Abstract
Alpha subunit of Escherichia coli ATP synthase was expressed with a C-terminal 6-His tag and purified. Pure alpha was monomeric, competent in nucleotide binding, and had normal N-terminal sequence. In F1-subunit dissociation/reassociation experiments it supported full reconstitution of ATPase, and reassociated complexes were able to bind to F1-depleted membranes with restoration of ATP-driven proton pumping. Therefore interaction between the stator delta subunit and the N-terminal residue 1-22 region of alpha occurred normally when pure alpha was complexed with other F1 subunits. On the other hand, three different types of experiment showed that no interaction occurred between pure delta and isolated alpha subunit. Unlike in F1, the N-terminal region of isolated alpha was not susceptible to trypsin cleavage. Therefore, during assembly of ATP synthase, complexation of alpha subunit with other F1 subunits is prerequisite for delta subunit binding to the N-terminal region of alpha. We suggest that the N-terminal 1-22 residues of alpha are sequestered in isolated alpha until released by binding of beta to alpha subunit. This prevents 1/1 delta/alpha complexes from forming, and provides a satisfactory explanation of the stoichiometry of one delta per three alpha seen in the F1 sector of ATP synthase, assuming that steric hindrance prevents binding of more than one delta to the alpha3/beta3 hexagon.
The cytoplasmic fragment of the b subunit (bsol) did not bind to isolated alpha. It might also be that complexation of alpha with beta subunits is prerequisite for direct binding of stator b subunit to the F1-sector.
ATP synthase is the terminal enzyme of oxidative phosphorylation and photophosphorylation, which synthesizes ATP from ADP and phosphate (Pi). The energy for ATP synthesis comes from transmembrane movement of protons down an electrochemical gradient, generated by substrate oxidation or by light capture. Initially, as the protons move through the interface between a and c subunits in the membrane-bound Fo-sector of the enzyme, the realized energy is transduced into mechanical rotation of a group of subunits (γεcring) which comprise the “rotor”. A helical coiled coil domain of γ projects into the central space of the α3β3 hexagon, in the membrane-extrinsic F1-sector. α3β3 hexagon contains three catalytic sites at α/β interfaces. In a manner that is not yet understood, rotation of γ vis-à-vis the three α/β subunit pairs brings about ATP synthesis at the three catalytic sites using a sequential reaction scheme (1). Detailed reviews of ATP synthase mechanism may be found in (2–5).
“Stator” subunits b2 and δ are present to prevent co-rotation of α3β3 with the rotor (6). The interaction between δ and α subunits is known to be of major importance for this function. Initially (7) it was shown by proteolysis experiments using the Escherichia coli enzyme that δ subunit bound to the N-terminal region (residues 1-19) of the α subunit in the F1-complex. More recently, quantitative binding experiments showed that this interaction is extremely tight (8,9), and utilizes a binding surface formed between two parallel helices on the N-terminal domain of the δ subunit (10). A 22-residue peptide which mimicked residues 1-22 of α subunit was found to bind with high affinity to the same surface on δ subunit (11), forming a 1/1 complex. A high resolution structure of the complex between the N-terminal domain of the δ subunit and the 22-residue peptide was obtained by NMR (12). It revealed in detail the nature of the binding surface on the δ subunit, confirmed previous suggestions from circular dichroism measurements and mutagenesis/deletion analyses (11, 13) that the 22-residue peptide is alpha-helical and amphipathic when bound to δ, demonstrated how the helical 22-residue peptide nestles between the two helices on δ which form the binding surface, and identified key side-chains in the peptide that mediate protein-protein interaction with δ.
In mitochondrial ATP synthase the subunit corresponding to E. coli δ is called “OSCP” (14). OSCP was recently found to have similar structure to E. coli δ (15,16) and parallel experiments to those first performed as described above in E. coli confirmed that it binds to the N-terminal region of α subunit of mitochondrial enzyme (16,17). Although a detailed structural model of the interaction of OSCP with mitochondrial α subunit N-terminus has not yet been derived at the time of writing, it appears that it will resemble that already established for E. coli. This is noteworthy, since in other aspects of stator structure and subunit composition, the two enzymes are remarkably different (6,14).
It is clear therefore that α/δ interaction is an important facet of stator structure and function in the E. coli enzyme and we envisaged that further understanding might be achieved by combination of the quantitative fluorimetric binding assays devised previously (8,10) with more extensive mutagenesis experiments, particularly since we now have a high-resolution NMR structure of the interaction surface. In a previous study of sequence determinants in the N-terminal 22-residues of α subunit for binding δ we used a set of synthetic peptides, each containing a specific mutation or deletion together with purified δ subunit for binding assays (13). In that study we established also that the effect of the mutations on binding of peptide to δ subunit in vitro was faithfully reflected in effects on growth of cells in vivo and on enzyme function as measured by ATP hydrolysis and ATP-driven proton pumping. However, synthesis of mutiple mutant peptides proved both expensive and cumbersome. Mutations can be introduced easily into E. coli ATP synthase α subunit (18), still there is the problem that mutations that impact on α/δ interaction are almost certain to interfere with assembly of ATP synthase in the cell, reducing or eliminating the yield of source material for study.
In another study of stator function, direct binding of the soluble cytoplasmic portion of the b subunit to F1 was also detected using fluorometric assays (19). The site of binding of b on F1 is not yet known precisely (see ref. 6 for review); however, Trp residues introduced at positions α-55 and α-406 gave substantial fluorescence signal enhancement in our assays, indicating α subunit as a possible site of interaction.
We decided therefore to express isolated α subunit in an uncA− (ATP synthase α-subunit deletion) strain, purify it and use it together with pure δ subunit and pure cytoplasmic portion of b subunit for in vitro studies of α/δ and α/b interaction. Expression and purification of wild-type and mutant isolated α subunit in native functional form was achieved and is described. The results of binding experiments are also described, and lead to insights into assembly of the stator in ATP synthase.
EXPERIMENTAL PROCEDURES
Preparation of E. coli membranes; Assay of ATPase activity of membranes and F1; Measurement of proton pumping in membrane vesicles
E. coli membranes were prepared as in (20). Prior to the experiments, membranes were washed twice by resuspension and ultracentrifugation in 50 mM TrisSO4 pH 8.0, 2.5 mM MgSO4. ATPase activity was measured in 1 ml assay buffer containing 10 mM NaATP, 4 mM MgCl2, 50 mM TrisSO4, pH 8.5 at 37°C. Reactions were started by addition of membranes or F1 and stopped by addition of SDS to 3.3% final concentration. Pi released was assayed as in (21). All reactions were shown to be linear with time and protein concentration. ATP-driven proton pumping was measured by following the quench of acridine orange fluorescence as described in (22).
E. coli strains
Wild-type strain was pBWU13.4/DK8 (23) for membrane preparations or SWM1 (24) for purified F1. New mutant strains were constructed as below.
Purification of F1; Dissociation of purified F1 into component subunits; Reassociation of F1; Preparation of F1-depleted membranes; Binding of F1 to F1-depleted membranes
F1 was purified as in (25). Dissociation of F1 into subunits and reassociation back into F1 followed in principle the methods of Dunn and Futai (26). For dissociation F1 was precipitated in 66% saturated ammonium sulfate, redissolved to 4 mg/ml in buffer containing 50 mM succinic acid, 1 M NaCl, 0.25 M NaNO3, 1 mM EDTA and 1 mM DTT1, 1 mM PMSF, adjusted to pH 6.0 with Tris, and dialysed against the same buffer overnight at 4°C. The mixture was then flash frozen in dry ice/ethanol and stored at −70°C for at least 3 days. For reassociation the thawed mixture was dialysed overnight at room temperature against buffer containing 50 mM succinic acid, 10% glycerol, 1 mM DTT, 2 mM MgCl2, 1 mM PMSF, 2 mM ATP, 0.1 mM EDTA, adjusted to pH 6.0 with Tris. Membranes were stripped of F1 by KSCN extraction as in (22) and F1 was re-bound to these membranes as in (8).
Purification of δ subunit: Purification of bST34-156; Preparation of δ-depleted F1
Purification of isolated α subunit
Isolated α subunit containing a C-terminal 6-His tag was purified from strain pSWM131/DK8 (see below) as follows. Cells were grown in 1l LB-Ampicillin medium in Fernbach flasks with shaking at 35°C to an optical density of 0.5 – 0.6 at 600 nm. The incubator temperature was reduced to 28°C, IPTG was added to 0.5 mM and shaking continued for 3 hr. Cells were harvested by centrifugation and resuspended in ice-cold 50 mM TrisCl, pH 8.0, 10% glycerol, 50 mM NaCl, 10 mM imidazole, 1 mM 2-mercaptoethanol and 8.5% sucrose to which protease inhibitors (leupeptin, 4 mg/liter; pepstatin A, 4 mg/liter; chymostatin, 1 mg/liter; and PMSF, 1 mM) were added immediately before use. All subsequent steps were carried out at 4°C. Cells were recentrifuged, resuspended (10 g cells/20 ml buffer) and frozen overnight at −70°C. After thawing, the cell slurry was passed twice through a French press at 20,000 psi, centrifuged at 10,000 g for 10 min, then centrifuged for 2 hr at 38,000 rpm in a Beckman 60Ti rotor. Supernatants were rotated for 1 hr in a 50 ml conical tube with 3 ml of packed NiNTA-agarose resin which had been washed in water and resuspended in 50 mM TrisHCl pH 8.0. The NiNTA resin was transferred to a column (1 cm × 10 cm) and washed with 75 ml of buffer containing 50 mM TrisHCl pH 8.0, 10% glycerol, 50 mM NaCl, 10 mM imidazole and 1 mM 2-mercaptoethanol plus protease inhibitors (same concentrations as above). Isolated α subunit was then eluted by washing with the same buffer containing 100 mM imidazole but no protease inhibitors.
SDS-gel electrophoresis; N-terminal sequence analysis; Gel filtration experiments; Trp content analysis
SDS-gel electrophoresis was carried out as in (27). For N-terminal sequencing 300 pmol of sample was run on 10–20% gradient gels in Tris-glycine buffer and blotted to Millipore Immobilin P PVDF membranes in 1 mM CAPS/NaOH pH 11.0, 10% methanol. The blot was briefly stained with Ponceau S in CH3COOH, and air-dried. N-terminal analysis was carried out by standard automated Edman procedure at the Protein Chemistry Laboratory, University of Texas Medical Branch, Galveston, Texas. Typical yields of detected amino acids for the first five cycles were 30 – 40 pmol/cycle. Gel filtration analysis was carried out in 50 mM HEPES/NaOH pH 7.0, 5 mM MgSO4, using a 1.5 × 100 cm column of Sephacryl S-300 or S-100. Size markers used were cytochrome c, carbonic anhydrase, ovalbumin, hemoglobin, phosphorylase b, alcohol dehydrogenase, β-amylase, and F1. Trp content was determined as follows. Samples were dissolved in 6.3 M guanidine hydrochloride solution at 35 – 50 μg/ml, excitation was at 295 nm and the fluorescence emission spectrum at 310 – 410 nm was recorded after 15 min. Trp content of isolated α was calculated by comparing the fluorescence at 350 nm, corrected for buffer control, with that of wild-type F1 which contains 9 Trp (mol/mol).
Nucleotide binding assays
Radioactive nucleotide binding assays were carried out as follows. Isolated wild-type α subunit was preequilibrated in 50 mM TrisSO4 pH 8.0 by elution through 1 ml centrifuge columns of Sephadex G-50, then incubated in 100 μl at 0.2 mg/ml for 1 hour at room temperature with 100 μM concentration of [α-32P]ATP in buffer containing 50 mM TrisSO4, pH 8.0, 0.1 mM DTT, 2.5 mM MgCl2. After centrifuge column elution to separate bound from free nucleotide, protein was assayed in the eluates and nucleotide content was estimated by liquid scintillation counting. Fluorimetric analysis of nucleotide binding was as follows. Isolated α subunit (αR365W/αW513F) was added to 2 ml cuvettes at 100 nM concentration, room temperature in 50 mM HEPES/NaOH pH 7.0 with either 1 mM EDTA or 2.5 mM MgSO4. ATP or ADP were titrated in and the fluorescence signal quench (excitation at 295 nm, emission at 340 nm) was measured. Correction for volume, buffer and inner filter effects (the latter using wild-type F1) were applied. Calculation of nucleotide binding stoichiometry is described in legend to Figure 1.
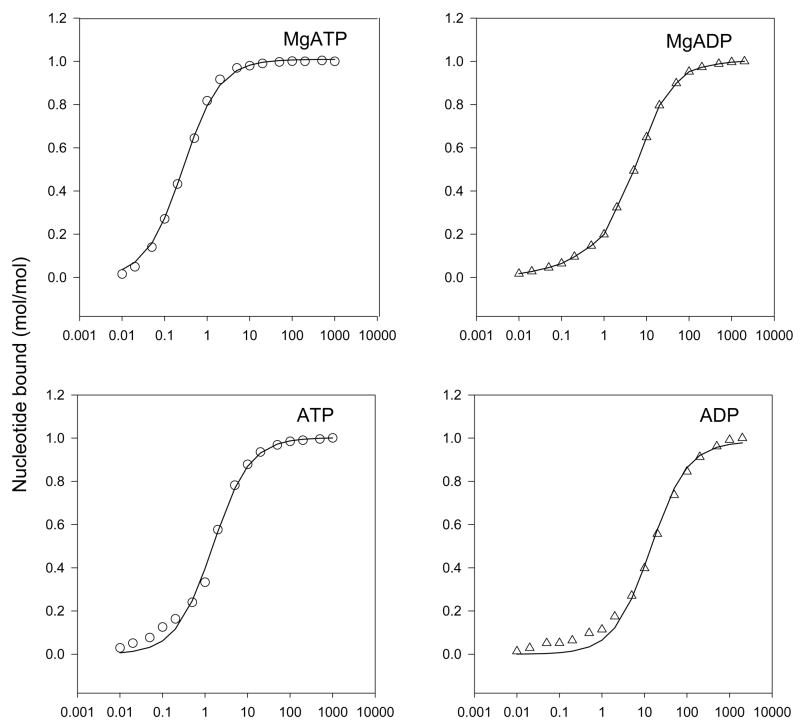
Figure 1. Nucleotide binding to isolated α subunit in absence and presence of Mg2+.
Fluorometric titration was carried out in 2 ml of 50 mM HEPES/NaOH, pH 7.0 with 100 nM α subunit (αR365W/W513F). λexc = 295 nm, λem = 340 nm. MgSO4 was added to 2.5 mM concentration (upper panels) and EDTA concentration was 1 mM (lower panels). After correction for volume and inner filter effects, n and Kd values were calculated using the equation: [Sb]/[Et] = n · [Sf]/([Sf] + Kd), where [S] = [nucleotide], [E] = [α subunit], and subscripts b, f, and t refer to bound, free and total respectively. Upper panels, presence of Mg2+, left MgATP, right MgADP. Lower panels, absence of Mg2+, left ATP, right ADP.
Fluorescence assays to detect binding of δ or bST34-156 to isolated α subunit
NiNTA binding assays to detect binding of δ or bST34-156 to isolated α subunit
NiNTA-agarose slurry was washed in water and resuspended in 50 mM HEPES/NaOH pH 7.0. Purified isolated α subunit, δ subunit and bST34-156 were each preequilibrated in the same buffer by centrifuge column elution. 0.5 mg samples of each were mixed in different permutations in the same buffer with added MgSO4 (2.5 mM) in a total volume of 5 ml and incubated at room temperature for 30 min. Then imidazole was added to final concentration of 10 mM followed by 0.5 ml of NiNTA-agarose slurry. The samples were rotated 1 hr at 4°C then poured into columns (0.5 × 10 cm). Columns were washed with 5 ml buffer containing 10 mM imidazole, then sequentially with three 500 μl aliquots of buffer containing 100 mM imidazole. Proteins in the eluates were identified by SDS-gel electrophoresis.
Trypsin proteolysis of δ-depleted F1 and isolated α subunit
The conditions used followed in principle those described by Dunn et al. (7). The buffer was 50 mM TrisSO4 pH 8.0, 10% sucrose, 10% glycerol, 2 mM EDTA, 10 mM NaATP. Final δ-depleted F1 and isolated α concentration was 2.3 mg/ml. Trypsin-TPCK (Sigma) was dissolved in buffer at 0.1 mg/ml immediately prior to experiments and was added to a final 2.3 μg/ml. Reactions were at room temperature and were stopped by addition of PMSF (final = 0.1 mg/ml) followed immediately by SDS-gel buffer solution and boiling. Samples were run on 10% SDS-gels for 2 hr at 24 milliamp, this long time being necessary to satisfactorily separate α, α′, and β subunits in the δ-depleted F1.
Construction of strain for expression and purification of wild-type isolated α subunit of E. coli ATP synthase
PCR was carried out using plasmid pBWU13.4 (23) as template to generate a 1590 bp fragment containing the gene for the ATP synthase α subunit with a C-terminal 6-His tag. The forward primer was: CAGAATTCTGGAGGATTTCGCATGCAACTGAACTCCACCGAAATC. The reverse primer was: TTATCAAGCTTTTAGTGATGGTGATGGTGATGCCAGGATTGGGTTGCTTTGAAGG. The expected fragment contains an EcoR1 site upstream of initial ATG for cloning and deletes the natural EcoR1 site at codon 4. An Sph1 site is included at codon 1. Behind the TAA stop is an HinD3 site for cloning. The PCR product was cloned in Sma1-cut pUC118 and correct inserts identified by restriction digests and DNA sequencing. Then an EcoR1-HinD3 fragment was cloned into plasmid pJB3 (28) to form plasmid pSWM131. This plasmid was transformed into strain JP2 (18) in which the uncA (α subunit gene) is deleted, and also into strain DK8 (29) in which all the ATP synthase structural genes are deleted.
Construction of plasmid pSWM132 containing the unc operon with an α subunit gene containing a C-terminal 6-His tag
This was accomplished by moving a Csp45I-Kpn1 fragment from the initial plasmid after cloning the PCR fragment in pUC118 (above) directly into plasmid pBWU13.4. DNA sequencing of the final plasmid, named pSWM132, showed that it contained the α subunit gene plus the C-terminal His tag, plus an extra 9 bases carried over from the pUC118 polylinker as expected. As seen in Table 1, this did not affect complementation by the modified α subunit.
TABLE 1. Assays of function in E. coli strain pSWM132/DK8 expressing ATP synthase in which α subunit contains a C-terminal 6-His tag.
| Strain | Growth on Succinate | Membrane ATPase activitya | Proton pumping activityb |
|---|---|---|---|
| Wild-typec | +++ | 3.2 ± 0.38 | 89 |
| α with C-His tagc | +++ | 2.9 ± 0.30 | 87 |
| Null controlc | - | ≤0.01 | 0 |
Measured at 30°C and expressed as μmol ATP hydrolysed/min/mg membrane protein.
Measured using acridine orange and expressed as per cent quench of acridine orange fluorescence in membrane vesicles upon addition of 1 mM MgATP.
Wild-type, pBWU13.4/DK8; α with C-His tag, pSWM132/DK8; Null control, pUC118/DK8. Data are means of six experiments obtained from two independent membrane preparations of each mutant, and are expressed as means ± standard deviation. (Null controls had negligible activity).
Mutant forms of isolated α subunit
αW513F.2 The PCR was repeated as above with the reverse primer containing a codon change TGG to TTT at codon 513 (which normally immediately precedes the stop codon). The final plasmid was named pSWM134. αL55W/αW513F. The method of mutagenesis was as in (30). The template was the EcoR1-HinD3 fragment from plasmid pSWM134 cloned in M13mp19, the oligonucleotide used was as in (8). αR365W/αW513F and αF406W/αW513F. The mutations were moved from original plasmids described in (19,31) on Xho1-Csp45I fragments into pSWM134. All mutant plasmids were checked by DNA sequencing and were transformed into JP2 and pBWU13.4 as above for expression of isolated α subunit.
Materials
[α-32P]ATP was from Perkin Elmer Life Sciences; TPCK-trypsin was from Sigma; IPTG was from Calbiochem; NiNTA resin was from Qiagen.
RESULTS
Complementation tests of in vivo function of E. coli ATP synthase α subunit with C-terminal 6-His tag
As will be described in the next section, we constructed a gene containing the E. coli ATP synthase α subunit with a C-terminal 6-His tag to facilitate purification. It was necessary before proceeding further to ascertain that the His-tag did not abolish function. To show this we took the plasmid pBWU13.4 (23) which encodes all of the E. coli ATP synthase structural genes and replaced the gene for wild-type α subunit with one containing the C-terminal His-tag. We then expressed the new plasmid (pSWM132) in strain DK8 (in which all ATP synthase structural genes are deleted). Function of ATP synthase containing an α subunit with C-terminal His-tag was assayed as shown in Table 1. The data show that the α subunit with the C-terminal His tag assembles normally in the ATP synthase complex and functions normally to support oxidative phosphorylation with succinate as substrate, ATP hydrolysis, and ATP-driven proton pumping across the membrane.
Expression and purification of isolated wild-type E. coli ATP synthase α subunit with a C-terminal 6-His tag
Construction of strain pSWM131/JP2 for expression of wild-type α subunit with a C-terminal 6-His tag is described in detail in EXPERIMENTAL PROCEDURES. Briefly, PCR was used to generate a fragment (1590 bp) containing the whole uncA (α subunit) gene with 6 His codons at the C-terminal end, which was initially cloned into Sma1-cut pUC118, then moved into plasmid pJB3 (28) to form pSWM131 which was transformed into the α-deletion strain JP2. (It should be noted that strain DK8 containing a deletion of all the unc (ATP synthase) structural genes could also be used as host with identical results).
Details of the expression and purification of the isolated α subunit may be found in EXPERIMENTAL PROCEDURES. The purified α subunit was stored at −70°C and was stable for months. Typical yields were 4 to 6 mg of α subunit per liter of cells, with one liter equivalent to 1.5 to 2 g wet wt of cells. Several mutant versions of α subunit were also made as detailed in EXPERIMENTAL PROCEDURES and described below, and gave the same yields as wild-type.
Structural characterization of the isolated α subunit
On SDS-gels the isolated α subunit appeared as a single band, running slightly slower than authentic α subunit in purified F1 as expected (data not shown). N-terminal analysis was carried out on two different preparations of purified α subunit and in both cases the N-terminal sequence was found to be MQLNS-, i.e. it corresponded exactly to that expected from the gene sequence. Therefore no cleavage of N-terminal residues had occurred during expression or purification. The fact that the subunit was purified by NiNTA-agarose chromatography demonstrates that the C-terminal sequence was also intact. Gel filtration was carried out (see EXPERIMENTAL PROCEDURES) to determine the aggregation state of isolated α subunit. It was found that α eluted from Sephacryl S-300 columns as a single peak with an estimated molecular size of 55,730 daltons (actual value expected from sequence = 55,999) demonstrating that it existed entirely in monomeric form. Wild-type α subunit contains just one Trp residue in the sequence, at the C-terminal residue 513. Trp analysis was carried out on two different preparations of purified subunit (see EXPERIMENTAL PROCEDURES) with the following results: Preparation one, Trp content = 0.94 Trp (mol/mol). Preparation two, Trp content = 1.17 Trp (mol/mol). These results are each means of duplicate assays, and indicate that the α subunit was of high purity.
Nucleotide binding to isolated α subunit
X-ray structures of F1-ATPase show that the α subunit contains a nucleotide binding domain which binds one Mg-nucleotide per α at the so-called noncatalytic sites (32). Earlier work had demonstrated that isolated α subunit which had been purified from F1-ATPase after depolymerisation of the latter in chaotropic salts bound ATP tightly at a stoichiometry of one ATP per mol of α subunit (26). Here, to assess nucleotide binding capability, we first incubated isolated α subunit with radioactive [α-32P]ATP then passed the mixture through centrifuge columns to separate bound from free nucleotide. Results are shown in Table 2. It was evident from these results that full nucleotide binding was displayed by the purified isolated α subunit, supporting the idea that the subunit had assumed native folded structure.
TABLE 2. Stoichiometry of binding of [α-32P]ATP to isolated wild-type α subunit.
Isolated α subunit was pre-equilibrated with 50 mM Tris-SO4 pH 8.0 by centrifuge column elution, then incubated in 100 μl volume at 0.2 mg/ml concentration with [α-32P]ATP (100 μM) in buffer containing 50 mM TrisSO4, pH 8.0, 0.1 mM DTT, 2.5 mM MgCl2 for 1 hour at room temperature followed by 10 min at 4°C. The whole sample was then passed through a 1 ml centrifuge column pre-equilibrated in 50 mM Tris-SO4 pH 8.0. Protein and nucleotide were assayed in the eluates.
| ATP bound (mol/mol α subunit) | |
|---|---|
| α subunit preparation one | 1.13 ± 0.17 (SD) n = 4 |
| α subunit preparation two | 1.05 ± 0.05 (SD) n = 4 |
ATPase activity was undetectable in isolated α subunit (turnover number with 10 mM MgATP at pH 7.5, 30°C, was < 1 × 10−3 s−1), consistent with previous work (26).
Our laboratory has previously demonstrated (31) that the substitution αR365W2 in E. coli F1-ATPase allowed fluorometric assay of the binding of nucleotides to the noncatalytic, α subunit-located sites. The introduced Trp lies very close to the bound nucleotide and undergoes a large quench of the fluorescence signal upon nucleotide binding. It was interesting therefore to introduce this substitution into the isolated α subunit, to determine whether it could be used in a similar manner in that context, and more particularly to determine whether isolated α subunit binds Mg-nucleotides differently from uncomplexed (“free”) nucleotide, which has not previously been done. First we made the mutation αW513F in order to remove the single native Trp and then we introduced the αR365W mutation. Both αW513F and αR365W/αW513F mutant α subunits were expressed and purified with the same yields and purity as wild-type. In addition, in complementation tests (performed as in Table 1) both mutants gave similar results to wild type (data not shown).
Figure 1 shows the results of fluorometric nucleotide binding assays carried out with αR365W/αW513F α subunit and free or Mg-complexed nucleotides. Quench of fluorescence was very rapid and in all cases maximal quench of fluorescence of the single Trp present at residue α-365 was by 80–85%. The calculated stoichiometry of binding at saturating nucleotide concentration from twelve experiments was 0.99 mol/mol of α subunit (range = 0.86 – 1.07). Calculated Kd values were as follows: MgATP, 0.31 μM; free ATP, 1.23 μM; MgADP, 4.4 μM, free ADP, 14.1 μM. (Results are means of triplicate titrations). These results confirm earlier data of Dunn and Futai (26) in showing that ATP generally binds ten times more tightly than ADP, and additionally show that presence of Mg2+ tightens binding by about 4- fold. In X-ray structures of F1-ATPase (32) Mg-nucleotide complexes are located in the noncatalytic (α-subunit) binding sites.
Isolated α subunit reassociates with other F1 subunits to form a complex with full ATPase activity, which will bind to F1-depleted membranes and restore ATP-driven proton pumping activity
Wild-type E. coli F1-ATPase can be dissociated into its individual subunits by chaotropic salts (26) and then reassociated to reform the complex after removal of these salts by dialysis and addition of Mg2+ ions. This process can be conveniently monitored following ATPase activity which is destroyed by dissociation and restored by reassociation. Membranes containing ATP synthase can be depleted of the F1 sector by incubation in KSCN followed by centrifugation, and F1 can then be re-bound to the washed membranes by incubation in the presence of Mg2+ ions (22). This process can be conveniently monitored by the initial destruction of ATP-driven proton pumping activity and then its restoration.
Here we first took wild-type F1 and confirmed that upon dissociation it lost all ATPase activity, which was regained after reassociation (Table 3 line 1). When the reassociated F1 was added to F1-depleted membranes, it bound and gave a normal ATP-driven proton pumping response (Table 3 line 1). If an excess of isolated α subunit (30 mol α/mol F1) was added to the dissociated subunits mixture, and then reassociation was carried out, the results were similar (Table 3 line 2). Under these latter conditions, 91% of the α-subunits in the subunit mixture immediately before reassociation would be from the added isolated α subunit, and 9% from the original F1 enzyme. In E. coli enzyme the minimal unit for expression of normal ATPase activity is the α3β3γ complex (26,33)3. Thus these experiments strongly suggest that added isolated α subunit was fully able to reassociate with β and γ subunits present in the dissociated F1. Furthermore, there must have been a normal interaction between the N-terminal region of the α subunits and the δ subunit in the reassociated F1, since membrane-binding and ATP-driven proton pumping could not occur otherwise. Also, and for the same reason, the ε-subunit must also have been recruited into the reassociated F1 complex because it also is required for membrane-binding (34).
TABLE 3. Effects of isolated α-subunit on reassociation of F1-ATPase activity and rebinding of resultant F1-ATPase to F1-depleted membranes.
Purified F1-ATPase was dissociated into individual subunits and reassociated as described in detail in EXPERIMENTAL PROCEDURES. Where indicated an excess of isolated α subunit (30 mol α subunit per mol F1) was added prior to the reassociation phase. ATPase activity was assayed on the original, the dissociated and the reassociated enzymes. Then the reassociated enzymes were rebound to F1-depleted membranes as described in EXPERIMENTAL PROCEDURES, and rebinding of F1 to the membranes was monitored by assay of ATP-driven proton pumping.
| F1 species | ATPase activitya | ATP-driven proton pumpingb | |||
|---|---|---|---|---|---|
| Original | Dissociated | Reassociated | Original | Reassociated | |
| Wild-typec | 25.2 | 0.08 | 22.3 | 81 | 79 |
| Wild-type plus α | 24.2 | 79 | |||
| αS373Fc | ≤ 0.01 | ≤ 0.01 | ≤ 0.01 | 0 | 0 |
| αS373F plus α | 22.0 | 72 | |||
Measured at 30°C and expressed as μmol ATP hydrolysed/min/mg original F1 protein.
Measured using acridine orange and expressed as per cent quench of acridine orange fluorescence in membrane vesicles upon addition of 1 mM MgATP.
Wild-type from strain SWM1 (24); αS373F from strain AW5 (50). Note that AW5 F1 also contains the βY331W mutation, but this mutation has been shown not to affect ATPase, proton pumping or membrane-binding of F1.
Data are means of triplicate experiments
In a second series of experiments we used a mutant form of F1 containing a “dead” α subunit, namely αS373F. Table 3 line 3 shows the results when this F1 was dissociated, reassociated, and rebound to membranes. In concordance with much previous work on this mutant, no ATPase nor proton pumping activity was seen. However if isolated wild-type α-subunit was added before the reassociation phase of the experiment, now it was seen that ATPase activity was generated and that the reassociated F1 was able to rebind to F1-depleted membranes and support proton pumping (Table 3 line 4). Generation of ATPase could only have come about if the added wild-type α-subunit reassociated with the (normal) β and γ subunits from the mutant F1, replacing the mutant α subunits. Moreover the high ATPase activity seen shows that the proportion of “hybrid” enzymes (e.g. containing one normal α/two mutant α or two normal α/one mutant α) in the reassociated F1 population was low, as expected from the fact that the mutant α subunit represented only 9% of total α present. The activity of such hybrid enzymes is much lower than normal (35). Thus we can conclude confidently that the added isolated α-subunit was able to interact normally with the δ subunit from the original F1 through its N-terminal region in these experiments, because if it did not, then rebinding of F1 to the membranes would not have occurred and proton pumping would not have been seen.
All the experiments reported in Table 3 were carried out with wild-type isolated α subunit. A parallel series of experiments established that αW513F mutant α-subunit gave similar results to wild-type (data not shown).
Binding of δ subunit to isolated α subunit
When pure δ subunit is mixed with δ-depleted F1 (α3β3γε complex) it binds with 1/1 stoichiometry and the fluorescence signal due to the single natural Trp in δ at residue δ-28 is enhanced by 50% at 325 nm (8). The same fluorescence response is seen when δ forms a 1/1 complex with the synthetic peptide “αN1-22” which has the sequence of the first 22 residues of α subunit (11,13). This has provided a sensitive method by which to probe the interaction of δ with α in the F1 catalytic unit and with the N-terminal region of α.
Here we performed the same type of experiment, under the same conditions, but using pure isolated α subunit, either titrating α with pure δ or vice versa. We used both the wild-type isolated α and also the mutant αW513F; in the latter case the only Trp present was the δ-28 residue. Where wild-type α was used, we confirmed that δ did not quench the fluorescence of the single natural Trp at residue α-513 by use of δW28L mutant. We also tested δ′, the fragment of δ that contains just the N-terminal domain. We varied concentrations of α and δ, included Mg2+ or excluded it with EDTA, added 1 mM ATP, ADP, MgATP, MgADP, varied the pH, added different salts, added isolated β subunit and/or “bST34-156”, the soluble cytoplasmic portion of the b subunit (19,28). Under no condition were we able to detect any significant enhancement of the fluorescence of δ Trp fluorescence in presence of isolated α. Addition of αN1-22 peptide to the mixture of δ and isolated α at the end of the experiment always did elicit the expected fluorescence response. These experiments showed that isolated α does not bind to pure δ subunit, and does not compete for binding with αN1-22.
A second type of experiment was used to detect whether α/δ binding occurred. Isolated α subunit was incubated with NiNTA-agarose slurry in presence of δ subunit (±bST34-156). The slurry was transferred to small columns and bound protein was eluted with 100 mM imidazole. Then SDS-gel electrophoresis was performed to identify the NiNTA-bound protein. Isolated α subunit bound quantitatively to the resin. However, neither δ nor bST34-156 were found to bind along with α to any extent above a minor background level of binding seen even in absence of α in control experiments.
In a third type of experiment, gel filtration was used. The column was 100 cm × 1.5 cm containing Sephacryl S-100 equilibrated in 50 mM HEPES/NaOH pH 7.0, 5 mM MgSO4. Pure isolated α eluted in a single sharp peak at 91.5 ml, pure δ eluted in a single sharp peak at 103.2 ml. An equimolar mixture of α and δ gave the same two sharp peaks at the same elution volumes and no other peak was evident.
Therefore, three types of experiment failed to detect binding of δ to isolated α subunit. It should be borne in mind that the peptide αN1-22 binds to δ with a Kd of 0.13 μM (11), and δ binds to δ-depleted F1 with Kd of 1.4 nM (8), thus all three negative experiments are meaningful.
Trypsin proteolysis of isolated α-subunit and δ-depleted F1
Dunn et al. (7) demonstrated that trypsin removed the first 15 residues from α subunit in intact F1, forming an α′, and that the resultant F1 was unable to bind δ subunit. Under conditions described in (7) further proteolysis of α′ was limited and the resultant F1 retained full ATPase activity. This experiment therefore represented a potential avenue to study whether the N-terminal region of pure isolated α-subunit was available for proteolysis or whether it was in some way sequestered. However, the experiment was not straightforward because as Senda et al. (36) had reported, isolated α-subunit, unlike α present in F1 complex, readily undergoes multiple cleavages by trypsin, producing much smaller polypeptides than the α′ seen by Dunn et al. In addition, we decided to use δ-depleted F1 rather than intact F1. This should provide a better comparison with isolated α subunit because in neither case is there any δ bound to the N-terminal region of α.
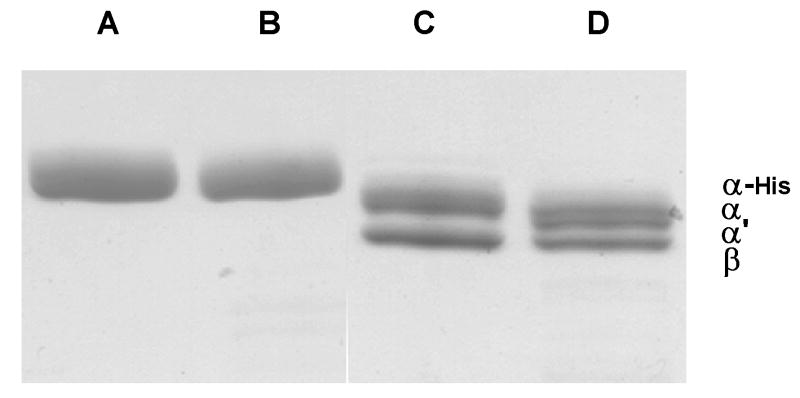
We found that the α subunit in δ-depleted F1 was cleaved at the N-terminus to produce α′ as originally described by Dunn et al. (7) in their work with intact F1, but that cleavage occurred much more quickly with δ-depleted F1. Thus short times of proteolysis were necessary. Figure 2 shows a typical experiment. It is seen that after just two minutes of digestion with trypsin the α′ species was clearly evident in the δ-depleted F1, yet in the same experiment with isolated α subunit, α′ was not at all evident. If the experiment was continued for longer time (or if higher trypsin concentration were used) the full-size α band in the δ-depleted F1 decreased in intensity further, however cleavage of the other subunits including α′ did begin to occur. The behavior of the full-size α band in isolated α was different. With increased time or higher trypsin, it also began to decrease in intensity, but never, in multiple different experiments under varied conditions, was the α′ band seen. It may also be noted that no evidence of the α′ band can be seen in the figures presented by Senda et al. (36).
Figure 2. Trypsin proteolysis of the N-terminal region of the α subunit in δ-depleted F1 and in the isolated α subunit.
Trypsin proteolysis was carried out as described in EXPERIMENTAL PROCEDURES. Briefly, δ-depleted F1 or isolated α subunit (2.3 mg/ml) were incubated with trypsin (2.3 μg/ml) for 2 min at room temperature. Reaction was stopped by addition of PMSF with immediate boiling in SDS-gel buffer and samples were run on SDS-gels. A. 1 μg isolated α subunit, zero time (PMSF added before trypsin). B. 1 μg isolated α subunit, 2 min incubation with trypsin. C. 2 μg δ-depleted F1, zero time. D. 2 μg δ-depleted F1, 2 min incubation with trypsin.
These data demonstrate that the N-terminal region that is available for proteolysis by trypsin in the F1 complex is not available for proteolysis in isolated α subunit. We also tested chymotrypsin, S. aureus V8 protease and proteinase K, and none of them generated an α′ band from isolated α subunit.
Does bST34-156 bind to isolated α subunit?
“bsol” is the generic name for the cytoplasmic fragment of the b subunit which may be expressed and purified in soluble form (37). bsol binds to the δ subunit through its C-terminal end (38,39). In recent work we found that one version of bsol called bST34-156, containing residues b-34 through to the C-terminal b-156 (28) binds directly to F1 in Mg2+-sensitive manner with Kd around 100 nM (19), thus demonstrating quantitatively that binding of the cytoplasmic portion of the b subunit directly to F1 contributes to stator resistance. To establish binding parameters (13) we used F1 enzymes that each had Trp inserted at a unique position in α or β subunits, and measured the enhancement of fluorescence seen upon titration with bST34-156. Two Trp residues that gave substantial enhancement of fluorescence were αL55W and αF406W. We therefore combined each of these mutations with αW513F in the construct for expression of isolated α subunit and purified the two mutant α subunits, namely αL55W/W513F and αF406W/W513F.
Upon titration with bST34-156 neither of these mutant α-subunits showed any change in fluorescence signal. Addition of wild-type or δW28L pure δ subunit, addition of nucleotide, or a wide range of other varied conditions, failed to elicit any fluorescence response. The data indicated therefore that bST34-156 did not bind to isolated α subunit. We described experiments above in which isolated α subunit failed to bind δ subunit in NiNTA-agarose “pull down” experiments. Those experiments showed that isolated α subunit also failed to bind bST34-156.
DISCUSSION
General
The original aim of this work was to express and purify α subunit of the F1 portion of ATP synthase from E. coli, demonstrate its native state and functional integrity, and then proceed to use it as a covenient material for mutagenic and biophysical analysis of binding of δ subunit to the N-terminal region of α. Additionally there was the possibility that binding of the cytoplasmic portion of the b subunit (“bsol”) to isolated α might also occur and be subject to similar analyses, which could illuminate the mode of direct binding of b subunit to F1.
Expression and purification of wild-type and mutant α subunits containing a C-terminal 6-His tag to facilitate rapid purification was accomplished, and assays established functional integrity and native structure. Isolated α was found to be monomeric in solution with the expected molecular size, Trp analysis confirmed its purity, and nucleotide binding analyses confirmed its ability to bind ATP, ADP, MgATP and MgADP. Kd values were established for nucleotide binding, and showed that the Mg-nucleotides bound ~4 times more tightly than the Mg-free forms, and that ATP and MgATP bound 10 times more tightly than ADP and MgADP. It was shown that isolated α subunit was fully competent to reassociate with other F1 subunits to form a complex with normal ATPase activity, and that this complex was able to rebind to F1-depleted membranes to restore ATP-driven proton pumping.
To our initial surprise isolated α subunit did not bind to pure δ subunit or to δ′, the N-terminal domain of δ. This was disappointing since it meant that the original idea of using isolated α to characterize α/δ interaction was untenable. However, N-terminal analysis confirmed that the normal N-terminus of α was present, i.e that no proteolytic “clipping” had occurred during expression or purification, and proteolytic cleavage experiments showed that in isolated α the N-terminal region was not exposed. This latter finding was in contrast to the situation in intact F1 and in δ-depleted F1. On further reflection therefore it was apparent that the results indicated important features of the assembly of the stator with F1 in vivo, and indeed that they might have been anticipated.
Assembly of δ, α, and other F1 subunits in E. coli ATP synthase
Earlier work in the laboratory of L. Heppel established that E. coli F1 can be dissociated in vitro into its component subunits α, β, γ, δ, and ε, and reassociated (26,34,40). Among complexes that can readily be formed in vitro are α3β3γ, α3β3γδ, α3β3γε, and α3β3γδε, but not α3β3 (see also ref. 33 and footnote 3).
Beginning with (41) there was an early flurry of work on assembly of E. coli ATP synthase in vivo, which was well-summarized by Brusilow (42). This early work focussed on assembly of the Fo sector and control of its proton permeability. Intriguingly, δ subunit was inferred to play a role in enhancing proton permeability of Fo synthesized and assembled in vivo (43) although the basis for this effect has not yet been explained. More recent work has also focussed on Fo assembly, demonstrating YidC- and Sec/SRP-involvement for insertion of subunits a, b and c (44–46). There is little experimental data regarding the order of assembly of F1 subunits in vivo, although in ref. 41 it was speculated that the F1 complex was built up incrementally on an Fo template, following a pathway clearly different from that followed in in vitro reconstitution experiments. However it is also well-established that in E. coli strains carrying various mutant forms of ATP synthase, ATPase activity due to partial or intact F1 complexes can accumulate in the cytoplasmic fraction.
The order of genes in the ATP synthase operon in E. coli is uncIBEFHAGDC. The genes are transcribed into a polycistronic message and translated by polysomes (42). Thus there is the possibility of in cis assembly of F1 subunits as discussed in (42). Specifically in relation to the δ and α subunits whose genes (uncH and uncA) are contiguous on the message, given the high affinity of binding seen between δ and the αN1-22 synthetic peptide (0.13 μM, ref. 11) one could predict that newly-translated δ and α subunits might immediately form an αδ complex with 1/1 stoichiometry. However, if this did happen, it is difficult to see how the stoichiometry of one δ to three α in the final F1 (α3β3γδε) could be achieved. It is relevant that in (47) it was reported that α is translated at a rate somewhat greater than δ, opening up the possibility that not all newly-synthesized α would bind δ. On the other hand if the N-terminal region of newly-translated α subunit were sequestered in some way then binding of δ would be temporarily prevented, providing a particularly efficient assembly pathway. The data presented in this paper show that this is indeed the case.
We propose that during assembly of ATP synthase in vivo, before δ/α interaction can occur α subunit must first be incorporated with β into the α3β3 hexagon (this probably requires also the γ subunit); only then does the N-terminal region of α become available for binding δ. After the first δ binds, steric hindrance presumably prevents attachment of a second or third δ. How the N-terminal region is sequestered in isolated α subunit can only be guessed at from current data. Presumably it involves binding of residues α1-22 to the six-stranded beta-barrel domain of α that encompasses residues α24-95. One can postulate that the N-terminal region of α binds to a surface of the six-stranded beta-barrel domain that normally interacts with β subunit in the α3β3 hexagon, and that when the α3β3 hexagon forms the N-terminal region of α is displaced by β and becomes available for binding to δ. As has been reviewed by Weber (6), there is evidence that binding to δ itself leads to an increase in ordered helical structure in the N-terminal residues 1-22 of α. Prior to binding to δ, the N-terminal region of α might exist in a more extended conformation that could reach some way down the surface of the beta-barrel domain on isolated α.
Direct binding of b subunit to F1 may also require the α3β3 hexagon
As discussed in INTRODUCTION we recently established by a fluorimetric technique that a “bsol” construct named bST34-156 showed direct binding to F1 with high affinity (19). Diez et al. (48) and Krebstakies et al. (49) have also reached the same conclusion, the latter study actually using full-length b subunit. In regard to the possible location of the binding site(s) on F1 for b subunit, there are several publications on this point, but as reviewed by Weber (6), they are not in agreement. Indeed they suggest several different binding sites, either to α or to β, in the upper or lower portions of the α3β3 hexagon, or to the α/β clefts associated with catalytic or noncatalytic nucleotide binding sites.
In this paper we tested whether isolated α subunit could bind bST34-156. We had found in (19) that two Trp residues, introduced singly into α subunit of intact and otherwise Trp-free F1 at residue α-55 (located close to the noncatalytic α/β cleft in the upper region of F1) or at residue α-406 (located close to the catalytic cleft in the lower region of F1) both gave enhancement of fluorescence signal when bST34-156 was added. However, we found here that when the same Trp residues were inserted in isolated α subunit, addition of bST34-156 was without effect on fluorescence. In NiNTA-agarose “pull-down” experiments, no binding of bST34-156 to isolated α was seen. From these data we would suggest tentatively that direct binding of b subunit cytoplasmic region to F1 may require prior formation of the α3β3 complex, just as binding of δ appears to do.
Acknowledgments
We thank Sarah Lockwood for outstanding technical assistance.
Supported by: NIH grant GM25349 to AES.
Footnotes
Abbreviations used: DTT, dithiothreitol; PMSF, phenylmethylsulfonyl fluoride; IPTG, isopropropylthiogalactoside, αN1-22, synthetic peptide consisting of residues 1-22 of ATP synthase α subunit.
E. coli residue numbers used throughout.
We have carried out extensive experimental attempts to purify a stable complex of α and β subunits from the E. coli enzyme without success (LaReau, R.D. and Senior, A.E., unpublished results).
References
- 1.Senior AE, Weber J. Happy motoring with ATP synthase. Nature Struct Mol Biol. 2004;11:110–112. doi: 10.1038/nsmb0204-110. [DOI] [PubMed] [Google Scholar]
- 2.Weber J, Senior AE. ATP synthesis driven by proton transport in F1Fo-ATP synthase. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 3.Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by F1Fo-ATP synthase. Biochim Biophys Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 4.Noji H, Yoshida M. The rotary machine in the cell, ATP synthase. J Biol Chem. 2001;276:1665–1668. doi: 10.1074/jbc.R000021200. [DOI] [PubMed] [Google Scholar]
- 5.Leslie AGW, Walker JE. Structural model of F1-ATPase and the implications for rotary catalysis. Phil Trans R Soc Lond B. 2000;355:465–472. doi: 10.1098/rstb.2000.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber J. ATP synthase: Subunit-subunit interactions in the stator stalk. Biochim Biophys Acta. 2006;1757:1162–1170. doi: 10.1016/j.bbabio.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn SD, Heppel LA, Fullmer CS. The NH2-terminal portion of the α subunit of Escherichia coli F1-ATPase is required for binding the δ subunit. J Biol Chem. 1980;255:6891–6896. [PubMed] [Google Scholar]
- 8.Weber J, Wilke-Mounts S, Senior AE. Quantitative determination of binding affinity of δ subunit in Escherichia coli F1-ATPase: effects of mutation, Mg2+ and pH on Kd. J Biol Chem. 2002;277:18390– 18396. doi: 10.1074/jbc.M201047200. [DOI] [PubMed] [Google Scholar]
- 9.Häsler K, Panke O, Junge W. On the stator of rotary ATP synthase: the binding strength of subunit δ to (αβ)3 as determined by fluorescence correlation spectroscopy. Biochemistry. 1999;38:13759–13765. doi: 10.1021/bi991236m. [DOI] [PubMed] [Google Scholar]
- 10.Weber J, Wilke-Mounts S, Senior AE. Identification of the F1- binding surface on the δ subunit of ATP synthase. J Biol Chem. 2003;278:13409– 13416. doi: 10.1074/jbc.M212037200. [DOI] [PubMed] [Google Scholar]
- 11.Weber J, Muharemagic A, Wilke-Mounts S, Senior AE. F1Fo- ATP synthase. Binding of δ subunit to a 22-residue peptide mimicking the N- terminal region of α subunit. J Biol Chem. 2003;278:13263–13626. doi: 10.1074/jbc.C300061200. [DOI] [PubMed] [Google Scholar]
- 12.Wilkens S, Borchardt D, Weber J, Senior AE. Structural characterization of the interaction of the δ and α subunits of the Escherichia coli F1Fo-ATP synthase. Biochemistry. 2005;44:11786–11794. doi: 10.1021/bi0510678. [DOI] [PubMed] [Google Scholar]
- 13.Weber J, Muharemagic A, Wilke-Mounts S, Senior AE. Analysis of sequence determinants of F1Fo-ATP synthase in the N-terminal region of α subunit for binding of δ subunit. J Biol Chem. 2004;279:25673–25679. doi: 10.1074/jbc.M402738200. [DOI] [PubMed] [Google Scholar]
- 14.Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Wilkens S, Dunn SD, Chandler J, Dahlquist FW, Capaldi RA. Solution structure of the N-terminal domain of the δ subunit of the Escherichia coli ATP synthase. Nature Struct Biol. 1997;4:198–201. doi: 10.1038/nsb0397-198. [DOI] [PubMed] [Google Scholar]
- 16.Carbajo RJ, Kellas FA, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D. Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an α subunit. J Mol Biol. 2005;351:824–838. doi: 10.1016/j.jmb.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Hundal T, Norling B, Ernster L. Lack of ability of trypsin-treated mitochondrial F1-ATPase to bind the oligomycin-sensitivity conferring protein (OSCP) FEBS Lett. 1983;184:677–701. doi: 10.1016/0014-5793(83)81038-2. [DOI] [PubMed] [Google Scholar]
- 18.Rao R, Pagan J, Senior AE. Directed mutagenesis of the strongly conserved lysine 175 in the proposed nucleotide binding domain of α subunit from Escherichia coli F1-ATPase. J Biol Chem. 1988;263:15957–15963. [PubMed] [Google Scholar]
- 19.Weber J, Wilke-Mounts S, Nadanaciva S, Senior AE. Quantitative determination of direct binding of b subunit to F1 in Escherichia coli F1Fo-ATP synthase. J Biol Chem. 2004;279:11253–11258. doi: 10.1074/jbc.M312576200. [DOI] [PubMed] [Google Scholar]
- 20.Senior AE, Langman L, Cox GB, Gibson F. Oxidative phosphorylation in Escherichia coli. Characterization of mutant strains in which F1-ATPase contains abnormal β subunits. Biochem J. 1983;210:395–403. doi: 10.1042/bj2100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taussky HH, Shorr E. A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem. 1953;202:675–685. [PubMed] [Google Scholar]
- 22.Perlin DS, Cox DN, Senior AE. Integration of F1 and the membrane sector of the proton ATPase of Escherichia coli. J Biol Chem. 1983;258:9793–9800. [PubMed] [Google Scholar]
- 23.Ketchum CJ, Al-Shawi MK, Nakamoto RK. Intergenic suppression of the γM23K uncoupling mutation in F1Fo-ATP synthase by βGlu- 381 substitutions: the role of the β380DELSEED386 segment in energy coupling. Biochem J. 1998;330:707–712. doi: 10.1042/bj3300707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao R, Al-Shawi MK, Senior AE. Trinitrophenyl-ATP and -ADP bind to a single nucleotide site on isolated β subunit of Escherichia coli F1- ATPase. J Biol Chem. 1988;263:5569–5573. [PubMed] [Google Scholar]
- 25.Weber J, Lee RSF, Grell E, Wise JG, Senior AE. On the location and function of tyrosine β311 in the catalytic site of Escherichia coli F1- ATPase. J Biol Chem. 1992;267:1712–1718. [PubMed] [Google Scholar]
- 26.Dunn SD, Futai M. Reconstitution of a functional coupling factor from the isolated subunits of Escherichia coli F1-ATPase. J Biol Chem. 1980;255:113–118. [PubMed] [Google Scholar]
- 27.Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Dunn SD, Chandler J. Characterization of the b2δ complex from Escherichia coli F1Fo-ATP synthase. J Biol Chem. 1998;273:8646–8651. doi: 10.1074/jbc.273.15.8646. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Brusilow WSA, Simoni RD. In vivo evidence for the role of the ε subunit as an inhibitor of the proton translocating ATPase of Escherichia coli. J Bacteriol. 1984;160:1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandeyar M, Weiner M, Hutton C, Batt C. A novel method for site-specific mutagenesis. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 31.Weber J, Wilke-Mounts S, Grell E, Senior AE. Tryptophan fluorescence provides a direct probe of nucleotide binding in the noncatalytic sites of Escherichia coli F1-ATPase. J Biol Chem. 1994;269:11261–11268. [PubMed] [Google Scholar]
- 32.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 33.Al-Shawi MK, Parsonage D, Senior AE. Adenosine triphosphatase and nucleotide binding activity of isolated β subunit preparations from Escherichia coli F1Fo-ATP synthase. J Biol Chem. 1990;265:5595–5601. [PubMed] [Google Scholar]
- 34.Sternweis P. The ε subunit of Escherichia coli coupling factor 1 is required for its binding to the cytoplasmic membrane. J Biol Chem. 1978;253:3123– 3128. [PubMed] [Google Scholar]
- 35.Rao R, Senior AE. Properties of hybrid F1-ATPase enzymes suggest that a cyclical catalytic mechanism involving three catalytic sites occurs. J Biol Chem. 1987;262:17450–17454. [PubMed] [Google Scholar]
- 36.Senda M, Kanazawa H, Tsuchiya T, Futai M. Conformational change of the α subunit of Escherichia coli F1-ATPase: ATP changes the trypsin sensitivity of the subunit. Arch Biochem Biophys. 1983;220:398–404. doi: 10.1016/0003-9861(83)90429-0. [DOI] [PubMed] [Google Scholar]
- 37.Dunn SD. The polar domain of the b subunit of Escherichia coli F1Fo- ATPase forms an elongated dimer that interacts with the F1-sector. J Biol Chem. 1992;267:7630–7636. [PubMed] [Google Scholar]
- 38.McLachlin DT, Bestard JA, Dunn SD. The b and δ subunits of the Escherichia coli F1Fo-ATP synthase interact via residues in their C-terminal regions. J Biol Chem. 1998;273:15162–15168. doi: 10.1074/jbc.273.24.15162. [DOI] [PubMed] [Google Scholar]
- 39.Dunn SD, McLachlin DT, Revington M. The second stalk of Escherichia coli F1Fo-ATP synthase. Biochim Biophys Acta. 2000;1458:356–363. doi: 10.1016/s0005-2728(00)00086-4. [DOI] [PubMed] [Google Scholar]
- 40.Sternweis PC, Smith JB. Characterization of the purified membrane attachment (δ) subunit of the proton translocating adenosine triphosphatase from Escherichia coli. Biochemistry. 1977;16:4020–4025. doi: 10.1021/bi00637a013. [DOI] [PubMed] [Google Scholar]
- 41.Cox GB, Downie JA, Langman L, Senior AE, Ash G, Fayle DRH, Gibson F. Assembly of the adenosine triphosphatase complex in Escherichia coli: Assembly of Fo is dependent on the formation of specific F1 subunits. J Bacteriol. 1981;148:30–42. doi: 10.1128/jb.148.1.30-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brusilow WSA. Assembly of the Escherichia coli F1Fo-ATPase, a large multimeric membrane-bound enzyme. Mol Microbiol. 1993;9:419–424. doi: 10.1111/j.1365-2958.1993.tb01703.x. [DOI] [PubMed] [Google Scholar]
- 43.Monticello RA, Brusilow WSA. Role of the δ subunit in enhancing proton conduction through the Fo of the Escherichia coli F1Fo-ATPase. J Bacteriol. 1994;176:1383–1389. doi: 10.1128/jb.176.5.1383-1389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi L, Jiang F, Chen M, Cain B, Bolhuis A, Dalbey RE. YidC Is strictly required for membrane insertion of subunits a and c of the F1F0 ATP synthase and SecE of the SecYEG translocase. Biochemistry. 2003;42:10537–10544. doi: 10.1021/bi034309h. [DOI] [PubMed] [Google Scholar]
- 45.Yi L, Celebi N, Chen M, Dalbey RE. Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1Fo-ATP synthase. J Biol Chem. 2004;279:39260– 39267. doi: 10.1074/jbc.M405490200. [DOI] [PubMed] [Google Scholar]
- 46.Kol S, Turrell BR, de Keyzer J, van der Laan M, Nouen N, Driessen AJM. YidC mediated membrane insertion of assembly mutants of subunit c of the Escherichia coli F1Fo-ATPase. J Biol Chem. 2006;281:29762–29768. doi: 10.1074/jbc.M605317200. [DOI] [PubMed] [Google Scholar]
- 47.Brusilow WSA, Klionsky DJ, Simoni RD. Differential polypeptide synthesis of the proton-translocating ATPase of Escherichia coli. J Bacteriol. 1982;151:1363–1371. doi: 10.1128/jb.151.3.1363-1371.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diez M, Börsch M, Zimmermann B, Turina P, Dunn SD, Gräber P. Binding of the b subunit in Escherichia coli F1Fo-ATP synthase. Biochemistry. 2004;43:1054–1064. doi: 10.1021/bi0357098. [DOI] [PubMed] [Google Scholar]
- 49.Krebstakies T, Zimmermann B, Gräber P, Altendorf K, Börsch M, Greie JC. Both rotor and stator subunits are necessary for efficient binding of F1 to Fo in functionally assembled Escherichia coli F1Fo-ATP synthase. J Biol Chem. 2005;280:33338–33345. doi: 10.1074/jbc.M506251200. [DOI] [PubMed] [Google Scholar]
- 50.Weber J, Wilke-Mounts S, Senior AE. Cooperativity and stoichiometry of substrate binding to the catalytic sites of Escherichia coli F1- ATPase. J Biol Chem. 1994;269:20462–20467. [PubMed] [Google Scholar]