target DNA and consensus oligonucleotides for several transcription factors. Reactions contained 5 μg of nuclear proteins from PMA-treated astrocytes and a 150-fold molar excess of unlabeled competitor. A 50-fold excess of unlabeled target DNA was sufficient to completely abolish binding. The numbers are for the following competitors: (1) OCT1; (2) CRE; (3) AP1; (4) AP2; (5) NFκB; (6) STAT1/2; (7) STAT3/4; (8) unlabeled −624/−555-bp FGF-2 promoter fragment; (9) no competitor; and (10) no protein extract. (C) Binding to −555/−500-bp promoter region containing GFRE. Nuclear proteins (5 μg) from subconfluent astrocytic cultures treated with 5.0 × 10−9 M EGF for 24 h were used in binding reactions with the −555/−500-bp fragment of the FGF-2 promoter. The target DNA was labeled with [γ-32P]ATP and T4 kinase, and 3000 cpm were used for each reaction as described in MATERIALS AND METHODS. The arrows indicate three DNA–protein complexes; fp, free probe. Binding reactions were carried out with or without unlabeled target DNA or various consensus oligonucleotides for known transcription factors. The numbers are for the following competitors: (1 and 2) no competitor; (3) unlabeled −555/−500 bp FGF-2 promoter fragment; (4) STAT 3/4; (5) STAT Ω; (6) NFκB; (7) AP1; (8) AP2; (9) CRE; and (10) OCT 1. Competitors were used in a 150-fold molar excess. A 50-fold excess of unlabeled target DNA was sufficient to completely abolish binding.
Within the GFRE a strong protection against DNase I was detected between −540 and −524 and between −520 and −510 nt. Scanning of the autoradiographs revealed that protection of the −520/−510-nt subregion was increased by 80% by treatment of astrocytes with forskolin and PMA (Figure 6B) or EGF. In EMSA, nuclear extracts from subconfluent EGF-treated astrocytes formed three complexes with the −555/−500-bp GFRE probe (Figure 7C). Formation of all three complexes was prevented by an excess of unlabeled −555/−500-bp probe DNA (Figure 7C) or its −560/−530- or −533/−509-bp fragments. In contrast, competitor oligonucleotides with unrelated growth factor-responsive sequences did not affect protein binding to GFRE (Figure 7C). The GFRE contains several T/A-rich regions containing sequences similar to STAT binding sites [TT(N)4–6AA] (Seidel et al., 1995; Ihle, 1996). However, in most of these sequences the core spacing (N)8 is 2 bp longer than allowed for STAT binding (Seidel et al., 1995). Binding to GFRE was not competed out by double-stranded oligonucleotides with consensus sequences for different STATs (Figure 7C). Also, preincubation of nuclear extracts with 1–5 μg of antibodies against STAT1–6 did not affect protein binding to either the −555/−500- or the −624/−555-bp region.
To characterize factors that interact with the PKC/cAMP-responsive region, nuclear extracts from confluent astrocytes nonstimulated or treated with PMA were resolved on denaturing SDS-polyacrylamide gels, transferred to nitrocellulose membrane, renatured, and probed with 32P-labeled −624/−555-bp fragment of FGF-2 promoter (Figure 8A). Nuclear proteins from confluent astrocytes did not bind to DNA, regardless of whether the cells were treated with PMA (Figure 8A). The nuclear extracts from subconfluent nonstimulated astrocytes showed only a weak −624/−555-bp binding activity at ∼120 kDa. In extracts from subconfluent PMA-treated astrocytes, the −624/−555-bp probe detected four distinct bands (165, 112, 100, and 90 kDa). They were more abundant than in the nuclei of subconfluent astrocytes treated with EGF or FGF-2. The nuclear proteins that interact with the GFRE were also characterized by Southwestern analysis. A 32P-labeled −555/−500-bp fragment of the FGF-2 promoter did not detect protein binding in nuclear extracts from confluent astrocytes that were untreated or treated with growth factors (Figure 8B). In extracts from subconfluent astrocytes, the GFRE probe detected a doublet of 80- to 90-kDa proteins. Incubation of subconfluent cultures with FGF-2 or EGF induced additional 40-, 50-, and 60-kDa bands and a 100-kDa band that replaced the 80- to 90-kDa doublet. No protein binding was detected with with 32P-labeled −453/−274-bp or −274/+11-bp FGF-2 promoter fragments. Thus binding to −624/−555 bp (Figure 8A) and to −555/−500 bp (Figure 8B) was sequence specific.
Figure 8.
Southwestern analysis of protein binding to PMA/PKC-responsive (A) and growth factor-responsive (B) regions. Thirty-five micrograms of nuclear proteins isolated from confluent (high-density cultures) or subconfluent (low-density) astrocytes incubated in control serum-free medium (−) or treated with 0.5 nM 18-kDa FGF-2, 5 nM EGF, or PMA (100 nM) for 24 h were resolved on SDS-PAGE gels and electroblotted to nitrocellulose membranes. The membranes were probed with 32P-labeled (1 × 106 cpm/ml) −624/−555-bp (A) or −555/−500-bp (B) FGF-2 promoter fragment. The approximate molecular masses of the detected proteins were estimated by comparing with prelabeled protein standards. No protein binding was detected with 32P-labeled −453/−274-bp or −274/+11-bp FGF-2 promoter fragments.
DISCUSSION
Increased Expression of the FGF-2 Gene Underlies Induction of FGF-2 Protein
In the brain, the expression of FGF-2 in reactive astrocytes is associated with cellular degeneration (Joy et al., 1997). Induction of FGF-2 can be reconstituted in vitro in cultured human astrocytes by reducing cell density (Moffett et al., 1996; Joy et al., 1997). Here we report that growth factors, known to be synthesized and secreted by damaged brain tissue, can increase the FGF-2 content in subconfluent human astrocytes. As found previously (Joy et al., 1997), FGF-2 proteins were absent in culture media conditioned by those cells. FGF-2 accumulates predominantly in the nuclei of astrocytic cells (Stachowiak et al., 1996a; Joy et al., 1997), and its nuclear content is increased by growth factors or direct stimulation of cAMP or PKC signaling pathways (Stachowiak et al., 1997b; also see Figure 2). These findings and the presence of FGF-2 receptors in the cell nucleus (Stachowiak et al., 1996a,b, 1997a) support a direct nuclear action of FGF-2 in human glial cells (Stachowiak et al., 1997b).
The up-regulation of FGF-2 mRNA by growth factors and agents that stimulate adenylate cyclase or PKC and the inhibition of nuclear accumulation of FGF-2 by the antisense FGF-2 oligonucleotide (also see Stachowiak et al., 1994) indicate increased FGF-2 synthesis. The increase in the steady-state levels of FGF-2 mRNA in stimulated astrocytes could result from an increase in mRNA stability (Murphy et al., 1990) as well as from increased transcription of the FGF-2 gene (Stachowiak et al., 1994; Moffett et al., 1996). The sensitivity of the nuclear runon assay does not allow a reliable estimation of transcriptional activity of the endogenous FGF-2 gene (Moffett and Stachowiak, unpublished observations). Therefore, we used transfection of FGF-2 promoter–luciferase reporter constructs to demonstrate that the induction of FGF-2 mRNA reflects increased transcription of the FGF-2 gene. The sequences between −650 and +314 bp of the FGF-2 gene were sufficient to support basal FGF-2 promoter activity in human astrocytes and to confer activation by growth factors, PKC, and cAMP, similar to the stimulation of the endogenous FGF-2 gene. An additional 1150 bp upstream from this region (−1800 to −651 bp) did not influence regulation of FGF-2 promoter activity in cultured human astrocytes. So far, studies of the FGF-2 gene have revealed two regulatory regions. One region overlaps with the transcription start site and maps to −20 to +50 bp of the FGF-2 gene (Shibata et al., 1991; Ueba et al., 1994). In the FGF-2 gene, the function of this core promoter is repressed by the wild-type p53 and may be increased by the natural mutant form of p53 in some glioma cells (Ueba et al., 1994). This core promoter has also been implicated in the regulation of the FGF-2 gene by Egr-1 in rat astrocytes (Biesiada et al., 1996). As shown in this study, however, the FGF-2 core promoter is not sufficient to confer stimulation to the FGF-2 gene promoter by growth factors, cAMP, or PKC in human astrocytes. The sequences that are both essential and sufficient to confer the activation by growth factors and PKC to the minimal core FGF-2 promoter in human astrocytes are located within an upstream −650/−453-bp promoter region. The GFRE that confers growth factor stimulation and basal promoter activity to the downstream FGF-2 promoter was mapped between −555 and −512 bp, and the region essential for PKC/cAMP stimulation was mapped at −624/−556 bp. In vitro protein-binding assays demonstrated sequence-specific binding of nuclear proteins both to GFRE and the PKC/cAMP-responsive region. Within the GFRE, nuclear extracts protected almost the entire −555/−512-bp sequence. Proteins are likely to bind to GFRE sites in an interactive manner, because the formation of all protein complexes with the full-length GFRE was prevented by its short fragments and competitors. Hence, mutations of protein-binding GFRE subregions might not reveal how their proteins regulate promoter activity. A more effective approach might be to identify and manipulate the individual GFRE-binding factors. The GFRE lacks homology with known transcription factor target DNA sites, and the astrocytic GFRE-binding proteins do not interact with common growth factor-responsive sequences. Thus, the 40-, 50-, 60-, and 100-kDa proteins detected with the GFRE probe on Southwestern blots may represent novel cytokine trans-activating factors. We have recently isolated two cDNA clones of GFRE-binding proteins from the human brain library. Both clones show no homology to sequences in the gene bank and may represent new proteins (Moffett et al., 1997).
PDGF, FGF-2, and EGF activate receptor tyrosine kinase and IL-1β activates proteins that stimulate nonreceptor tyrosine kinase activity (Fantl et al., 1993; Rossi, 1993). Activation of tyrosine kinases is translated through a cascade of proteins into the activation of serine/threonine kinases, which phosphorylate and activate transcriptional factors. Two such protein kinases, PKA and PKC, are stimulated by a variety of growth factors (Fantl et al., 1993). PMA and forskolin, which bypass growth factor receptors in stimulation of PKC or PKA, respectively, up-regulated FGF-2 protein, mRNA, and the FGF-2 gene promoter. However, the sequence essential for the PKC/cAMP stimulation was located upstream from the GFRE. Thus, the PKC and cAMP signaling pathway and their target proteins, which bind to the −624/−555-bp promoter region, may not be involved in promoter activation by growth factor receptors.
Within the −624/−556-bp sequence essential for the PKC/cAMP stimulation, PMA- and forskolin-stimulated protein binding was detected between −620 and −600 nt. Neither this sequence nor the entire −624/−556-bp promoter region shares homology with known cAMP- or PKC-regulatory elements. Furthermore, proteins that interact with this region do not bind to common cAMP- or PKC-responsive sequences (Figure 7B), and their apparent molecular sizes (165, 112, and 90 kDa; Figure 8A) were larger than known CRE-binding proteins, AP1, or AP2 factors (Walton and Rehfuss, 1992). Thus, identification of the trans-activating factors that interact with the PKC/cAMP-responsive region of the FGF-2 gene promoter and the proteins that interact with the GFRE is now a priority. Given the roles of FGF-2 in the regulation of growth and the cell cycle of glial cells (Kniss and Burry, 1988; Morrison 1991; Joy et al., 1997; Stachowiak et al., 1997), identification of these factors should shed further light on the mechanisms of reactive and neoplastic transformation of human astrocytes and the plasticity of brain tissue.
Two-Stage Regulation of FGF-2 Gene Expression by Cell Contact and Growth Factors or PKC
In confluent astrocytes the FGF-2 gene is inactive because of cell contact-induced inhibition (Moffett et al., 1996; Joy et al., 1997). Even treatment with 10% FBS, EGF, or PMA did not induce nuclear FGF-2 protein or FGF-2 mRNA (Moffett et al., 1996; Joy et al., 1997) (our unpublished observations), although, as shown in the present study, the same treatments increased the contents of FGF-2 protein and mRNA in subconfluent astrocytes (Figures 1–3). Thus, cell density is a gating factor in the activation of FGF-2 expression by growth factors or PKC. The GFRE and the region essential for PMA stimulation are located within a larger FGF-2 promoter region (−650/−512 bp) previously found to mediate promoter regulation by cell contact (Moffett et al., 1996). Consistent with this colocalization, the activity of the FGF-2 gene promoter is subject to a two-tier activation. An initial induction occurs in response to reduced cell–cell contact (Moffett et al., 1996). Only after the FGF-2 gene shifts from an inactive to a basal activity state after the release from cell contact inhibition does it become responsive to further stimulation by growth factors. This hierarchic activation correlates with a two-stage regulation of protein binding to GFRE and the region required for PKC/cAMP simulation. The results of the in vitro protein–DNA binding assays suggest that in contact-inhibited astrocytes neither the GFRE- nor the PKC/cAMP-responsive sequences interact with transcriptional factors.
The 80- and 90-kDa GFRE-binding proteins detected in the nuclei of subconfluent, nonstimulated astrocytes by Southwestern analysis are candidate factors for mediating initial promoter induction. The growth factor stimulation appears to be mediated by inducing a sequence-specific interaction of 40-, 50- to 60-, and 100-kDa proteins with the GFRE, which occurs after the astrocytes are released from contact inhibition. In addition to growth factors, the second-stage activation can be elicited by signals from PKC and cAMP pathways that converge on factors associated with the sequence upstream from the GFRE. However, proteins that interact with the PKC/cAMP-responsive region in subconfluent astrocytes may not contribute to the initial promoter induction, given that its deletion does not affect basal promoter activity in subconfluent astrocytes. The mechanisms through which cell density and growth factors affect protein binding to GFRE are unknown. They could involve changes in protein concentration as well as their ability to interact with the GFRE.
Thus, in brain tissue, the initial stimulus that induces the FGF-2 gene in astrocytes could be a reduction in cell contact associated with cell death or involution caused by physiological changes in cell activity or by pathological degeneration. Subsequently, the FGF-2 gene may be further activated by its own protein or other growth factors and independently by hormones or neurotransmitters that access the cAMP/PKC pathways. The stimulation lasts until a new confluent state is attained. Inactivation of the FGF-2 gene trans-activating factors by cell contact, even when other stimulants may still be present, provides a safety mechanism that seizes FGF-2 expression and cell proliferation after the vacated extracellular space is filled. In contrast, this safety mechanism is disrupted in the neoplastic astrocytes in which nuclear proteins interact with the upstream FGF-2 promoter region independent of cell density, and the FGF-2 gene is constitutively expressed (Moffett et al., 1996; Joy et al., 1997; Stachowiak et al., 1997b). This constitutive expression may promote proliferation that is insensitive to cell contact inhibition.
The two-stage regulation of FGF-2 may not be limited to glial cells. The expression of FGF-2 protein and mRNA in endothelial cells is affected by cell density as in the astrocytes (Yu et al., 1993). Hence, the hierarchic regulation of the FGF-2 gene by cell contact and cytokines may represent a general mechanism through which anchorage signals and signals from soluble molecules are integrated to control cellular growth.
ACKNOWLEDGMENTS
This study was supported by National Science Foundation grant IBN-9728923, National Institutes of Health grant HL-49376-01A1, the March of Dimes, and Arizona Disease Control Research Commission grant 1-209 (to M.K.S.). E.K. was supported by the Undergraduate Biology Research Program at the University of Arizona. J.M. was supported by National Science Foundation Research Experience for Undergraduates (supplement to grant 94-011226). R.Z.F. was supported by National Institutes of Health grant DK-18811.
Abbreviations used:
- as
antisense
- CRE
cAMP-responsive element
- EGF
epidermal growth factor
- EMSA
electrophoretic mobility shift assay
- FGF-2
fibroblast growth factor-2
- GFRE
growth factor-responsive element
- HG
human astrocytic cultures prepared from frontal cortex
- IL-1β
interleukin-1β
- NHA
normal human astrocyte
- nt
nucleotide
- PDGF
platelet-derived growth factor
- PKA
protein kinase A
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- QG
human astrocytic cultures prepared from frontal lobe
- RSV
Rous sarcoma virus
- s
sense
- STAT
signal transducer and activator of transcription
REFERENCES
- Abraham JA, Whang JL, Tunolo A, Mergia A, Friedman J, Gospodarowicz D, Fiddes JC. Human basic fibroblast growth factor: nucleotide sequence and genomic organization. EMBO J. 1986;5:2523–2528. doi: 10.1002/j.1460-2075.1986.tb04530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson ED. Growth factors and their receptors in development. Dev Genet. 1993;14:159–164. doi: 10.1002/dvg.1020140302. [DOI] [PubMed] [Google Scholar]
- Araujo DM, Cotman CW. bFGF in astroglial, microglial, and neuronal cultures: characterization of binding sites and modulation of release by lymphokines and trophic factors. J Neurosci. 1992;12:1668–178. doi: 10.1523/JNEUROSCI.12-05-01668.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenander AT, de Vellis J. Development of the nervous system. In: Siegel GJ, Agranoff BW, Albers RW, editors. Basic Neurochemistry: Molecular, Cellular, and Medical Aspects. 4th ed. P.B. Molinoff, New York: Raven Press; 1989. pp. 479–506. [Google Scholar]
- Ban EM, Sarlieve LL, Haour FG. Interleukin-1 binding sites on astrocytes. Neuroscience. 1993;52:725–733. doi: 10.1016/0306-4522(93)90421-b. [DOI] [PubMed] [Google Scholar]
- Biesiada E, Razanadi M, Levin ER. Egr-1 activates basic fibroblast growth factor transcription. J Biol Chem, 1996;271:18576–18581. doi: 10.1074/jbc.271.31.18576. [DOI] [PubMed] [Google Scholar]
- Binger SH, Humphrey PA, Wong AJ, Vogelstein B, Mark J, Friedman HS, Binger DD. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenographs. Cancer Res. 1990;50:8017–8022. [PubMed] [Google Scholar]
- Birecree E, King LE, Jr, Nanney LB. Epidermal growth factor and its receptors in the developing human nervous system. Brain Res. 1991;60:145–154. doi: 10.1016/0165-3806(91)90043-i. [DOI] [PubMed] [Google Scholar]
- Boulikas T. A critical compilation of DNA binding sites for protein transcription factors from vertebrates. Crit Rev Gene Express. 1994;4:117–321. doi: 10.1615/critreveukargeneexpr.v4.i2-3.10. [DOI] [PubMed] [Google Scholar]
- Da Cunha A, Jefferson JJ, Tyor WR, Glass JD, Jannotta FS, Vitkovic L. Control of astrocytosis by interleukin-1 and transforming growth factor-beta 1 in human brain. Brain Res. 1993;631:39–45. doi: 10.1016/0006-8993(93)91183-s. [DOI] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski ER, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng LF, Yu ACH, Lee YL. Astrocytic response to injury. Prog Brain Res. 1992;94:353–365. doi: 10.1016/s0079-6123(08)61764-1. [DOI] [PubMed] [Google Scholar]
- Engele J, Bohn MC. Effects of acidic and basic FGF on glial precursor cell proliferation: age dependency and region specificity. Dev Biol. 1992;152:363–372. doi: 10.1016/0012-1606(92)90143-5. [DOI] [PubMed] [Google Scholar]
- Fantl WJ, Johnson DE, Williams LT. Signaling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- Finklestein SP, Apostoloides PJ, Caday CG, Prosser J, Philips MF, Klagsbrun M. Increased bFGF immunoreactivity at the site of focal brain wounds. Brain Res. 1988;460:253–259. doi: 10.1016/0006-8993(88)90370-8. [DOI] [PubMed] [Google Scholar]
- Florkiewicz RZ, Baird A, Gonzalez MA. Multiple forms of bFGF: differential nuclear and cell surface localization. Growth Factors. 1991;4:265–275. doi: 10.3109/08977199109043912. [DOI] [PubMed] [Google Scholar]
- Florkiewicz RZ, Sommer A. Human basic fibroblast growth factor gene encodes four peptides: three initiate translation from non-AUG codons. Proc Natl Acad Sci USA. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frautschy SA, Walicke P, Baird A. Localization of basic fibroblast growth factor and its mRNA after CNS injury. Brain Res. 1991;553:291–299. doi: 10.1016/0006-8993(91)90837-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas DJ, Schmitz A. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3151–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goc A, Stachowiak MK. Bovine tyrosine hydroxylase gene promoter regions involved in basal and angiotensin II-stimulated expression in nontransformed adrenal medullary cells. J Neurochem. 1994;62:834–843. doi: 10.1046/j.1471-4159.1994.62030834.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Lee JWK, Cotman CW. Basic FGF in adult rat brain: cellular distribution and response to ethorhinal lesion and fimbria-fornix transection. J Neurosci. 1992;12:345–355. doi: 10.1523/JNEUROSCI.12-01-00345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe C, Meisinger C. The multifunctionality of FGF-2 in the adrenal medulla. Anat Embryol. 1997;195:103–111. doi: 10.1007/s004290050029. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Lynch M, Rydel RE, Sanchez J, Joseph-Silverstein J, Mosactelli D, Rifkin DB. In vitro neurite extension by granule neurons is dependent upon astroglia-derived fibroblast growth factors. Dev Biol. 1988;125:280–289. doi: 10.1016/0012-1606(88)90211-4. [DOI] [PubMed] [Google Scholar]
- Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- Joy A, Moffett J, Neary K, Shapiro J, Coons S, Mordechai E, Stachowiak EK, Stachowiak MK. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene. 1997;14:171–183. doi: 10.1038/sj.onc.1200823. [DOI] [PubMed] [Google Scholar]
- Kniss DA, Burry RW. Serum and basic fibroblast growth factor stimulates quiescent astrocytes to re-enter the cell cycle. Brain Res. 1988;439:281–288. doi: 10.1016/0006-8993(88)91485-0. [DOI] [PubMed] [Google Scholar]
- Lee KAW, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- Liu HM, Chen HH. Correlation between fibroblast growth factor expression and cell proliferation in experimental brain infarct: studied with proliferating cell nuclear antigen immunohistochemistry. J Neuropathol Expt Neurol. 1994;53:118–126. doi: 10.1097/00005072-199403000-00002. [DOI] [PubMed] [Google Scholar]
- Liu HM, Yang HB, Chen RM. Expression of basic fibroblast growth factor, nerve growth factor, platelet-derived growth factor and transforming growth factor-beta in human brain abscess. Acta Neuropathol. 1994;88:143–150. doi: 10.1007/BF00294507. [DOI] [PubMed] [Google Scholar]
- Mason IJ. The ins and outs of fibroblast growth factors. Cell. 1994;78:547–552. doi: 10.1016/0092-8674(94)90520-7. [DOI] [PubMed] [Google Scholar]
- Mayer E, Dunnett SB, Fawcett JW. Mitogenic effect of basic fibroblast growth factor on embryonic ventral mesencephalic dopamine neuron precursors. Dev Brain Res. 1993;72:253–258. doi: 10.1016/0165-3806(93)90190-l. [DOI] [PubMed] [Google Scholar]
- McMillian MK, Thai L, Hong J-S, O’Callaghan PO, Pennypacker KR. Brain injury in a dish: a model for reactive gliosis. Trends Neurosci. 1994;17:138–142. doi: 10.1016/0166-2236(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Miskimins WK, Roberts MP, McCleland A, Ruddle FH. Use of a protein blotting procedure and specific DNA probe to identify nuclear proteins that recognize the promoter region of the transferin receptor gene. Proc Natl Acad Sci USA. 1985;82:6741–6744. doi: 10.1073/pnas.82.20.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett J, Kratz E, Florkiewicz R, Stachowiak MK. Promoter regions involved in density-dependent regulation of basic fibroblast growth factor gene expression in human astrocytic cells. Proc Natl Acad Sci USA. 1996;93:2470–2475. doi: 10.1073/pnas.93.6.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett J, Yee C, Myers J, Shapiro W, Stachowiak MK. Cloning and characterization of trans-acting factors that regulate FGF-2 gene expression in human astrocytes. Soc Neurosci Abstr. 1997;23:1707. [Google Scholar]
- Morrison RS. Suppression of basic fibroblast growth factor expression by antisense oligonucleotides inhibits the growth of transformed human astrocytes. J Biol Chem. 1991;263:728–734. [PubMed] [Google Scholar]
- Murphy PR, Guo JZ, Friesen HG. Messenger RNA stabilization accounts for elevated basic fibroblast growth factor transcript levels in a human astrocytoma cell line. Mol Endocrinol. 1990;4:196–201. doi: 10.1210/mend-4-2-196. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986;45:35–33. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Phi-Van L, Sartling WH. Association of DNA with nuclear matrix. Prog Mol Subcell Biol. 1990;11:1–11. [Google Scholar]
- Pieper RO, Futscher BW, Dong Q, Ellis TM, Erickson LC. Comparison of O-6-methylguanine DNA methyltransferase (MGMT) mRNA levels in Mer+ and Mer− human tumor cell lines containing the MGMT gene by polymerase chain reaction technique. Cancer Comm. 1990;2:13–20. doi: 10.3727/095535490820874812. [DOI] [PubMed] [Google Scholar]
- Powell PP, Klagsbrun M. Three forms of rat basic fibroblast growth factor are made from a single mRNA and localize to the nucleus. J Cell Physiol. 1991;148:202–210. doi: 10.1002/jcp.1041480204. [DOI] [PubMed] [Google Scholar]
- Puchacz E, Stachowiak EK, Florkiewicz RZ, Lukas RJ, Stachowiak MK. Basic fibroblast growth factor regulates tyrosine hydroxylase and proenkephalin mRNA levels in adrenal chromaffin cells. Brain Res. 1993;610:39–52. doi: 10.1016/0006-8993(93)91214-d. [DOI] [PubMed] [Google Scholar]
- Rossi B. IL-1 transduction signals. Eur Cytokine Network. 1993;4:181–187. [PubMed] [Google Scholar]
- Sawadogo M, Van Dyke MW, Gregor PD, Roeder RG. Multiple forms of the human gene-specific transcription factor-USF. I. Complete purification and identification of USF from HeLa cell nuclei. J Biol Chem. 1988;263:11985–11993. [PubMed] [Google Scholar]
- Shibata F, Baird A, Florkiewicz RZ. Functional characterization of the human basic fibroblast growth factor gene promoter. Growth Factors. 1991;4:277–287. doi: 10.3109/08977199109043913. [DOI] [PubMed] [Google Scholar]
- Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Westermark B, Heldin C-H, Claesson-Welsh L. Effect of receptor kinase inactivation on the rate of internalization and degradation of PDGF and the PDGF β-Receptor. J Cell Biol. 1991;112:469–478. doi: 10.1083/jcb.112.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak EK, Maher PA, Tucholski J, Mordechai E, Joy A, Moffett J, Coons S, Stachowiak MK. Nuclear accumulation of fibroblast growth factor receptors in human glial cells—association with cell proliferation. Oncogene. 1997a;14:2201–2211. doi: 10.1038/sj.onc.1201057. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK. Nuclear localization of functional FGF receptor-1 in human astrocytes suggests a novel mechanisms for growth factor action. Mol Brain Res. 1996a;38:161–165. doi: 10.1016/0169-328x(96)00010-1. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK. Nuclear accumulation of fibroblast growth factor receptors is regulated by multiple signals in adrenal medullary cells. Mol Biol Cell. 1996b;7:1299–1317. doi: 10.1091/mbc.7.8.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MK, Moffett J, Joy A, Puchacz E, Florkiewicz RZ, Stachowiak EK. Regulation of bFGF gene expression and subcellular distribution of bFGF protein in adrenal medullary cells. J Cell Biol. 1994;127:203–223. doi: 10.1083/jcb.127.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MK, Moffett J, Maher PA, Tucholski J, Stachowiak EK. Growth factor regulation of cell growth and proliferation in the nervous system—a new intracrine nuclear mechanism. Mol Neurobiol. 1997b;15:257–284. doi: 10.1007/BF02740663. [DOI] [PubMed] [Google Scholar]
- Ueba T, Nosaka T, Takahashi JA, Shibata F, Florkiewicz RZ, Vogelstein B, Oda Y, Kikuchi H, Hatanaka M. Transcriptional regulation of basic fibroblast growth factor gene by p53 in human glioblastoma and hepatocellular carcinoma cells. Proc Natl Acad Sci USA. 1994;91:9009–9013. doi: 10.1073/pnas.91.19.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats AC. Alternative translation of human fibroblast growth factor-2 occurs by internal entry of ribosomes. Mol Cell Biol. 1995;18:35–33. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovi AL, Reynolds BA, Fraser DD, Weiss S. bFGF regulates the proliferative fate of unipotent (neuronal) and bipotent (neuronal/astroglial) EGF-generated CNS progenitor cells. Neuron. 1993;11:951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- Wagner JA. The fibroblast growth factors: an emerging family of neural growth factors. Curr Top Microbiol Immunol. 1991;169:95–118. doi: 10.1007/978-3-642-75747-1_6. [DOI] [PubMed] [Google Scholar]
- Walton KM, Rehfuss RP. Molecular mechanisms of cAMP-regulated gene expression. Mol Neurobiol. 1992;4:197–210. doi: 10.1007/BF02780341. [DOI] [PubMed] [Google Scholar]
- Westermann R, Unsicker K. Basic fibroblast growth factor (FGF-2) and rat C6 glioma cells: regulation of expression, absence of release, and response to exogenous FGF-2. Glia. 1993;3:510–521. doi: 10.1002/glia.440030610. [DOI] [PubMed] [Google Scholar]
- Woodward WR, Nishi R, Meshul CK, Williams TE, Coulombe M, Eckstein FP. Nuclear and cytoplasmic localization of basic fibroblast growth factor in astrocytes and CA2 hippocampal neurons. J Neurosci. 1992;12:142–152. doi: 10.1523/JNEUROSCI.12-01-00142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JHJ, Ruit KG, Wang YX, Parks WC, Snider WD, Deuel TF. PDGF A-chain gene is expressed by mammalian neurons during development and in maturity. Cell. 1991;64:209–216. doi: 10.1016/0092-8674(91)90222-k. [DOI] [PubMed] [Google Scholar]
- Yu Z-X, Biro S, Fu Y-M, Sanchez J, Smale G, Sasse J, Ferrans VJ, Cassells W. Localization of basic fibroblast growth factor in bovine endothelial cells: immunohistochemical and biochemical studies. Exp Cell Res. 1993;204:247–259. doi: 10.1006/excr.1993.1031. [DOI] [PubMed] [Google Scholar]