Abstract
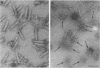
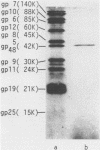
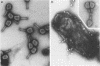
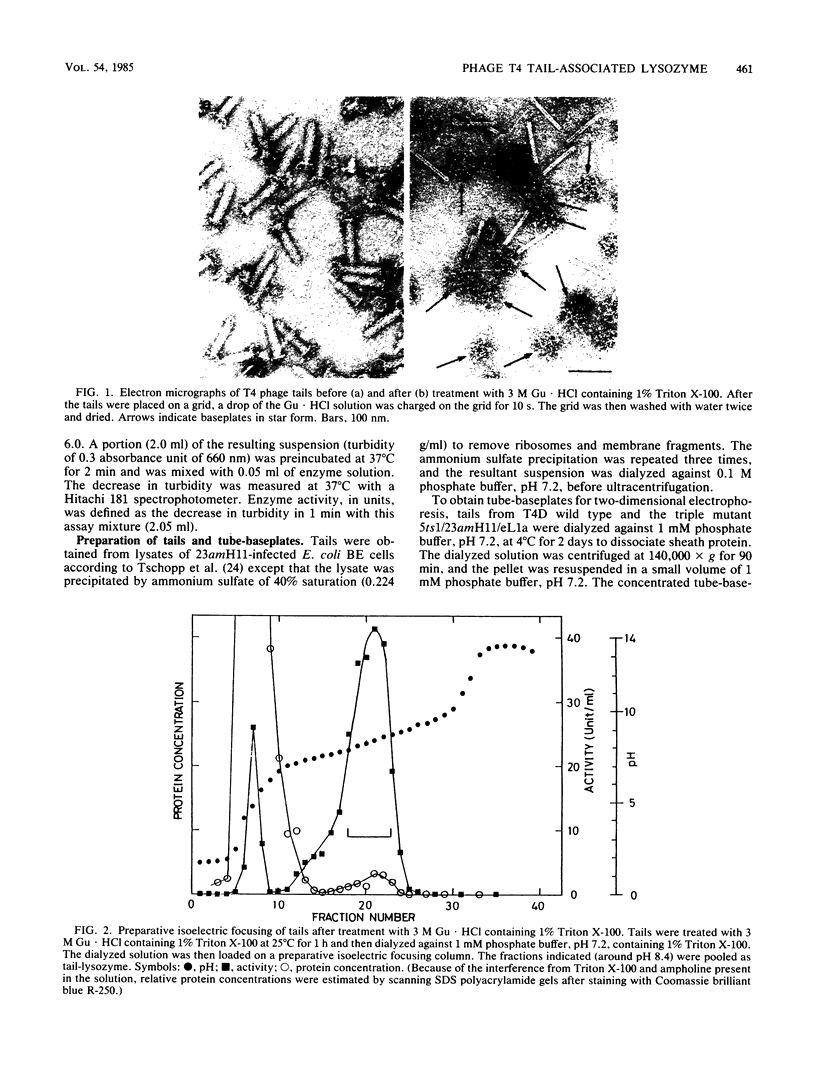
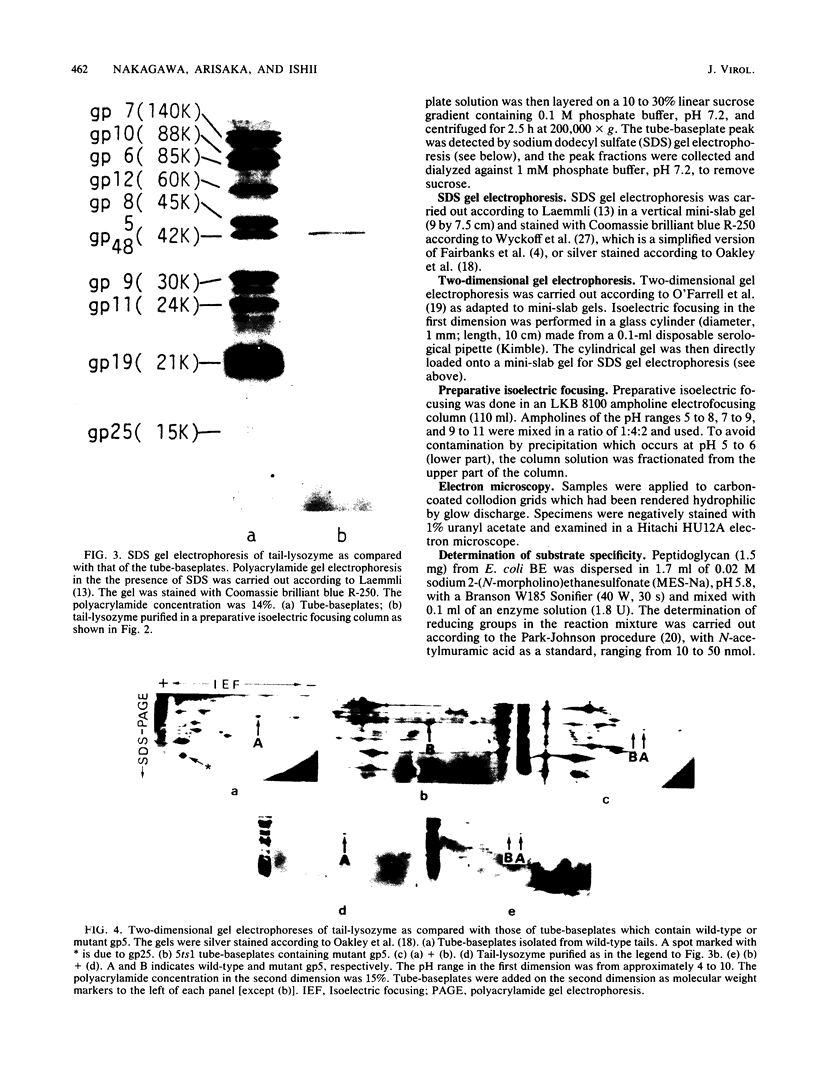
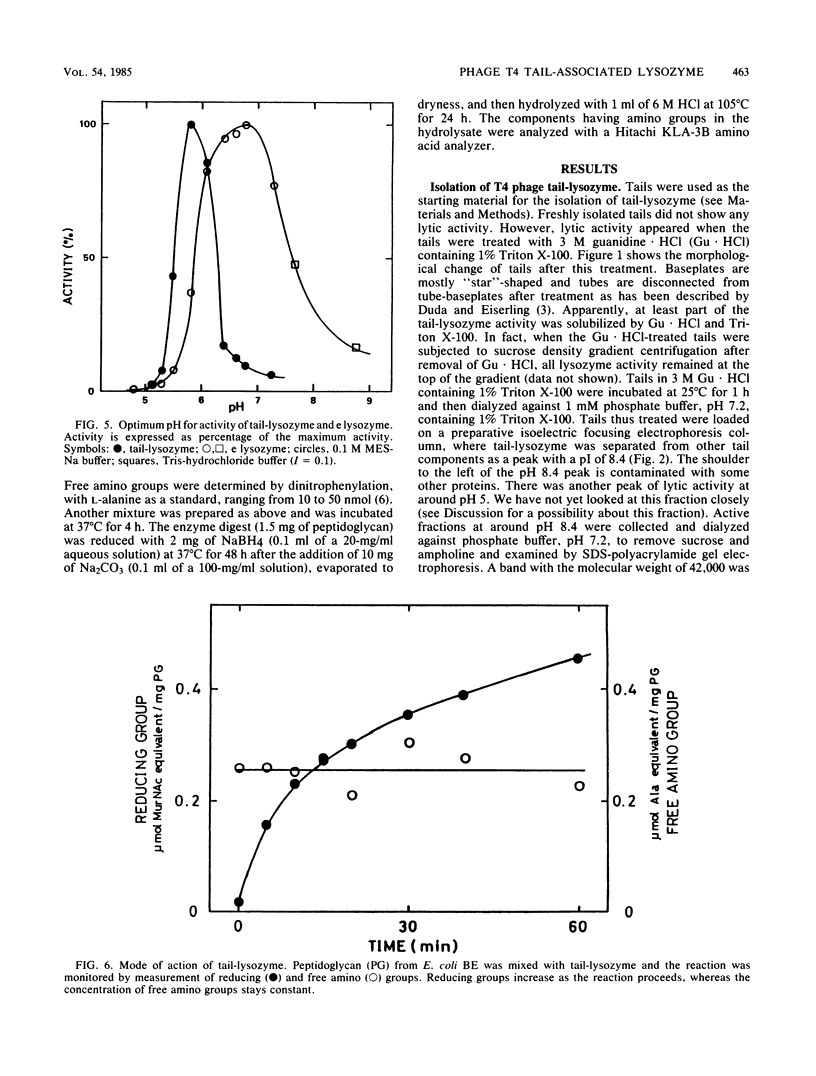
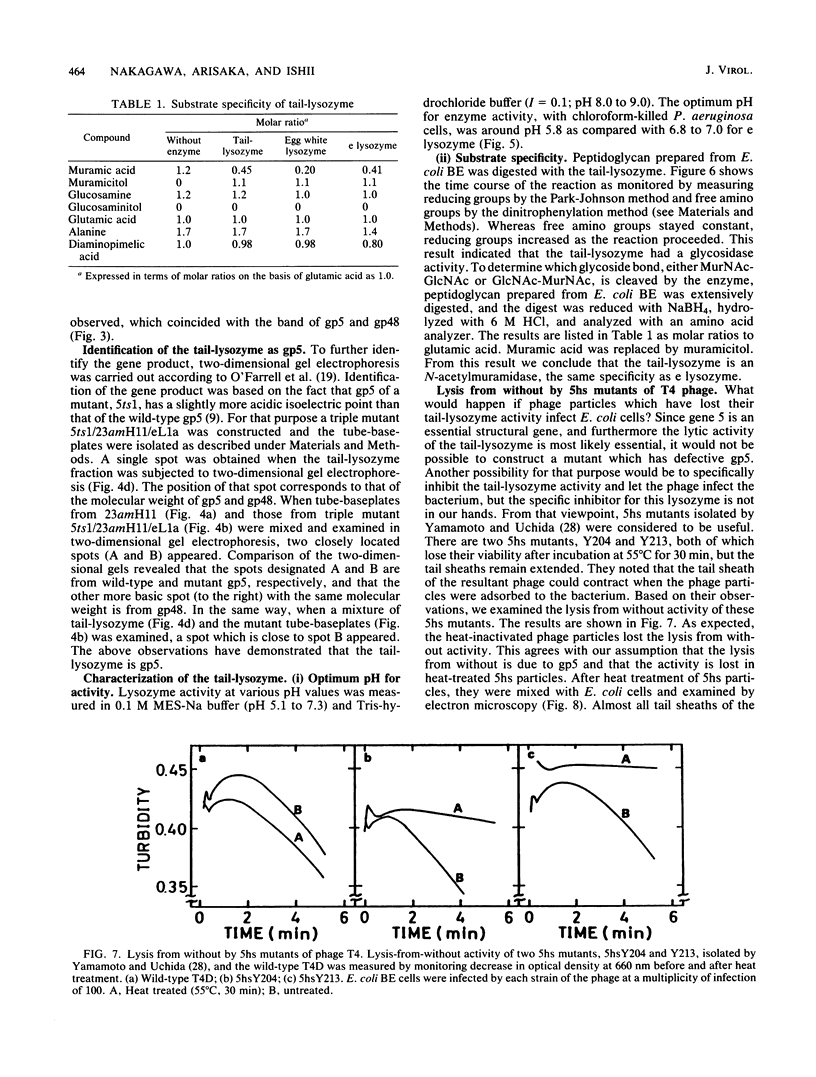
Direct evidence has been obtained that the tail-associated lysozyme of bacteriophage T4 (tail-lysozyme) is gp5, which is a protein component of the hub of the baseplate. Tails were treated with 3 M guanidine hydrochloride containing 1% Triton X-100, and the tail-lysozyme was separated from other tail components by preparative isoelectric focusing electrophoresis as a peak with a pI of 8.4. The molecular weight as determined from sodium dodecyl sulfate electrophoresis was 42,000. The tail-lysozyme was unambiguously identified as gp5 when the position of the lysozyme was compared with that of gp5 of tube-baseplates from 5ts1/23amH11/eL1ainfected Escherichia coli cells by two-dimensional gel electrophoresis. The tail-lysozyme has N-acetylmuramidase activity and the same substrate specificity as gene e lysozyme; the optimum pH is around 5.8, about 1 pH unit lower than for the e lysozyme. We assume that the tail-lysozyme plays an essential role in locally digesting the peptidoglycan layer to let the tube penetrate into the periplasmic space. The tail-lysozyme is presumably also responsible for "lysis from without."
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black L. W., Hogness D. S. The lysozyme of bacteriophage lambda. I. Purification and molecular weight. J Biol Chem. 1969 Apr 25;244(8):1968–1975. [PubMed] [Google Scholar]
- Crowther R. A., Lenk E. V., Kikuchi Y., King J. Molecular reorganization in the hexagon to star transition of the baseplate of bacteriophage T4. J Mol Biol. 1977 Nov 5;116(3):489–523. doi: 10.1016/0022-2836(77)90081-x. [DOI] [PubMed] [Google Scholar]
- Duda R. L., Eiserling F. A. Evidence for an internal component of the bacteriophage T4D tail core: a possible length-determining template. J Virol. 1982 Aug;43(2):714–720. doi: 10.1128/jvi.43.2.714-720.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Furukawa H., Kuroiwa T., Mizushima S. DNA injection during bacteriophage T4 infection of Escherichia coli. J Bacteriol. 1983 May;154(2):938–945. doi: 10.1128/jb.154.2.938-945.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba H., Nanno M., Okada Y. Identification and partial purification of a lytic enzyme in the bacteriophage phi 6 virion. FEBS Lett. 1979 Jul 15;103(2):234–237. doi: 10.1016/0014-5793(79)81334-4. [DOI] [PubMed] [Google Scholar]
- Iwashita S., Kanegasaki S. Smooth specific phage adsorption: endorhamnosidase activity of tail parts of P22. Biochem Biophys Res Commun. 1973 Nov 16;55(2):403–409. doi: 10.1016/0006-291x(73)91101-7. [DOI] [PubMed] [Google Scholar]
- KATZ W., WEIDEL W. [Isolation and characterization of the lysozyme bound to T2 phage]. Z Naturforsch B. 1961 Jun;16B:363–368. [PubMed] [Google Scholar]
- Kao S. H., McClain W. H. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J Virol. 1980 Apr;34(1):95–103. doi: 10.1128/jvi.34.1.95-103.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kells S. S., Haselkorn R. Bacteriophage T4 short tail fibers are the product of gene 12. J Mol Biol. 1974 Mar 15;83(4):473–485. doi: 10.1016/0022-2836(74)90508-7. [DOI] [PubMed] [Google Scholar]
- Labedan B., Goldberg E. B. Requirement for membrane potential in injection of phage T4 DNA. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4669–4673. doi: 10.1073/pnas.76.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loeb M. R. Bacteriophage T4-mediated release of envelope components from Escherichia coli. J Virol. 1974 Mar;13(3):631–641. doi: 10.1128/jvi.13.3.631-641.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M. F. Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol. 1973 Nov 15;80(4):613–635. doi: 10.1016/0022-2836(73)90200-3. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- PARK J. T., JOHNSON M. J. A submicrodetermination of glucose. J Biol Chem. 1949 Nov;181(1):149–151. [PubMed] [Google Scholar]
- Simon L. D., Anderson T. F. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology. 1967 Jun;32(2):298–305. doi: 10.1016/0042-6822(67)90278-4. [DOI] [PubMed] [Google Scholar]
- Szewczyk B., Skórko R. Lysozyme activity of bacteriophage T4 ghosts. Biochim Biophys Acta. 1981 Nov 13;662(1):131–137. doi: 10.1016/0005-2744(81)90233-3. [DOI] [PubMed] [Google Scholar]
- Szewczyk B., Skórko R. Purification and some properties of bacteriophage T4 particle-associated lysozyme. Eur J Biochem. 1983 Jul 1;133(3):717–722. doi: 10.1111/j.1432-1033.1983.tb07521.x. [DOI] [PubMed] [Google Scholar]
- Tschopp J., Arisaka F., van Driel R., Engel J. Purification, characterization and reassembly of the bacteriophage T4D tail sheath protein P18. J Mol Biol. 1979 Feb 25;128(2):247–258. doi: 10.1016/0022-2836(79)90128-1. [DOI] [PubMed] [Google Scholar]
- Tsugita A., Inouye M. Purification of bacteriophage T4 lysozyme. J Biol Chem. 1968 Jan 25;243(2):391–397. [PubMed] [Google Scholar]
- Tsukagoshi N., Schäfer R., Franklin R. M. Structure and synthesis of a lipid-containing bacteriophage. An endolysin activity associated with bacteriophage PM2. Eur J Biochem. 1977 Aug 1;77(3):585–588. doi: 10.1111/j.1432-1033.1977.tb11702.x. [DOI] [PubMed] [Google Scholar]
- Wyckoff M., Rodbard D., Chrambach A. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate-containing buffers using multiphasic buffer systems: properties of the stack, valid Rf- measurement, and optimized procedure. Anal Biochem. 1977 Apr;78(2):459–482. doi: 10.1016/0003-2697(77)90107-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Uchida H. Organization and function of bacteriophage T4 tail. I. Isolation of heat-sensitive T4 tail mutants. Virology. 1973 Mar;52(1):234–245. doi: 10.1016/0042-6822(73)90412-1. [DOI] [PubMed] [Google Scholar]
- Zorzopulos J., Kozloff L. M., Chapman V., DeLong S. Bacteriophage T4D receptors and the Escherichia coli cell wall structure: role of spherical particles and protein b of the cell wall in bacteriophage infection. J Bacteriol. 1979 Jan;137(1):545–555. doi: 10.1128/jb.137.1.545-555.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]