Abstract
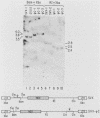
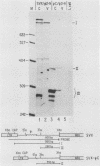
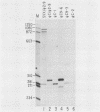
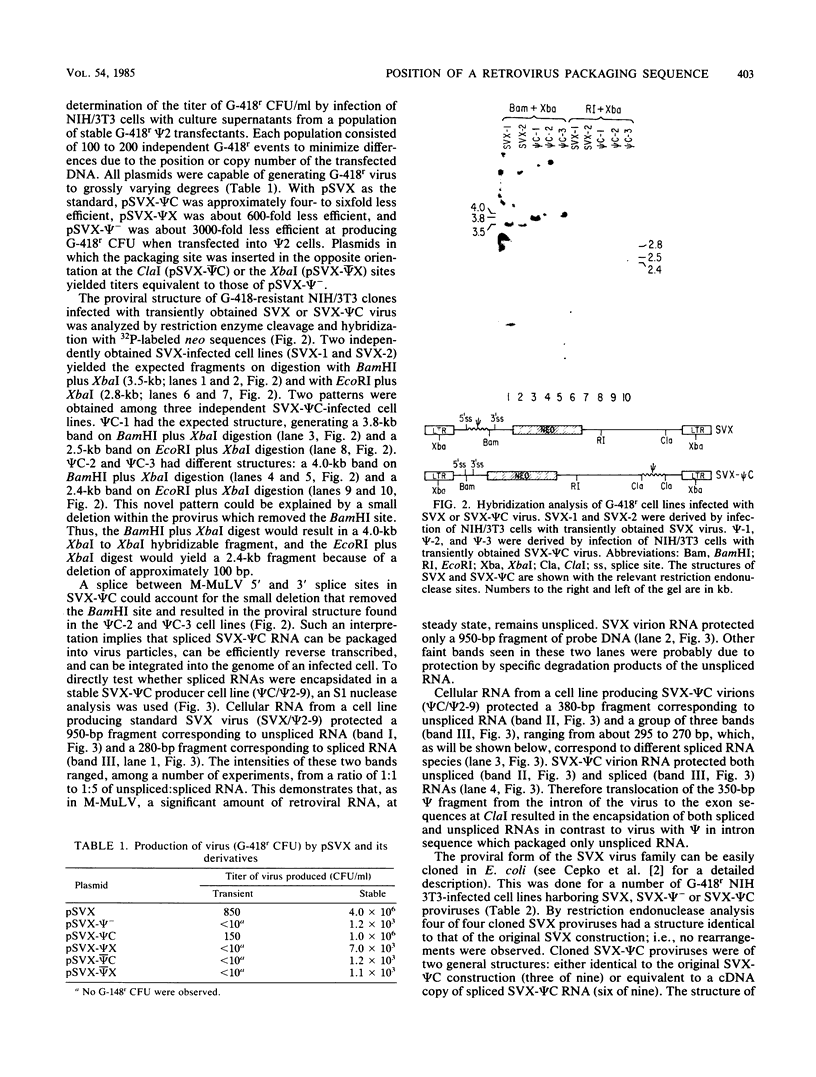
By using a retroviral construct derived from Moloney murine leukemia virus and capable of expressing the dominant selectable neo gene, we measured the effects of moving or deleting a sequence (psi) known to be required in cis for the packaging of genomic RNA into virus particles. When psi was at its wild-type position (in SVX virus) near the 5' end of the RNA, the titer of infectious virus production was 5 X 10(6) G-418-resistant CFU per ml. The titer was decreased approximately fivefold when psi was moved, in its proper orientation, to near the 3' end of the virus (SVX-psi C) and was decreased approximately 600-fold when psi was moved, in its proper orientation, into the U3 region of the long terminal repeat. When psi was deleted (SVX-psi-) or inserted in the opposite orientation at either of these two positions, the titer was decreased by 3000-fold relative to SVX. In SVX-psi C, psi was no longer in the intron (as it is in SVX and Moloney murine leukemia virus) but was moved to a region which is only exonic. This resulted in the encapsidation of both spliced and unspliced RNAs, their efficient reverse transcription, and their integration into the genome of an infected cell. A number of proviruses resulting from integration of either spliced or unspliced RNAs were cloned. Four of these clones were subjected to sequence analysis in the region of the splice sites, and it was determined which sites are used by these viruses and also which are used by Moloney murine leukemia virus.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benz E. W., Jr, Wydro R. M., Nadal-Ginard B., Dina D. Moloney murine sarcoma proviral DNA is a transcriptional unit. Nature. 1980 Dec 25;288(5792):665–669. doi: 10.1038/288665a0. [DOI] [PubMed] [Google Scholar]
- Cepko C. L., Roberts B. E., Mulligan R. C. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984 Jul;37(3):1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chumakov I., Stuhlmann H., Harbers K., Jaenisch R. Cloning of two genetically transmitted Moloney leukemia proviral genomes: correlation between biological activity of the cloned DNA and viral genome activation in the animal. J Virol. 1982 Jun;42(3):1088–1098. doi: 10.1128/jvi.42.3.1088-1098.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbère-Garapin F., Horodniceanu F., Kourilsky P., Garapin A. C. A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol. 1981 Jul 25;150(1):1–14. doi: 10.1016/0022-2836(81)90321-1. [DOI] [PubMed] [Google Scholar]
- Cone R. D., Mulligan R. C. High-efficiency gene transfer into mammalian cells: generation of helper-free recombinant retrovirus with broad mammalian host range. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6349–6353. doi: 10.1073/pnas.81.20.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell B., Stavnezer E., Friedrich R., Bishop J. M., Goodman H. M. Nucleotide sequence that binds primer for DNA synthesis to the avian sarcoma virus genome. J Virol. 1976 Aug;19(2):548–558. doi: 10.1128/jvi.19.2.548-558.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Jimenez A. A new selective agent for eukaryotic cloning vectors. Am J Trop Med Hyg. 1980 Sep;29(5 Suppl):1089–1092. doi: 10.4269/ajtmh.1980.29.1089. [DOI] [PubMed] [Google Scholar]
- Fan H., Verma I. M. Size analysis and relationship of murine leukemia virus-specific mRNA's: evidence for transposition of sequences during synthesis and processing of subgenomic mRNA. J Virol. 1978 May;26(2):468–478. doi: 10.1128/jvi.26.2.468-478.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Harada F., Dahlberg J. E., Panet A., Haseltine W. A., Baltimore D. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J Virol. 1977 Mar;21(3):1031–1041. doi: 10.1128/jvi.21.3.1031-1041.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugatsch T., Stacey D. W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983 Jul 30;128(2):505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E., Donoghue D. J., Baltimore D. Analysis of a 5' leader sequence on murine leukemia virus 21S RNA: heteroduplex mapping with long reverse transcriptase products. Cell. 1978 Mar;13(3):435–451. doi: 10.1016/0092-8674(78)90318-5. [DOI] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J Virol. 1983 May;46(2):538–546. doi: 10.1128/jvi.46.2.538-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Shields A., Witte W. N., Rothenberg E., Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978 Jul;14(3):601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- Sorge J., Ricci W., Hughes S. H. cis-Acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J Virol. 1983 Dec;48(3):667–675. doi: 10.1128/jvi.48.3.667-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge J., Wright D., Erdman V. D., Cutting A. E. Amphotropic retrovirus vector system for human cell gene transfer. Mol Cell Biol. 1984 Sep;4(9):1730–1737. doi: 10.1128/mcb.4.9.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5' long terminal repeat and the start of the gag gene. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]