Abstract
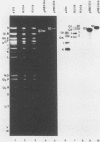
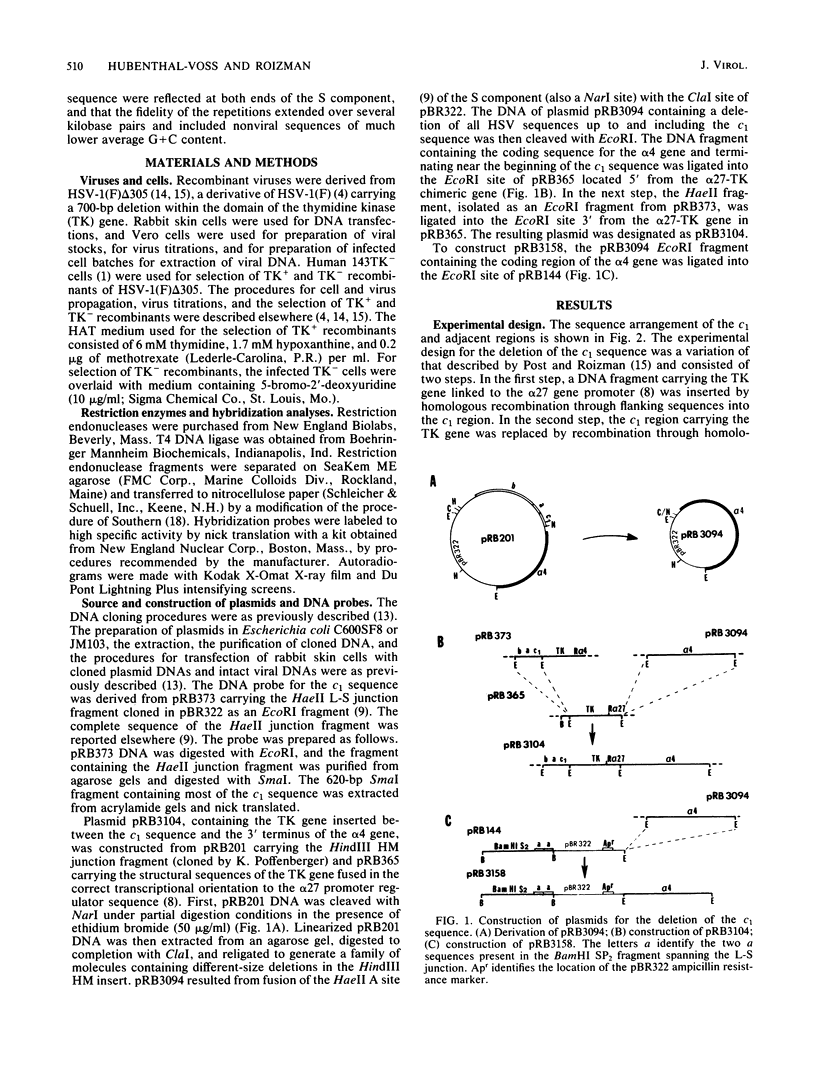
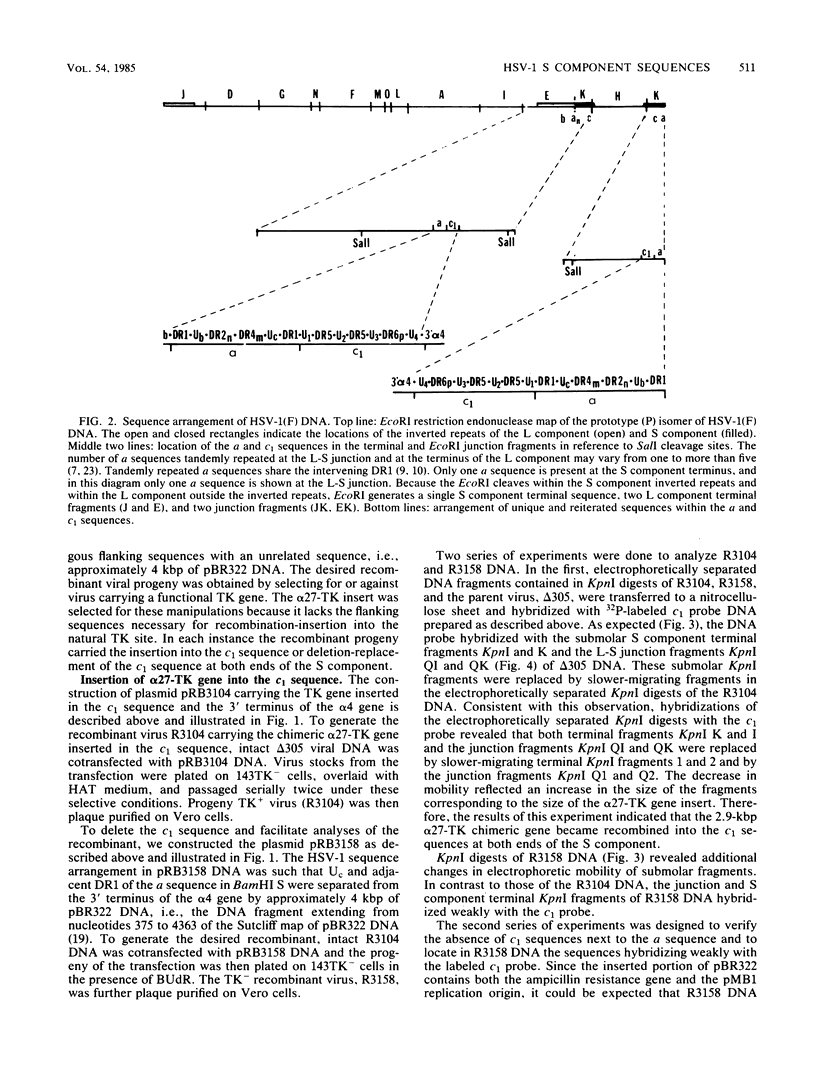
The herpes simplex virus 1 genome consists of two components, L and S, each containing unique sequences flanked by inverted repeats. Each of the 6.5-kilobase pair inverted repeats of the S component, designated a'c' and ca, contains an approximately 700-base pair sequence (designated c1) located between the a sequence and the 3' terminus of the alpha 4 gene. Like the a sequence, c1 consists of direct repeats and unique sequences. Its function is not known. To probe for its function, we constructed a plasmid containing a viral thymidine kinase (TK) gene inserted into the c1 sequence. The construct was recombined into the genome of a TK- virus by cotransfection with intact viral DNA and selection for TK+ virus. As predicted from previous studies (Knipe et al., Proc. Natl. Acad. Sci. U.S.A. 75:3896-3900, 1978), the TK gene was found to be present in both copies of the c1 sequence in the R3104 virus. To delete the c1 sequence we constructed a plasmid containing 4 kilobase pairs of pBR322 flanked by an a sequence and by structural sequences of the alpha 4 gene. In this instance the cells were transfected with the construct and R3104 DNA; the progeny of the transfection was plated in the presence of 5-bromo-2'-deoxyuridine, and the selection was for TK- virus (R3158). The pBR322 DNA sequences replaced the c1 at both termini of the S component in R3158 DNA, but a sequence homologous to c1 was present in proximity to the 3' terminus of the alpha 4 gene. The results indicate that the c1 region has no significant role in the replication of the virus in cell culture. The advantage of inserting the pBR322 sequence is that it permits efficient cloning of large herpes simplex virus 1 DNA fragments by simple ligation of digests and transformation of appropriate Escherichia coli strains. The effortless selection of recombinants carrying inserts in both copies of the c1 restates the usefulness of this technique for selection of insertion deletion recombinants and underscores the rapid emergence of sequence identity at both ends of the reiterated regions of the S component as previously reported (Knipe et al., Proc. Natl. Acad. Sci. U.S.A. 75:3896-3900, 1978).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Campione-Piccardo J., Rawls W. E., Bacchetti S. Selective assay for herpes simplex viruses expressing thymidine kinase. J Virol. 1979 Aug;31(2):281–287. doi: 10.1128/jvi.31.2.281-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D. M., Ruyechan W. T., Roizman B., Halliburton I. W. Molecular genetics of herpes simplex virus: demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3896–3900. doi: 10.1073/pnas.75.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979 Nov;32(2):429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Structural features of the herpes simplex virus alpha gene 4, 0, and 27 promoter-regulatory sequences which confer alpha regulation on chimeric thymidine kinase genes. J Virol. 1982 Dec;44(3):939–949. doi: 10.1128/jvi.44.3.939-949.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Conley A. J., Mocarski E. S., Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981 Jul;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Roizman B., Jacob R. J., Knipe D. M., Morse L. S., Ruyechan W. T. On the structure, functional equivalence, and replication of the four arrangements of herpes simplex virus DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):809–826. doi: 10.1101/sqb.1979.043.01.088. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Unstable heterozygosity in a diploid region of herpes simplex virus DNA. J Virol. 1984 Feb;49(2):356–362. doi: 10.1128/jvi.49.2.356-362.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Summers W. C. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J Virol. 1978 Aug;27(2):374–387. doi: 10.1128/jvi.27.2.374-387.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]