Abstract
The FGF signaling pathway plays essential roles in endochondral ossification by regulating osteoblast proliferation and differentiation, chondrocyte proliferation, hypertrophy, and apoptosis. FGF signaling is controlled by the complementary action of both positive and negative regulators of the signal transduction pathway. The Spry proteins are crucial regulators of receptor tyrosine kinase-mediated MAPK signaling activity. Sprys are expressed in close proximity to FGF signaling centers and regulate FGFR-ERK-mediated organogenesis. During endochondral ossification, Spry genes are expressed in pre-hypertrophic and hypertrophic chondrocytes. Using a conditional transgenic approach in chondrocytes in vivo, the forced expression of Spry1 resulted in neonatal lethality with accompanying skeletal abnormalities resembling thanatophoric dysplasia II, including increased apoptosis and decreased chondrocyte proliferation in the presumptive reserve and proliferating zones. In vitro chondrocyte cultures recapitulated the inhibitory effect of Spry1 on chondrocyte proliferation. In addition, overexpression of Spry1 resulted in sustained ERK activation and increased expression of p21 and STAT1. Immunoprecipitation experiments revealed that Spry1 expression in chondrocyte cultures resulted in decreased FGFR2 ubiquitination and increased FGFR2 stability. These results suggest that constitutive expression of Spry1 in chondrocytes results in attenuated FGFR2 degradation, sustained ERK activation, and upregulation of p21Cip and STAT1 causing dysregulated chondrocyte proliferation and terminal differentiation.
Introduction
During endochondral bone development, a cartilage template forms when mesenchymal cells condense and differentiate into chondrocytes. These chondrocytes then undergo a program of proliferation, hypertrophy, calcification, and cell death. Several families of signaling molecules have been identified that act at multiple points in skeletal development. FGFR3 has been extensively investigated during endochondral ossification. Mice with a loss-of-function mutation in Fgfr3 have abnormal long bone growth (Colvin et al., 1996; Deng et al., 1996), whereas mice with constitutively activating mutations of Fgfr3 exhibit dwarfism (Chen et al., 1999; Chen et al., 2001; Iwata et al., 2001). In humans, Fgfr3 mutations result in several skeletal disorders, including achondroplasia (Rousseau et al., 1994), hypochondroplasia (Bellus et al., 1995), and thanatophoric dysplasia type I and type II (TDII) (Rousseau et al., 1996; Rousseau et al., 1995; Tavormina et al., 1995). Although the STAT1-p21Cip pathway is implicated in achondroplasia mediated by Fgfr3 mutations and in Fgf2 transgenic mice (Sahni et al., 2001; Su et al., 1997), the ERK pathway is also a major pathway transmitting FGFR-mediated signals for chondrocyte proliferation, differentiation, and apoptosis (Ornitz and Marie, 2002). Transgenic mice expressing constitutively active MEK1 in chondrocytes exhibit the inhibition of hypertrophic differentiation of chondrocytes and a delay of endochondral ossification without affecting chondrocyte proliferation (Murakami et al., 2004). This effect is independent of the Stat1 pathway, however constitutively active MEK1 rescues the Fgfr3−/− phenotype. Targeted overexpression of CNP in chondrocytes counteracts dwarfism in a mouse model of achondroplasia by reversing the decrease in extracellular matrix synthesis in the growth plate by inhibition of FGFR-mediated ERK signaling (Yasoda et al., 2004). Thus, FGF signaling plays crucial roles in limb specification and endochondral bone formation through the STAT1-p21Cip and ERK pathways.
Spry family members are feedback inhibitors of FGFR-mediated activation of the ERK pathway, as well as other receptor tyrosine kinases (RTKs) (Mason et al., 2006; Tsang and Dawid, 2004). In Drosophila, null mutations in Spry cause hyperactivation of the FGF pathway and ectopic secondary tracheal branching (Hacohen et al., 1998). Overexpression of Spry during primary branch outgrowth causes the opposite effect, inhibiting the FGF inductive pathway and blocking secondary branching. Mammalian Spry proteins colocalize with FGFs, most notably Fgf8 and Fgf3, during early vertebrate embryogenesis in regions of the midbrain-hindbrain boundary, limb buds, and craniofacial primordia (Minowada et al., 1999). FGF signaling induces Spry expression in these tissues, and thus Spry acts as a feedback inhibitor of the pathway that induces its expression (Mason et al., 2006; Minowada et al., 1999). Conditional deletion of Spry1 with HoxB7-Cre results in mouse embryos with supernumerary ureteric buds, resulting in the development of multiple ureters and multiple kidneys (Basson et al., 2005). This aberrant development is due to increased sensitivity of the Wolffian duct to GDNF/RETS signaling. In addition, targeted deletion of Spry2 causes defects of the inner ear (Shim et al., 2005) and diastema development (Klein et al., 2006), with no other discernable phenotype (Klein et al., 2006; Shim et al., 2005). Targeted deletion of Spry4 also causes defects in tooth development, and syndactyly (Taniguchi et al., 2007). Retroviral overexpression of mouse Spry2 in developing wing buds of chick embryos retards chondrocyte differentiation and limb bud formation (Minowada et al., 1999). Mice with both Spry2 and Spry4 null mutant alleles deleted show multiple craniofacial and skeletal defects, although the mechanisms for these defects have not been addressed (Taniguchi et al., 2007). Despite recent advances in understanding the roles of Spry proteins on the regulation of FGF receptor signaling during embryogenesis, little is known about Spry’s regulation of endochondral ossification.
Fgfr3 is expressed in the growth plate in an increasing gradient from reserve to proliferating chondrocytes, and it is weakly expressed in hypertrophic chondrocytes (Peters et al., 1993); whereas Fgfr2 expression is apparent in skeletogenic mesenchymal condensations at early stages of development, and is localized to the perichondrium and nascent epiphyseal plate at late stages of development. Fgfr1 is expressed in the perichondrium and hypertrophic chondrocytes (Ornitz, 2001; Ornitz and Marie, 2002). The close association of Spry expression with FGF signaling centers prompted us to examine the expression of Spry family members in the developing long bones of late gestation mouse embryos. Our data indicate that Spry1, Spry2, and Spry4 are expressed in prehypertrophic and hypertrophic chondrocytes.
Given the potential for functional redundancy between Spry family members, and possible compensatory mechanisms that may function due to targeted deletion of a single family member, we chose a transgenic mouse approach to gain insight into Spry function in growth plate development. We generated conditional transgenic mice in which mSpry1 was over-expressed in Col2α1-expressing chondrocytes, and analyzed the morphological changes of long bones and the possible mechanisms that may be involved. Results reported here indicate that Spry1 dysregulates the duration of FGFR-mediated ERK activation in chondrocytes through attenuating FGFR ubiquitinylation. Thus, overexpression mSpry1 in chondrocytes results in a thanatophoric dysplasia type II-like phenotype similar to that of constitutively active Fgfr3 mutants and FGF2 transgenic mice.
MATERIALS AND METHODS
Histology, immunohistochemistry, immunofluorescence, and in situ hybridization
For histological analysis, embryos were fixed in 4% paraformaldehyde, embryo limbs were embedded in OCT, and serial 7 μm frozen sections were prepared using standard procedures. For immunohistochemistry, antibodies to phospho-MEK1/2 (Cell Signaling), Spry1, Spry2, Spry4, p21Cip (Santa Cruz), and STAT1 (Cell Signaling) were used. For in situ hybridization, plasmids were linearized with the appropriate restriction enzymes and digoxygenin-labeled riboprobes were generated using a digoxygenin RNA labeling kit (Roche) according to the manufacturer’s protocol. Probes for aggrecan (Agc), Indian Hedgehog (Ihh), collagen10α1 (Col10α1), collagen 2α1 (Col2α1), PTHrP receptor (Ppr), and osteocalcin (OC), were provided by Volkhard Lindner (Maine Medical Center Research Institute). The full length Spry1/pCS2+ construct was linearized with EcoRI, and the RNA probe was generated using T7 promoter. For Spry2 and Spry4, cDNA was generated by PCR using full length Spry2/pCS2+ and Spry4/pCS2+ respectively, and cloned into TOPO vector (Invitrogen). For p21Cip, MMP-13, and MMP-9 cDNA was generated by RT-PCR, and cloned into TOPO vector (Invitrogen). Antisense/TOPO constructs were linearized by EcoRV, and RNA probes were generated using T7 promoter. Primers for PCR or RT-PCR were: 5′-AGGCCAGAGCTCAGAGTGGCAACGG-3′ (Forward) and 5′-CTCCCATCGCTGACCATCGCGTACAAC-3′ (Reverse) for Spry2, 5′-ATGGAGCCCCCGGTTCCACAG-3′ (Forward) and 5′-AGAAGTGCTTGTCTAGCTCTG-3′ (Reverse) for Spry4, 5′-GCACCATGTCCAATCCTGGTG-3′ (Forward) and 5′-TCAGGGTTTTCTCTTGAGAAG-3′ (Reverse) for p21Cip, 5′-GGCACCATGCATTCAGCTATCCTG-3′ (Forward) and 5′-CACCACAATATGGAATTTGTTGGCATGAC-3′ (Reverse) for MMP-13, 5′-CTGCTCCTGGCTCTCCTGGCTTTCGGCTG-3′ (Forward) and 5′-CTCGTCGTCGTCGAAATGGGCATCTC-3′ (Reverse) for MMP-9. The sections were pretreated with proteinase K and hybridized with the probes according to standard procedures. Sense probes were used as controls. The color was developed with BM purple (Roche), and counterstained with nuclear fast red.
Cell culture and differentiation
293T cells, as well as 293T cells stably expressing Cre-recombinase (Cre8 cells), were grown in DMEM 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere of 5% CO2. The CAGGFP-Spry1 plasmid was transfected into 293T or Cre8 cells using Genejuice (Novagen) transfection reagent according to the manufacturer’s instructions. The prechondrocytic cell line, ATDC5 was maintained in F12/DMEM (1:1), 5% FBS maintenance medium at 37°C in a humidified atmosphere of 5% CO2, and differentiation was induced by addition of 10 μg/ml insulin (Sigma) to the maintenance medium. Primary chondrocytes were isolated by collagenase digestion of long bone growth plates of 5–7 day-old wild-type or CAGGFP-Spry1; CAGGCre-ER™ (Hayashi and McMahon, 2002), bitransgenic mice. The growth plates were carefully dissected free from connective tissue in PBS, and the cartilage was minced into 1 mm3 fragments. The cartilage fragments were placed into 10 ml 0.05% trypsin solution in serum free DMEM medium, and incubated for 25 min at room temperature with moderate agitation. The cartilage fragments were allowed to sediment and the trypsin solution was removed and replaced with 10 ml of 0.3% collagenase in serum free DMEM, and incubated for 30 min at room temperature, followed by replacement with 10 ml of 0.06% collagenase in DMEM medium containing 10% FBS. The fragments and solution were transferred to 10 cm Petri dishes and incubated overnight in an incubator at 37°C in 5% CO2. The cell suspension was transferred to a 50 ml tube and vortexed briefly; the cells were then filtered through a 40 μm nylon mesh (BD, Falcon). The resulting cell suspension was centrifuged at 1,200 rpm for 5 min, and the cells were resuspended in 10 ml maintenance medium and seeded onto a 10 cm dish. Primary chondrocytes from wild-type mice were induced to differentiate by the addition of 10 μg/ml insulin and were harvested at various intervals for up to 18 days.
For growth assays, primary chondrocytes were seeded in triplicate into 12-well plates at a density of 5×104 cells/ml (1 ml/well) in the presence 1 μM tamoxifen (Sigma). Triplicate wells were harvested daily by trypsinization, and counted using a Coulter counter. Student’s t-test was used for statistical analysis. For immunoblotting, the cells were lysed in HNTG buffer (25 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EGTA), containing complete protease inhibitor cocktail (Roche) and phosphatase inhibitors. Equal amounts of cellular protein were subjected to SDS-PAGE on 10% acrylamide gels, and were electrophoretically transferred to nitrocellulose membranes as described (Neilson and Friesel, 1996). Membranes were blocked with 5% nonfat dry milk in TBS, probed with antibodies to Spry1, Spry2, Spry4 (Santa Cruz), phospho-ERK1/2 (Sigma), p42/44ERK1/2 (Cell Signal), p21Cip (Upstate), and the bound antibodies were visualized with horseradish peroxidase conjugated secondary antibodies (BioRad) and enhanced chemiluminescence (ECL, Amersham Biosciences). For immunoprecipitation, the cell lysates were incubated with FGFR2 rabbit antibody (Santa Cruz), precipitated by protein A/G beads (Santa Cruz), and eluted in SDS sample buffer, and subjected to immunoblotting with a ubiquitin antibody (Sigma).
Spry1 transgenic mouse model
The CAGCAT-Z vector (Sakai and Miyazaki, 1997) containing a chicken β-actin gene (CAG) promoter-loxP-chloramphenicol acetyltransferase (CAT) gene-loxP-LacZ was modified by replacing the CAT gene with the GFP open reading frame, and by replacing LacZ with a myc/his-tagged mouse Spry1 open reading frame generated by PCR using gene specific primers. The CAGGFP-Spry1 construct constitutively expresses GFP, and upon Cre-mediated recombination, Spry1 expression is induced, with concomitant loss of GFP marker expression. Transgene activation by Cre recombinase was validated in Cre8 cells in vitro by immunoblotting using anti-myc antibodies (Upstate Biotech). The purified, linearized construct was injected into the pronucleus of FVB oocytes by standard procedures (Maine Medical Center Research Institute Transgenic Mouse Facility). The resulting mice were screened by PCR of genomic DNA for GFP expression, and founder lines validated by genotyping using GFP-specific primers, 5′-CTCGAGCCACCATGAGTAAAGGAGAAGAAC-3′ (Forward) and 5′-CTTCACTATTTGTAGAGTTCATCCATGC-3′ (Reverse), which amplify a 733 bp fragment. CAGGFP-Spry1 founder lines were identified and crossed to Col2α1-Cre transgenic mice (Sakai et al., 2001) (Jackson Laboratory, ME). The presence of Cre recombinase was identified by PCR using primers; 5′-GCATTACCGGTCGATGCAACGAGTG-3′ (Forward) and 5′-GAACGCTAGAGCCTGTTTTGCACGTT-3′ (Reverse), which amplify a 370 bp fragment.
Recombination was confirmed by immunoblot analysis of tail lysates by using an anti-myc antibody. For some experiments, CAGGFP-Spry1 mice were crossed with CAGGCre-ER™ mice (A. McMahon, Harvard). All mice were housed in a pathogen-free environment under light, temperature, and humidity-controlled conditions. For timed pregnancies, the plug date was defined as E0.5. All experimental protocols were reviewed and approved by the Maine Medical Center Institutional Animal Care and Use Committee.
Skeletal preparation and histology
Skeletal preparations were performed as described (Ito et al., 2003). Briefly, timed pregnant females or newborn mice were euthanized by asphyxiation in CO2. Embryos and neonates were skinned, eviscerated, and fixed in 95% ethanol. The skeletons were stained with alcian blue, cleared in 1% KOH, and counterstained with alizarin red.
BrdU and TUNEL labeling
For BrdU incorporation, pregnant females were injected intraperitoneally with 1ml/100g body weight of BrdU (Roche) 2 hrs before embryo harvest. For some experiments, pregnant females were injected with BrdU 36 hrs before collecting the embryos. Immunohistochemical detection of BrdU was performed using a BrdU detection kit (Roche) according to the manufacturer’s instructions. The sections were counterstained with methyl green. BrdU-positive chondrocytes in the reserve and proliferating zones were quantified on a minimum of five sections per embryo using Scion Image software, and expressed as a percentage of total chondrocytes. Statistical analysis was performed using the Student’s t-test. For TUNEL labeling, the fluorescent in situ Cell Death Detection kit (Roche) was used according to the manufacturer’s instructions, and the number of apoptotic cells per section was quantified.
RT-PCR analysis
Total RNA was extracted from ATDC5 cells or cultured primary chondrocytes using Tri-Reagent (Sigma). The RNA was treated with a DNA-free kit (Ambion) according to the manufacturer’s instructions. cDNA was synthesized using 1μg of total RNA and random hexamers using the Cloned AMV First-strand synthesis kit (Invitrogen). The first strand cDNA product was subjected to PCR using PCR Supermix (Invitrogen). The primer sequences for PCR were: 5′-GGGCCTATTAGGACGGTCTC-3′ (Forward) and 5′-CACAGGTATCTGGAGCAGCA-3′ (Reverse) for Spry1; 5′-GGTCTCGGAGCAGTACAAGG-3′ (Forward) and 5′-GTAGGCATGCAGACCCAAAT-3′ (Reverse) for Spry2; 5′-AGGCCAGAGCTCAGAGTGGCAACGG-3′ (Forward) and 5′-CTCCCATCGCTGACCATCGCGTACAAC-3′ (Reverse) for Spry4; 5′-CATGACCCAGCGCTGCAAGG-3′ (Forward) and 5′-CCTGGAAAGCTCTCAGCCGG-3′ (Reverse)for Ihh; 5′-GTCACTGTACTCAAGACTGCAG-3′ (Forward) and 5′-GAAACATTGGCCAGAACAGGAC-3′ (Reverse) for FGFR3; 5′-TGCGACTTCAACAGCAACTC-3′ (Forward) and 5′-GATGGAAATTGTGAGGGAGA-3′ (Reverse) for GADPH.
RESULTS
The expression of Spry family members in developing long bones is limited to prehypertrophic and hypertrophic zones
To begin to understand the biological function of Spry genes in endochondral bone formation, we evaluated endogenous Spry gene expression and protein localization in limb cryosections from E12.5, E14.5, and E17.5 embryos. By in situ hybridization, Spry1 and Spry2 were not detectable in E12.5 limb chondrocytes (data not shown), but were detected in E14.5 and E17.5 in the prehypertrophic and hypertrophic chondrocytes of the long bones (Figure 1A), as well as in the vertebral column, ribs, scapula, and occipital bone (data not shown). Spry4 showed a similar expression pattern (data not shown). Immunohistochemistry with Spry specific antibodies gave similar results for Spry1; however, Spry2 was also detected by immunohistochemistry in the proliferating zone (Figure 1B). The reasons for these differences are unknown but may indicate differences in sensitivity of our in situ hybridization probes and the Spry2 antibodies or potential cross-reactivity of the Spry2 antibodies.
Figure 1.
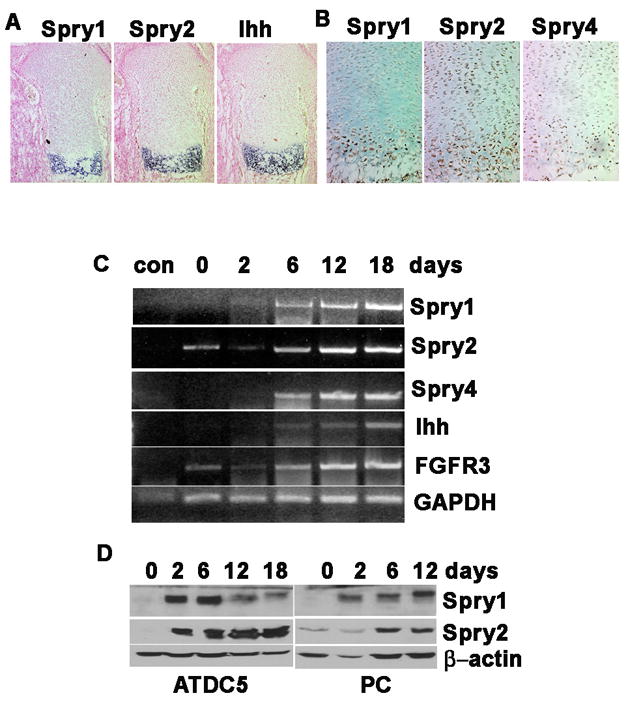
The expression of Spry family members was detected in prehypertrophic and hypertrophic chondrocytes. (A) In situ hybridization for Spry1, Spry2, and Ihh on E17.5 humerus cryosections. Spry expression overlaps with Ihh. (B) Immunohistochemistry for Spry1, Spry2, and Spry4 cryosections of E17.5 humeri. (C, D) ATDC-5 cells or primary chondrocytes were induced to differentiate in the presence of insulin. (B) The expression of Spry1, 2, 4, FGFR3, and Ihh in ATDC-5 was determined by RT-PCR on RNA samples collected on days 0, 2, 6, 12, and 18 days after induction of differentiation (Con, no RT control). (C) Immunoblot detection of Spry1 and Spry2 protein during differentiation ADTC-5 cells (ADTC-5) and primary chondrocytes (PC).
To determine whether the expression of Spry family members is regulated in a temporal manner during chondrocyte differentiation, we employed an in vitro assay using the prechondrocytic ATDC5 cell line and primary mouse chondrocytes induced to differentiate in response to insulin over 18 days. The RT-PCR results indicate that the expression of Spry1, Spry2, and Spry4 were increased in a temporal manner during differentiation of ATDC5 cells, beginning as early as 2 days after the addition of insulin and peaking around day 12 (Figure 1C). This increase paralleled that of the well known chondrocyte differentiation marker Ihh (Figure 1C). Immunoblotting revealed that Spry1 and Spry2 protein levels were increased in a temporal manner in ATDC5 cells. While Spry1 mRNA levels steadily increased during differentiation, Spry1 protein levels increased during the first six days of differentiation and then declined. The discrepancy between mRNA and protein levels at later stages of differentiation may reflect differences in Spry2 protein stability or degradation at specific stages of differentiation. Spry1 and Spry2 protein levels also increased during insulin-induced differentiation of primary mouse chondrocytes (Figure 1D). These results suggest that an increase in the expression of Spry family members may play a role in chondrocyte hypertrophy and maturation.
Conditional expression of Spry1 in chondrocytes
To address the role of Spry1 in chondrocyte development, we established mSpry1-inducible transgenic mouse lines, in which a myc/his tagged mSpry1 open reading frame was cloned into the CAGGFP vector where transgene expression is driven by a CMV enhancer and β-actin promoter. This vector was designated CAGGFP-Spry1. The open reading frame of GFP is placed downstream of the promoter and is flanked by two loxP sites, followed by transcriptional stop sequences to prevent Spry1 transgene expression until Cre-mediated excision of the Floxed GFP sequences (Figure 2). Mating with Col2α1-Cre (Col2-Cre) transgenic mice (Sakai et al., 2001) resulted in forced mSpry1 expression in the Col2α1-expressing chondrocytes. The efficacy of this approach was demonstrated by transient transfection of the CAGGFP-Spry1 plasmid into 293T cells or into Cre recombinase-expressing Cre8 cells. Immunofluorescence microscopy and immunoblotting revealed strong GFP expression without Cre recombinase (Figure 2B and 2C); whereas in Cre8 cells, Spry1myc/his was strongly expressed with concomitant loss of GFP expression (Figure 2B and 2C). To test Cre-mediated recombination in vivo, CAGGFP-Spry1 mice were crossed with Col2α1-Cre mice. Bitransgenic embryos were designated Spry1;Col2-Cre and demonstrated Spry1myc/his and GFP expression by immunoblotting whereas embryos carrying only the GFP transgene did not express detectable Spry1myc/his, as determined by immunoblotting (Figure 2D). Attempts to reveal Spry1 transgene protein expression by immunohistochemistry of embryonic long bone sections were unsuccessful, presumably because the level of Spry1 transgenic protein was below the limit of detection of the myc antibody. Three founder lines were identified that produced similar skeletal phenotypes, and one was chosen for further study.
Figure 2.
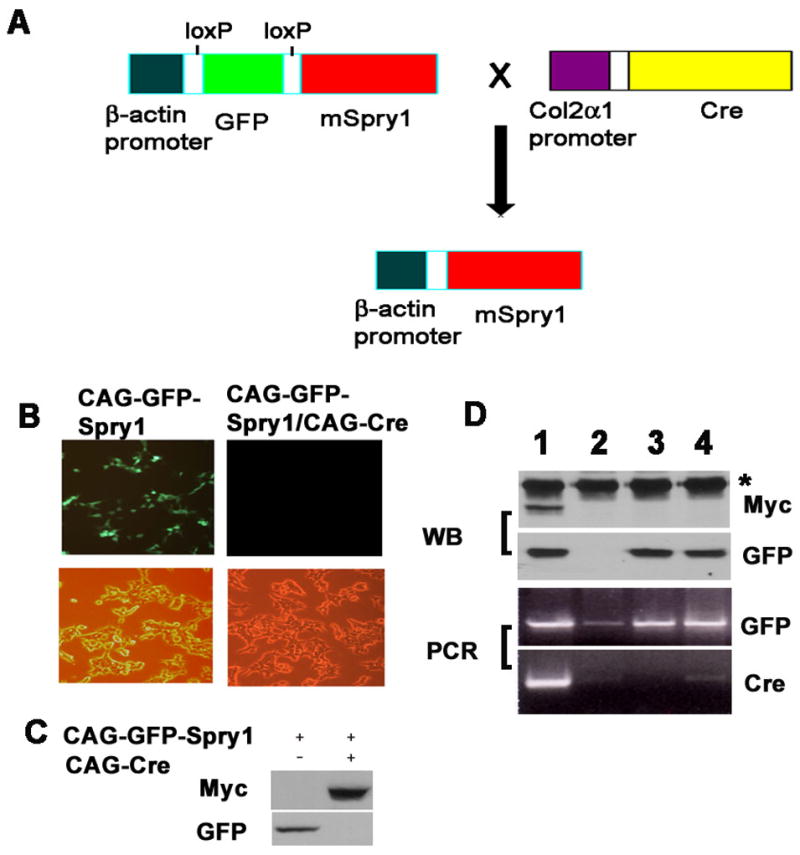
Strategy for the tissue-specific expression of Spry1 in transgenic mice. (A) Schematic representation for the tissue-specific, Cre-inducible expression of Spry1 in Col2a1-expressing chondrocytes. (B) In vitro recombination assay. Immunofluorescence and phase-contrast images of 293T cells transfected with CAGGFP-Spry1 alone or 293T cells stably transfected with a Cre-expressing plasmid (Cre-8 cells). (C) Immunoblot analysis of lysates from cells transfected with CAGGFP-Spry1 alone or together with Cre recombinase. Without Cre, GFP protein is detected, whereas Spry1 protein is not. When Cre recombinase is present, GFP expression is lost and Spry1 protein is now detected. (D) CAG-GFP-Spry1 mice were crossed with Col2a1-Cre mice, protein was extracted from the tails of E16.5 embryos and subjected to immunoblot analysis using anti-Myc antibodies and reprobed with anti-GFP antibodies (upper panel) (*, non-specific band). Genomic DNA was extracted from the yolk sacs and subjected to PCR screening for the presence of GFP and Cre sequences (lower panel). Only those samples that were PCR-positive for GFP and Cre expressed Spry1 protein (D, lane 1).
Spry1; Col2-Cre transgenic mice exhibit chondrodysplasia
During embryogenesis, we did not detect phenotypic or morphological differences between Spry1;Col2-Cre embryos and wild-type embryos before E12.5. However, by E14.5, Spry1;Col2-Cre embryos began to exhibit a dwarfing phenotype demonstrated by a shortened overall body length (data not shown). By E16.5, Spry1;Col2-Cre embryos had short limbs, shortened snouts, tails, and domed skulls consistent with a dwarfing phenotype (Figure 3A, 3B). The severity of the skeletal abnormalities in Spry1;Col2-Cre mice resulted in neonatal lethality within a few hours after birth, most likely due to respiratory difficulties. Alcian blue and alizarin red staining of skeletal preparations of newborn Spry1;Col2-Cre mice and their control littermates revealed that all skeletal elements derived by endochondral bone formation were reduced in size and often malformed in the Spry1;Col2-Cre mice (Figure 3C–F, 3H, and 3I). The bones at the base of the skull, such as the basioccipital bone and basisphenoid bone that are formed by endochondral ossification were remarkable smaller in the Spry1;Col2-Cre mice (Figure 3H, 3I). However, the size of calvarial bones were minimally affected (Figure 3D). In particular, the size of the frontal bone was comparable in control and Spry1;Col2-Cre mice (Figure 3D). The mandibular bones of Spry1;Col2-Cre mice showed normal ossification, but were shorter and wider than that of their control littermates (Figure 3G). This imbalance between normal intramembranous and aberrant endochondral ossification could account for the domed skull and short snout of Spry1;Col2-Cre mice. The ossification of the vertebrae was decreased significantly in Spry1;Col2-Cre mice (Figure 3F). The rib cage and the thoracic cavity were smaller, and the ribs were thin and irregular (Figure 3F), which most likely contributed to early postnatal death due to respiratory impairment.
Figure 3.
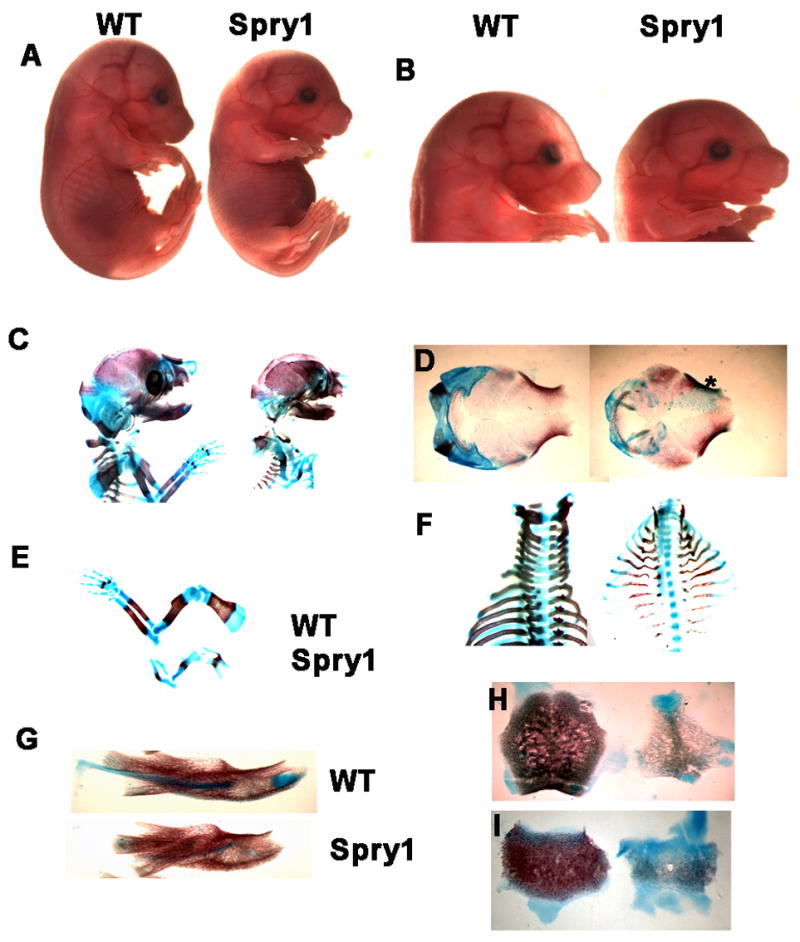
Spry1;Col2-Cre embryos exhibit skeletal dwarfism. (A, B) At E16.5, Spry1;Col2-Cre mice showed short limbs, shortened tail, domed skull, and short snouts. Original magnifications 1X (A), and 2.5X (B). (C-I) Skeletal preparations stained by Alcian blue and alizarin red. (C) Lateral view of upper body, showing ossification abnormalities in skull, vertebrae, and ribs. (D) Dorsal view of cranium (*area of abnormal ossification). (E) Overall chondrogenesis of the forelimbs was significant reduced in Spry1;Col2-Cre mice. (F) Dorsal view of vertebrae and ribs. Ribs were deformed and smaller, and ossification of the vertebrae was frequently absent in Spry1;Col2-Cre mice. However, the mandible (G) was shorter and wider due to the failure of Meckel’s cartilage to fully elongate. (H, J) The size and ossification of bones of the basioccipital bone (H) and basisphenoid bone (I) in Spry1;Col2-Cre mice were significantly reduced relative to control mice.
Overexpression of Spry1 in chondrocytes inhibits bone formation by inhibiting chondrocyte terminal differentiation and removal
Von kossa staining of sections of E16.5 humeri from Spry1;Col2-Cre transgenic mice and their Cre negative littermates reveals that there is some deposition of a mineralized matrix in the long bones of Spry1;Col2-Cre embryos, however there is a failure in the formation of trabecular bone (Figure 4A and B). Instead of giving way to the formation of ossification centers, hypertrophic chondrocytes persisted and accumulated a sparse mineralized matrix. In addition, in situ hybridization for the osteogenic marker gene, osteocalcin, showed that overexpression of Spry1 in chondrocytes resulted in a significant reduction in osteocalcin expression that correlated with inhibition of the formation of the primary ossification centers (Figure 4C). Histological examination showed that the normal pattern of chondrocyte differentiation in Spry1 transgenic mice was disrupted (Figure 4D, 4E). Cryosections of limbs were stained with hematoxylin eosin and alcian blue to examine the growth plate morphology and the distribution of sulphated glycosaminoglycans in the matrix. In control mice, chondrocytes of the long bone growth plate were organized into reserve, proliferating, and hypertrophic zones; however, the Spry1;Col2-Cre mice had disorganized growth plates that lacked the normal columnar organization of the proliferating and prehypertrophic zones and showed a greatly expanded hypertrophic zones (Figure 4D, 4E).
Figure 4.
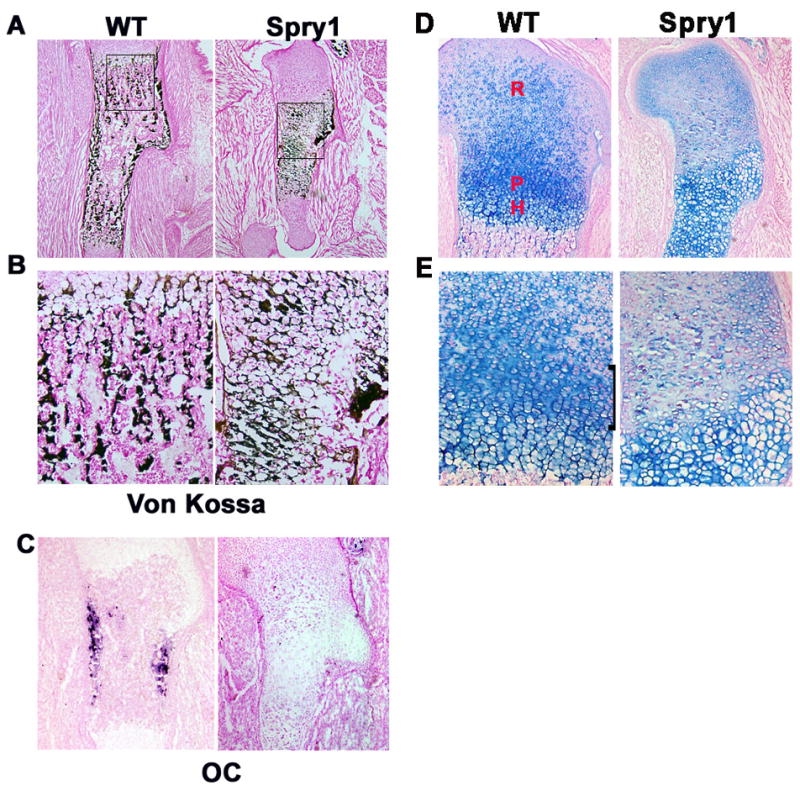
Spry1;Col2 embryos have reduced bone formation. (A) Von Kossa staining of long bone sections at E16.5 (50X). (B) Higher magnification from rectangular area of A. Note the persistence of hypertrophic chondrocytes and failure of formation of normal trabecular bone. (C) in situ hybridization for osteocalcin showed inhibition of osteogenesis in Spry1;Col2 growth plates. (D, E) Alcian Blue staining of E16.5 humerus sections. (D), higher magnification of D (E). Chondrocytes in CAG-GFP-Spry1 control mice formed an orderly columnar zone (black bracket), but the same region in Spry1;Col2-Cre mice was highly disorganized. R=resting, P=proliferating, H=hypertrophic zones.
To better understand the growth defects mediated by overexpression of Spry1 in Col2-Cre expressing cells, we performed in situ hybridization for stage-specific chondrocyte markers on serial sections of long bones from embryos of different gestational ages. In control embryos, aggrecan expression increased progressively from the reserve to the proliferating zones and was undetectable in the hypertrophic chondrocytes (Figure 5A). In Spry1;Col2-Cre mice, aggrecan expression was also expressed in an increasing gradient from the reserve to proliferating zones, but remained weakly expressed in the hypertrophic chondrocytes (Figure 5B). The pattern of expression of Ppr was similar between Spry1;Col2-Cre and control embryos (Figure 5C and D). Ihh was expressed in two distinct anatomical domains in control mice; corresponding to upper hypertrophic chondrocytes of long bone growth plates (Figure 5E) and overlapping with Col10α1 expression (Figure 5G). However, in Spry1;Col2-Cre mice, Ihh expression was confined to a single broad domain of hypertrophic chondrocytes (Figure 5F) which overlapped with the Col10α1 expression domain (Figure 5H). Mmp13 is expressed in the growth plates and primary ossification centers of the embryonic skeleton (Inada et al., 2004). In E15.5 Spry1;Col2-Cre embryos, Mmp13 expression was absent from hypertrophic chondrocytes and primary ossification centers compared to control mice (Figure 5I and J). However, by E16.5, Mmp13 was expressed in hypertrophic chondrocytes of Spry1;Col2-Cre embryos, albeit in a pattern distinct from control embryos (Figure 5K and L). The delay and alteration in the domains of expression of stage-specific marker gene expression suggests a delay or premature chondrocyte differentiation. To evaluate these possibilities, we examined Ihh and Col10α1 mRNA expression in E13.5 limbs to test whether the onset of terminal differentiation is also delayed (Minina et al., 2002). Robust expression of Ihh was found in the E13.5 limbs of control CAG-GFP-Spry1 embryos (Figure 5M); whereas in Spry1;Col2-Cre embryos, Ihh expression was slightly decreased (Figure 5N). Interestingly, expression of Col10α1, a marker of hypertrophy, was initiated at this stage in control but not in Spry1;Col2-Cre limbs (Figure 5O and P). Therefore, instead of accelerating chondrocyte differentiation, over-expression of Spry1 in nascent chondrocytes delayed chondrocyte differentiation. LacZ staining of CAGGFP-Spry1;ROSA26;Col2-Cre triple transgenic mice at E12.5–13.0 reveals an overall decreased in the LacZ staining compared to ROSA26;Col2-Cre control embryos suggesting an overall decrease in chondrocyte proliferation and differentiation by this stage (Figure 5Q and R). This difference was most pronounced in the presumptive proliferating zone.
Figure 5.
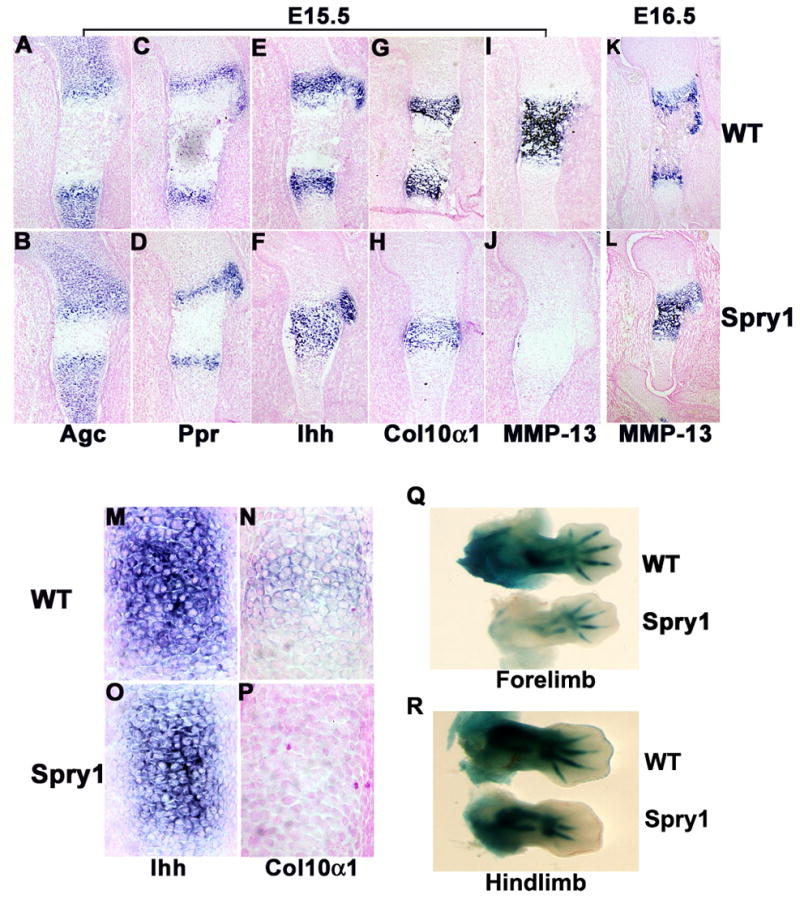
Overexpression of Spry1 in chondrocytes delayed chondrocyte differentiation and maturation resulting in accumulation of hypertrophic cartilage. (A-L) In situ hybridization for stage-specific chondrocyte differentiation markers on sections of E16.5 humeri from CAG-GFP-Spry1 control and Spry1;Col2-Cre embryos. The reserve chondrocyte marker aggrecan (Agc) (A, B), the prehypertrophic and hypertrophic chondrocyte marker Indian hedgehog (Ihh) (C, D), PTHrP receptor (Ppr) (E, F), hypertrophic chondrocyte marker Col10α1 (G, H, I, J), and the terminal differentiated marker MMP-13 (K, L) were employed. In situ hybridization was carried out on E13.5 forelimb sections from control (M, N) and Spry1;Col2-Cre (O, P) for Ihh (M, O) and Col10α1 (N, P). LacZ staining of forelimbs (Q) and hindlimbs (R) of ROSA26;Col2-Cre (WT) and Spry1;ROSA26;Col2-Cre (Spry1) embryos.
Forced expression of mSpry1 in chondrocytes decreases proliferation and increases apoptosis
The rate of chondrocyte proliferation and differentiation, along with the rate of chondrocyte hypertrophy and apoptosis directs the formation of the growth plate (Ornitz and Marie, 2002). To assess the effect of overexpression of Spry1 on chondrocyte proliferation, pregnant CAGGFP-Spry1 mice that were crossed with Col2α1-Cre transgenic mice were injected with BrdU at E16.5. After two hours, the embryos were collected and BrdU incorporation into long bone chondrocytes was detected by immunohistochemistry. The total number of chondrocytes in the presumptive reserve and proliferating zones of Spry1;Col2-Cre mice were markedly reduced compared to littermate controls (data not shown). Overall, the number of BrdU-positive cells in the epiphysis of Spry1 transgenic mice was significantly reduced compared to littermate controls (Figure 6A, 6B).
Figure 6.
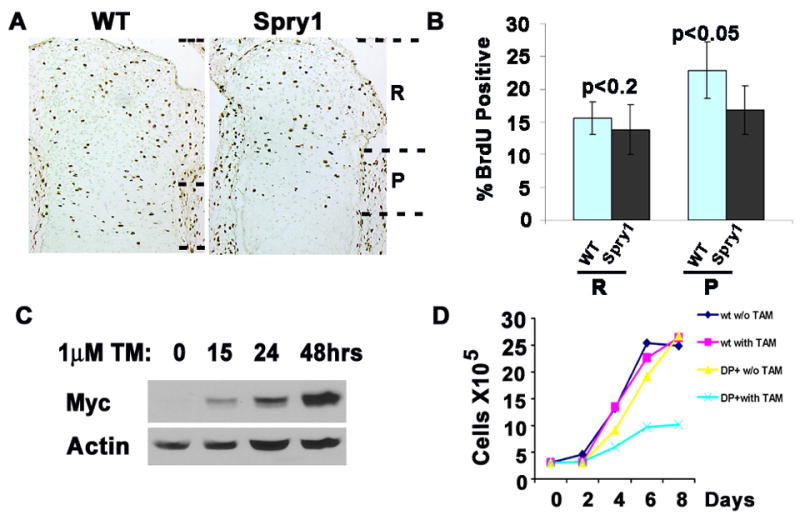
Overexpression of Spry1 in chondrocytes inhibited cell proliferation. (A, B) Short-term (2hr) BrdU incorporation into E16.5 mouse embryos shows the rate of chondrocyte proliferation. Immunochemistry showed decreased BrdU-labeled cells in the region of the presumptive proliferating columnar chondrocytes (P) of Spry1;Col2-Cre mice with little difference in the reserve chondrocytes (R) (A). Quantification of BrdU-positive cells in Spry1;Col2-Cre sections relative to controls (B). (C, D) Tamoxifen(TM)-induced Spry1 expression in primary chondrocytes inhibits cell growth. (C) Immunoblot analysis demonstrates tamoxifen-induced Spry1 protein expression. (D) Cell growth assay with and without tamoxifen-induced Spry1 expression in primary chondrocytes from CAG-GFP-Spry1;CAGGCre-ERTM embryos. Assays were performed in triplicate.
To gain additional insight into the effects of forced expression of mSpry1 on chondrocyte proliferation, we established primary chondrocyte cultures from long bone growth plates. Tamoxifen-induced expression of mSpry1 in primary chondrocytes from Spry1;CAGGCre-ER™ mice began as early as 15 hours after the addition of tamoxifen (Figure 6C). This expression correlated with a decrease in the number of tamoxifen-treated Spry1;CAGGCre-ER™ chondrocytes relative to tamoxifen-treated controls chondrocytes (Figure 6D). We did not observe any evidence that this decrease in cell number was due to increased apoptosis. These results indicate that forced expression of mSpry1 in chondrocytes inhibited growth plate columnar chondrocyte organization, in part by inhibiting chondrocyte proliferation.
To determine whether there were changes in rates of apoptosis in Spry1;Col2-Cre growth plates, we performed TUNEL labeling on sections of E16.5 humeri of Spry1;Col2-Cre and wild-type control mice (Figure 7). There was increased apoptosis in the presumptive reserve and proliferating zones of Spry1;Col2-Cre long bones but not in those of wild-type mice. These data suggest that the disruption in the normal columnar organization of the chondrocytes is likely due to decreased proliferation and increased apoptosis of chondrocytes that contribute to the presumptive reserve and proliferating zones.
Figure 7.
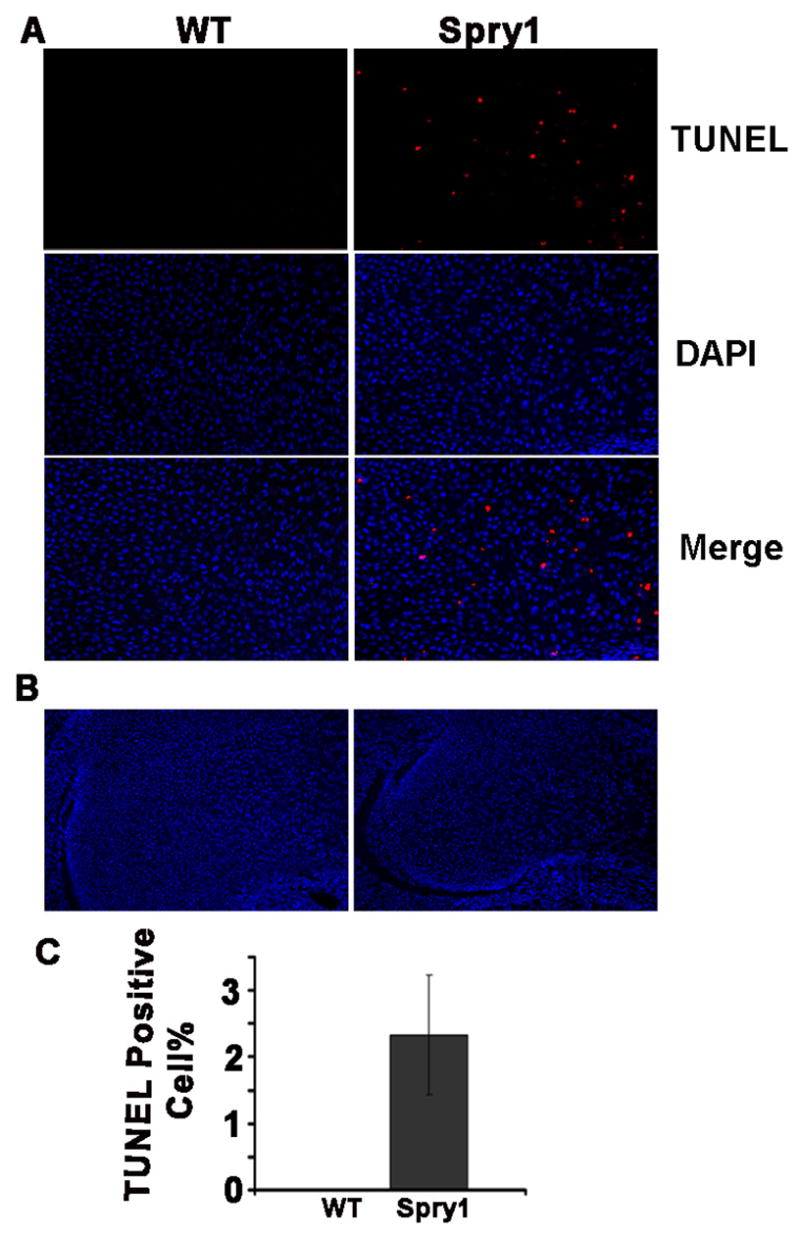
Overexpression of Spry1 in chondrocytes induced premature chondrocytes apoptosis. (A)TUNEL labeling of E16.5 humerus sections demonstrated increased apoptotic chondrocytes in the presumptive reserve and proliferating zones of Spry1;Col2-Cre growth plate relative to Cre-negative littermate controls. (B) Lower magnification shows the ephiphysis. (C) Quantification of TUNEL-positive cells as a percentage of total chondrocytes.
Overexpression of Spry1 in chondrocytes inhibits bone formation by inhibiting chondrocyte terminal differentiation and removal
The delay in terminal chondrocyte differentiation could affect hypertrophic chondrocyte elimination, which is required for formation of primary ossification centers. Therefore, we used long-term BrdU pulsed labeling to determine whether overexpression of Spry1 prolonged hypertrophic chondrocyte survival. Pregnant females were injected with BrdU at E15, and 36 hours later, the embryos at E16.5 were collected and BrdU incorporation into chondrocytes was quantified by immunohistochemistry. Figure 8A and 8B shows that the percentage of BrdU-positive cells in the hypertrophic zone of Spry1;Col2-Cre growth plates was significantly increased compared that of wild-type controls. Due to the growth arrest of hypertrophic chondrocytes, the BrdU labeled cells present in the hypertrophic zone are cells that moved down from the proliferating zone, and will accumulate if not removed through apoptosis. Thus, the increase in BrdU incorporation in the hypertrophic zone of Spry1;Col2-Cre transgenic mice reflects the slow elimination of these cells. To further examine this hypothesis, we performed TUNEL labeling to determine the rate of apoptosis of terminally differentiating hypertrophic chondrocytes. As expected from the BrdU results, most of the TUNEL labelled cells were localized to the chondro-osseous junction in wild-type but not in Spry1;Col2-Cre growth plates (Figure 8C, 8D),whereas increased apoptotic cells were observed in the chondrocytes distal to the chondro-osseous junction of Spry1;Col2-Cre growth plates but not in wild-type (Figure 7). Together, these results suggest that overexpression of Spry1 in chondrocytes delayed terminal chondrocyte differentiation causing excessive accumulation of hypertrophic chondrocytes, and consequently in the expansion of the hypertrophic zone and inhibition of endochondral ossification in Spry1;Col2-Cre mice.
Figure 8.
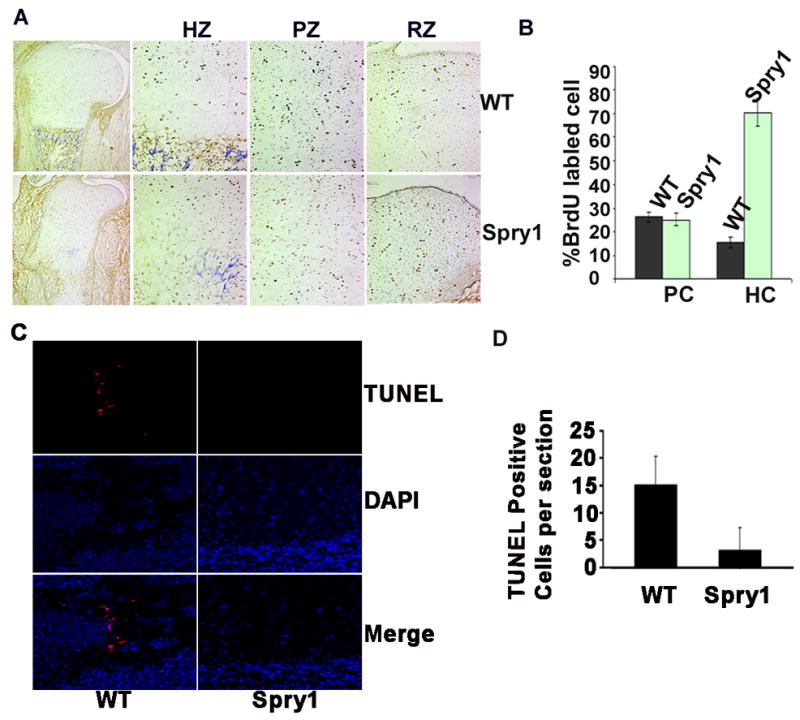
Overexpressing Spry1 in chondrocytes inhibited the elimination of hypertrophic chondrocytes. (A) BrdU pulse-labeling for 36 hrs showed an increase of BrdU positive cells in hypertrophic zone (HZ). (B) Quantification of BrdU-positive cells in A. (C) TUNEL labeling showed normal apoptotic chondrocytes in chondro-osseous junction in wild-type growth plates but not in Spry1;Col2-Cre growth plates. (D) Quantification of apoptotic cells in C.
Forced expression of mSpry1 in chondrocytes sustains ERK activation through attenuation of FGFR ubiquitinylation
The Spry family functions as feedback inhibitors of the FGFR-activated ERK pathway at or proximal to Raf1 in many cell types (Furthauer et al., 2002; Hanafusa et al., 2002; Iwanami et al., 2005; Yang et al., 2006). To examine whether overexpression of mSpry1 in chondrocytes regulates ERK signaling mediated by FGFR activation in a similar manner, we employed immunofluorescence microscopy on E16.5 humerus sections using phospho-Mek1/2 antibodies (Figure 9A). There was no significant difference in phospho-MEK1/2 immunostaining between reserve zone (RZ) and proliferating zone (RZ) chondrocytes of wild-type or Spry1;Col2-Cre mice. However, there was an increase in phospho-MEK1/2 immunostaining in the hypertrophic zone (HZ) of Spry1;Col2-Cre mice. Unexpectedly, time course analysis demonstrated that tamoxifen-induced expression of mSpry1 in primary chondrocytes from Spry1;CAGGCre-ER™ mice did not inhibit ERK activation mediated by FGF2 stimulation, but rather prolonged ERK activation (Figure 9B). We noted a consistent decrease in Spry1 protein levels in cultures stimulated with FGF2, which likely is the result of proteasomal degradation.
Figure 9.
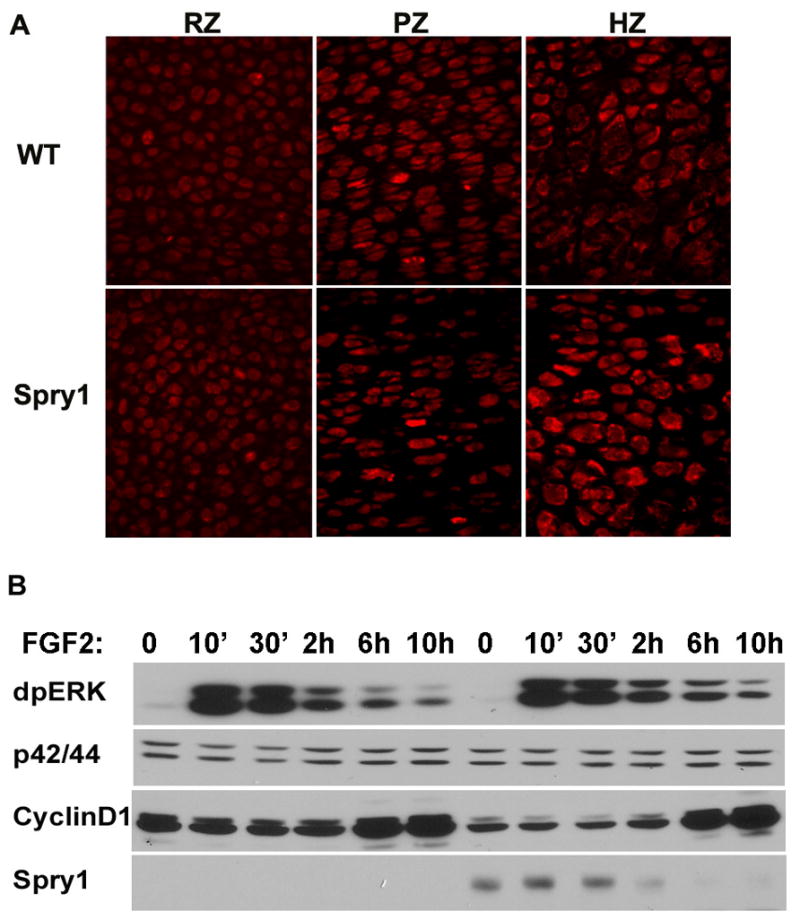
Overexpression of Spry1 in chondrocytes results in sustained ERK activation mediated by FGF2 stimulation. (A) Immunofluorescence for phospho-MEK1/2 on E16.5 humerus sections showed no significant inhibition of ERK signaling in the presumptive reserve and proliferating zones of Spry1;Col2 growth plates, whereas MEK1 phosphorylation was slightly enhanced in hypertrophic zone. (B) Immunoblot analysis of the time course of FGF2 stimulation of primary chondrocytes from Spry1;CAGGCre-ER™ embryos showed that overexpression of Spry1 sustained ERK activation and decreased basal cyclin D1 expression (B)
The dynamic up-regulation of D-type cyclins (cyclin D1, D2, and D3) in response to mitogenic stimulation is necessary for progression through the G1 phase of the cell cycle (Musgrove, 2006). It was reported that cyclin D1 antisense oligonucleotides inhibit the proliferation of rat chondrosarcoma cells and primary rat chondrocytes (Beier et al., 2001). It was of interest to determine whether sustained ERK activation mediated by mSpry1 overexpression in chondrocytes alters cyclin D1 expression. Similar to a previous report, the level of cyclin D1 protein in the growth plates is undetectable by immunohistochemistry (Sunters et al., 1998), data not shown), whereas cyclin D1 can be easily detected in cultures of primary chondrocytes (Li et al., 2006). Although overexpression of mSpry1 did not alter the dynamic pattern of cyclin D1 expression, it decreased the basal levels of cyclin D1 (Figure 9B). Thus, our results suggest that overexpression of mSpry1 somehow decreases cyclin D1, which may, in part, lead to a lower rate of chondrocyte proliferation.
Although Spry proteins inhibit ERK activation mediated by several RTKs, it was also reported that Spry2 prolonged EGFR-mediated ERK activation by sequestering c-Cbl away from EGFR and attenuating c-Cbl-mediated EGFR degradation (Haglund et al., 2005; Wong et al., 2002b; Wong et al., 2001). This prompted us to examine the effect of Spry1 expression on the degradation pathway of FGFRs. Immunofluorescence microscopy of E16.5 growth plates with FGFR3 antibodies revealed increased punctate staining in the presumptive reserve (Figure 10A) and proliferating zones (Figure 10B) of Spry1;Col2-Cre growth plates compared to controls. This punctate staining pattern likely represents FGFR3 accumulation in the endosomal compartment. We also performed immunofluorescence microscopy for FGFR2 on primary cultures of chondrocytes from Spry1;CAGGCre-ER™ with or without tamoxifen-induced Spry1 expression. Figure 10C demonstrates that there was a more rapid accumulation FGF2-induced FGFR2 immunoreactivity in the nucleus, particularly after 30 minutes in chondrocytes induced to express Spry1.
Figure 10.
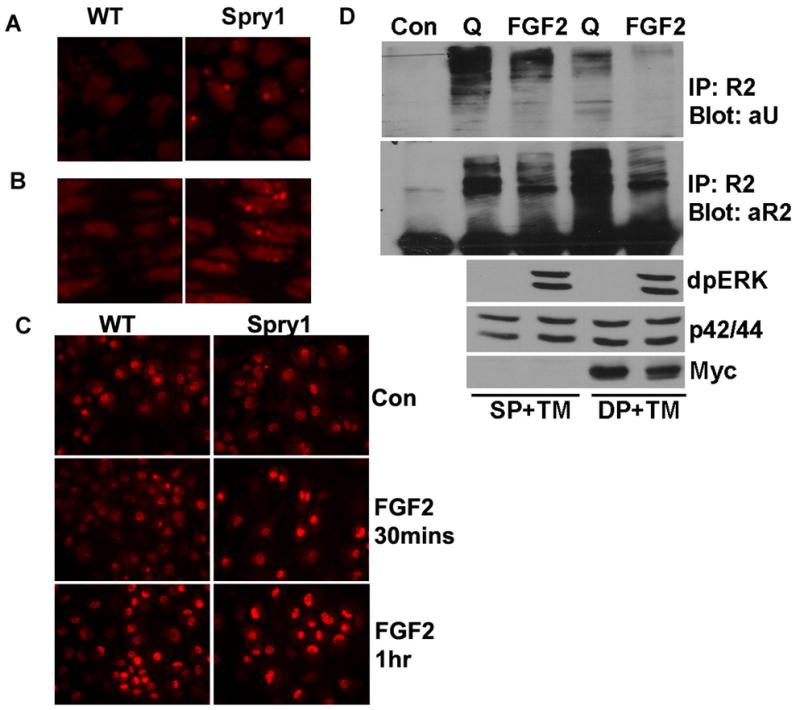
Overexpression of Spry1 resulted in accumulation of FGFRs in chondrocytes possibly through inhibiting degradation of FGFRs. Immunofluorescence microscopy showed an increase of FGFR3 immunoreactivity in the reserve zone (A) and proliferating zone (B) in Spry1;Col2-Cre growth plates. (C) Primary chondrocytes from Spry1;CAGGCre-ER™ embryos were induced to express Spry1 by the addition of 1 μM tamoxifen for 48 h, followed by the addition of FGF2 for the indicated times. Immunofluorescence staining for FGFR2 demonstrated a more rapid accumulation of FGFR2 in nucleus when Spry1 was overexpressed. (D) Primary chondrocytes from Spry1;CAGGCre-ER™ embryos (DP) or Cre-negative controls (SP) were treated with 1 μM tamoxifen (TM) for 48 h. Chondrocytes were then either left unstimulated (Q) or stimulated with FGF2 for 15 min. Immunoprecipitation with FGFR2 antibodies showed FGF2 stimulation induced degradation of FGFR2, and overexpression of Spry1 inhibited ubiquitination of FGFR2 in the quiescent state, and resulted in accumulation of FGFR2.
FGFR2, which directly or indirectly binds to c-Cbl and is degraded through the proteasome pathway (Kaabeche et al., 2004), is abundantly expressed in epiphyseal chondrocytes (Peters et al., 1993). FGFR2 is ubiquitinated under quiescent conditions and undergoes rapid degradation upon FGF2 stimulation (Figure 10D). Overexpression of Spry1 in cultures of primary chondrocytes significantly decreased FGFR2 ubiquitination both under quiescent conditions and after FGF2 stimulation, resulting in an accumulation of FGFR2 without FGF2 stimulation (Figure 10D).
Overexpression of mSpry1 in chondrocytes up-regulated p21Cip and STAT1
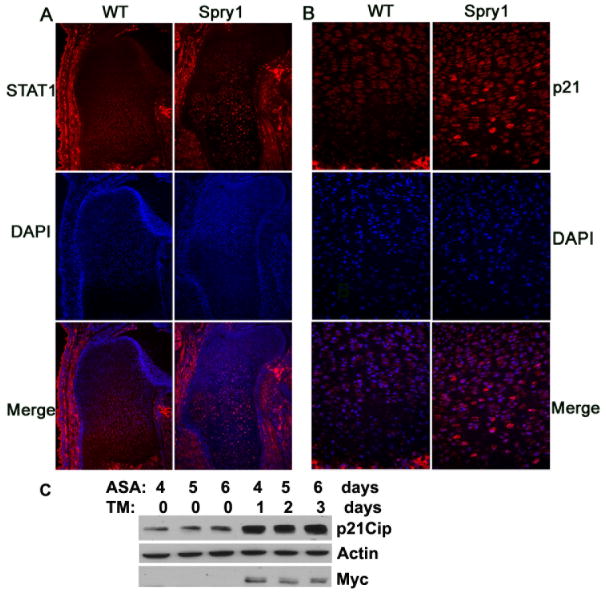
Cell cycle inhibitors play an important role in chondrogenesis (Ballock et al., 2000). STAT1 and p21Cip mediate increases in apoptosis and reduced chondrocyte proliferation in constitutively active Fgfr3 mouse mutants and Fgf2 transgenic mice (Sahni et al., 2001; Su et al., 1997). We questioned whether sustained ERK activation, as a result of forced expression of mSpry1 in chondrocytes, signals in a manner similar to activated FGFR3 (Sahni et al., 2001; Su et al., 1997). Therefore, we examined p21Cip and STAT1 expression in E16.5 long bone growth plates by immunofluorescence and in situ hybridization. The results showed that over-expression of Spry1 resulted in up-regulation of p21Cip and STAT1 expression in hypertrophic chondrocytes, and expanded the domain of p21Cip and STAT1 expression from the hypertrophic zone toward the reserve zone when compared to controls (Figure 11A, 11B, and data not shown). Consistently, tamoxifen-induced expression of mSpry1 in cultures of primary chondrocytes significantly up-regulated p21Cip compared to control chondrocytes (Figure 11C). Collectively, these data indicated that forced expression of Spry1 in chondrocytes inhibited chondrocyte proliferation, in part through a STAT1-p21Cip pathway.
Figure 11.
Overexpression of Spry1 up-regulates STAT1 and p21Cip in vivo and p21Cip in vitro. E16.5 humerus sections from Spry1;Col2-Cre embryos or their Cre-negative littermates were stained with STAT1 antibody (A) and p21Cip antibody (B). Sections were also stained with DAPI to visualize nuclei, and the two images were merged. (C) Primary chondrocytes from Spry1;CAGGCre-ER™ embryos were induced to differentiate by the addition of ascorbic acid (ASA), for three days, followed by the addition of 1 μM tamoxifen (TM). Cells were harvested on days 4, 5, and 6 and subjected to immunoblot analysis. Control and Spry1-expressing chondrocytes treated with ascorbic acid show increased p21Cip expression in the presence of induced Spry1 expression.
DISCUSSION
In this study, we demonstrated that: (i) Spry1 and Spry2 were expressed in prehypertrophic and hypertrophic chondrocytes of the developing long bones; (ii) forced expression of mSpry1 in chondrocytes of embryonic mice inhibited chondrocyte proliferation with coincident sustained ERK activation and decreased expression of cyclin D1; (iii) forced expression of mSpry1 in chondrocytes up-regulated the expression of p21Cip and STAT1; (iv) hypertrophic chondrocyte apoptosis was inhibited which lead to an increase in hypertrophic chondrocytes which inhibited endochondral bone formation. The results suggest that Spry1 and perhaps other Spry family members regulate FGFR signaling in a cell-type specific manner, and that dysfunction of Spry genes in chondrocytes and osteo-chondro-progenitor cells may play a role in some skeletal disorders.
Spry expression in developing chondrocytes
The expression of Spry transcripts during early embryogenesis and in adult tissues is documented (Chambers and Mason, 2000; de Maximy et al., 1999; Minowada et al., 1999; Zhang et al., 2001). However, the dynamic expression of Spry family members during chondrogenesis has not been determined. Our studies place Spry expression in mature and hypertrophic chondrocytes during the initial phases of endochondral ossification. Furthermore, our in vitro differentiation studies with ATDC5 cells and primary chondrocytes demonstrated that Spry expression is up-regulated during early stages of differentiation and coincides with the expression of differentiation markers such as Ihh. These in vitro differentiation studies support a model in which increased Spry expression may participate in initiation or progression of the chondrocyte differentiation program.
We observed extensive overlap in the expression of Spry1, Spry2, and Spry4 in the chondrocytes of developing growth plates. Spry family members are also co-expressed in other developmental contexts including regions of the brain, kidney, limb bud, and craniofacial primordia (Minowada et al., 1999; Zhang et al., 2001). The functional significance of this overlap has not been determined. Recent studies may provide some insight into the biological significance of co-expression of Spry family members in different tissues. Spry family members form homo- and hetero-oligomers through their C-terminal domains (Ozaki et al., 2005). These studies further revealed that while Spry homo-oligomers inhibited FGF-induced activation of the ERK pathway, Spry hetero-oligomers had greater inhibitory activity. In addition, these studies showed that Spry1 specifically interacts with Grb2, whereas Spry4 interacts specifically with Sos1 (Ozaki et al., 2005). The co-expression and hetero-oligomerization of different Spry family members during development may represent a combinatorial system for the strict regulation of the activation state of the ERK pathway from multiple signaling inputs. The observed increase in ERK activation in chondrocytes over expressing Spry1 may reflect a disruption in the normal distribution of Spry oligomers.
Sustained ERK activation contributes to defects in chondrogenesis
Retroviral-mediated expression of Spry2 in the prospective wing or limb buds of chick embryos causes a reduction of limb bud outgrowth and an inhibition of chondrocyte differentiation, although the mechanisms for this are not clear (Minowada et al., 1999). To examine the role of Spry in chondrogenesis in mice, we used Cre-loxP technology to over-express Spry1 in chondrocytes by using Col2(α1)-Cre to drive Spry1 expression. Interestingly, Spry1;Col2-Cre transgenic mice develop an thanatophoric dysplasia II-like dwarfism phenotype similar to that observed in the chick limb bud model (Minowada et al., 1999). Longitudinal growth of long bones is achieved through the proliferation of chondrocytes, chondrocyte hypertrophy, and endochondral ossification. BrdU incorporation showed that overexpression of mSpry1 in chondrocytes inhibited the proliferation rate of chondrocytes in the presumptive proliferative zone. Furthermore, growth of primary chondrocytes in culture was inhibited by forced expression of mSpry1. MAPK activation in response to growth factors and cytokines is required for cell proliferation in many cell types. The kinase MEK1/2 is immediately upstream of ERK, and its activation may be used as a marker of ERK activation. The intensity of p-MEK1/2 immunostaining was similar in the reserve and proliferating zones between control and Spry1;Col2-Cre embryos; whereas the MEK1/2 signal was increased in the hypertrophic zone in Spry1;Col2-Cre growth plates. Interestingly, transgenic mice that express a constitutively active form of MEK1 (c-MEK1) exhibit increased ERK phosphorylation and achondroplasia-like dwarfism (Murakami et al., 2004). In contrast to Spry1;Col2-Cre mice, c-MEK1 transgenic mice display incomplete chondrocyte hypertrophy, a smaller hypertrophic zone and delayed endochondral ossification. Furthermore, whereas chondrocyte proliferation was unaffected in c-MEK1 transgenic mice (Murakami et al., 2004), chondrocytes from Spry1;Col2 had reduced proliferative capacity. This suggests that Spry1 inhibits chondrocyte proliferation by a mechanism that may be independent of ERK activation.
The up-regulation of the cell cycle inhibitor p21Cip in chondrocytes is mediated in part by STAT1 activation, and correlates with reduced proliferation and increased apoptosis in the resting and proliferating zones of developing long bones (Sahni et al., 1999). FGF2 and FGF18 treatment of wild-type and Stat1-null chondrocytes results in similar up-regulation of p21Cip, which was inhibited by an inhibitor of the ERK pathway, indicating that activation of the ERK pathway by FGFs is independent STAT1 (Murakami et al., 2004). Spry1;Col2-Cre embryos exhibited an increase in STAT1 and p21Cip expression relative to control embryos. However there was no increase in STAT1 phosphorylation (X.Y., unpublished). Therefore, we propose that overexpression of Spry1 in chondrocytes inhibits their proliferation in a manner that may be independent of ERK signaling, but is mediated in part by up-regulation of p21Cip, in a manner that may be independent of STAT1. The mechanism by which Spry1 expression leads to increased p21Cip levels remains to be determined.
Spry1 regulates chondrocyte differentiation
Histological examination of long bones of E16.5 Spy1;Col2-Cre transgenic mice showed accumulation of hypertrophic chondrocytes that were uniformly larger than hypertrophic chondrocytes from Cre-negative CAG-GFP-Spry1 control mice. In addition, the chondrocytes of the proliferating and pre-hypertrophic zones were disorganized in Spry1;Col2-Cre growth plates when compared to Cre-negative littermate controls. Expression of Ihh and Col10α1, two markers of chondrocyte differentiation, were decreased in the limbs of Spry1;Col2-Cre transgenic as early as E13.5. However, by E15.5 the intensity of Ihh and Col10α1 expression was similar between Spry1;Col2-Cre embryos and Cre-negative littermates; but the domains of expression were different. In Cre-negative control mice, Col10α1 was expressed in the epiphyseal growth plates of long bones separated by the primary ossification center in which there was no Col10α1 expression. In contrast, Col10α1 expression was restricted to a single broad zone of expression with no intervening primary ossification center. The expression domains for Agc and Ppr were similar between Spry1;Col2-Cre and control E15.5 embryos. This suggests that expression of Spry1 in chondrocytes delays or inhibits the terminal stages chondrocytes differentiation and maturation.
Overexpression of mSpry1 in chondrocytes inhibits FGFR degradation and leads to sustained ERK activation
Attenuation of growth factor signaling is essential for the regulation of developmental processes and tissue homeostasis in most organisms. Spry family members were originally identified as feedback inhibitors of FGFR-ERK signaling in many cell types and in embryonic organogenesis (Mason et al., 2006). FGFR constitutively associates with the adaptor protein FRS2 through the juxtamembrane domain, and after stimulation by FGF ligands, become phosphorylated on six tyrosine residues that serve as docking sites for the SH2 domain-containing proteins, Grb2 and SHP2. This results in the recruitment and assembly of Grb2-Sos complexes at the membrane, where Ras is activated (Hanafusa et al., 2002). In addition, upon FGF stimulation, Spry1 and Spry2 are translocated to the plasma membrane where they become tyrosine phosphorylated on a conserved N-terminal tyrosine residue that acts as a docking site for Grb2, thus sequestering Grb2 away from Sos and resulting in attenuation of Ras activation (Hanafusa et al., 2002). Conversely, upon ligand stimulation of the EGFR that is internalized in complex with the SH2 domain-containing E3 ubiquitin ligase c-Cbl, Spry2 becomes tyrosine phosphorylated on the conserved N-terminal tyrosine residue (Y55) that serves as a docking site for c-Cbl. Tyrosine phosphorylated Spry2 then binds c-Cbl and sequesters it away from the EGFR, resulting in sustained EGFR and consequently sustained ERK activation (Haglund et al., 2005; Stang et al., 2004). Like EGFR, FGFR is also internalized upon ligand stimulation, although the mechanisms may be different. Ligand-stimulated activation of FGFR results in phosphorylation of FRS2 and c-Cbl, leading to formation of a c-Cbl-Grb2 complex that is recruited to FRS2. The recruitment of the c-Cbl-Grb2 complex by FRS2α results in the ubiquitinylation and degradation of FRS2α and FGFR (Wong et al., 2002a). FGF stimulation has also been reported to enhance Spry2 degradation via the proteasome (Hall et al., 2003).
Here we demonstrate that forced expression of Spry1 in chondrocytes causes decreased ubiquitinylation of FGFR2 that result in increased FGFR2 levels under both quiescent and FGF-stimulated conditions. FGFR3 levels were also increased in chondrocytes overexpressing Spry1. These data suggest that the increase in sustained ERK activation that we observe in Spry1 overexpressing chondrocytes may be due to Spry1-mediated attenuation of FGFR degradation, resulting in increased signaling to ERK. This mechanism probably involves Spry1-mediated sequestration of c-Cbl because the Spry-Cbl binding site is highly conserved among Spry family members. Spry1 likely sequesters c-Cbl away from FGFR-FRS2-Grb2 complexes thus preventing their ubiquitination and proteasome-mediated degradation. Because Spry binding to Cbl can occur without tyrosine phosphorylation of Spry (Fong et al., 2003; Wong et al., 2001), perhaps Spry1 preferentially binds to c-Cbl in chondrocytes, thereby causing (i) resulting Spry degradation, (ii) leaving Grb2 available to be recruited to FGFR-FRS2, (iii) resulting in decreased FGFR-FRS2 degradation and sustained ERK activation. Our data support this possibility. Future studies on the role of Spry in various cellular contexts will likely focus on the availability and affinity of Spry and its different binding partners.
Acknowledgments
This work was supported by NIH grants R01HL65301, R01DK73871 (to R.F.) and P20RR15555 (to R.F.) from the National Center for Research Resources. We thank the Kathleen Carrier of Histopathology Core Facility, supported by P20RR181789 to D. Wojchowski, for assistance with these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballock RT, Zhou X, Mink LM, Chen DH, Mita BC, Stewart MC. Expression of cyclin-dependent kinase inhibitors in epiphyseal chondrocytes induced to terminally differentiate with thyroid hormone. Endocrinology. 2000;141:4552–7. doi: 10.1210/endo.141.12.7839. [DOI] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD. Sprouty1 Is a Critical Regulator of GDNF/RET-Mediated Kidney Induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Beier F, Ali Z, Mok D, Taylor AC, Leask T, Albanese C, Pestell RG, LuValle P. TGFbeta and PTHrP control chondrocyte proliferation by activating cyclin D1 expression. Mol Biol Cell. 2001;12:3852–63. doi: 10.1091/mbc.12.12.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellus GA, McIntosh I, Smith EA, Aylsworth AS, Kaitila I, Horton WA, Greenhaw GA, Hecht JT, Francomano CA. A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat Genet. 1995;10:357–9. doi: 10.1038/ng0795-357. [DOI] [PubMed] [Google Scholar]
- Chambers D, Mason I. Expression of sprouty2 during early development of the chick embryo is coincident with known sites of FGF signalling. Mech Dev. 2000;91:361–4. doi: 10.1016/s0925-4773(99)00288-9. [DOI] [PubMed] [Google Scholar]
- Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517–25. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li C, Qiao W, Xu X, Deng C. A Ser(365)-->Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum Mol Genet. 2001;10:457–65. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–7. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- de Maximy AA, Nakatake Y, Moncada S, Itoh N, Thiery JP, Bellusci S. Cloning and expression pattern of a mouse homologue of drosophila sprouty in the mouse embryo. Mech Dev. 1999;81:213–6. doi: 10.1016/s0925-4773(98)00241-x. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–21. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- Fong CW, Leong HF, Wong ES, Lim J, Yusoff P, Guy GR. Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J Biol Chem. 2003;278:33456–64. doi: 10.1074/jbc.M301317200. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4:170–4. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–63. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Haglund K, Schmidt MH, Wong ES, Guy GR, Dikic I. Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation. EMBO Rep. 2005;6:635–41. doi: 10.1038/sj.embor.7400453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr Biol. 2003;13:308–14. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–8. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–18. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, Lopez-Otin C, Krane SM. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci U S A. 2004;101:17192–7. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr, Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–80. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Iwanami M, Hiromi Y, Okabe M. Cell-type specific utilization of multiple negative feedback loops generates developmental constancy. Genes Cells. 2005;10:743–52. doi: 10.1111/j.1365-2443.2005.00871.x. [DOI] [PubMed] [Google Scholar]
- Iwata T, Li CL, Deng CX, Francomano CA. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Hum Mol Genet. 2001;10:1255–64. doi: 10.1093/hmg/10.12.1255. [DOI] [PubMed] [Google Scholar]
- Kaabeche K, Lemonnier J, Le Mee S, Caverzasio J, Marie PJ. Cbl-mediated degradation of Lyn and Fyn induced by constitutive fibroblast growth factor receptor-2 activation supports osteoblast differentiation. J Biol Chem. 2004;279:36259–67. doi: 10.1074/jbc.M402469200. [DOI] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–90. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TF, Chen D, Wu Q, Chen M, Sheu TJ, Schwarz EM, Drissi H, Zuscik M, O’Keefe RJ. Transforming growth factor-beta stimulates cyclin D1 expression through activation of beta-catenin signaling in chondrocytes. J Biol Chem. 2006;281:21296–304. doi: 10.1074/jbc.M600514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Minina E, Kreschel C, Naski M, Ornitz D, Vortkamp A. Interaction of FGF, Ihh/Pthlh, and BMP Signaling Integrates Chondrocyte Proliferation and Hypertrophic Differentiation. Dev Cell. 2002;3:439. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–75. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Murakami S, Balmes G, McKinney S, Zhang Z, Givol D, de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18:290–305. doi: 10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA. Cyclins: roles in mitogenic signaling and oncogenic transformation. Growth Factors. 2006;24:13–9. doi: 10.1080/08977190500361812. [DOI] [PubMed] [Google Scholar]
- Neilson KM, Friesel R. Ligand-independent activation of fibroblast growth factor receptors by point mutations in the extracellular, transmembrane, and kinase domains. J Biol Chem. 1996;271:25049–57. doi: 10.1074/jbc.271.40.25049. [DOI] [PubMed] [Google Scholar]
- Ornitz DM. Regulation of chondrocyte growth and differentiation by fibroblast growth factor receptor 3. Novartis Found Symp. 2001;232:63–76. 76–80, 272–82. doi: 10.1002/0470846658.ch6. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–65. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Miyazaki S, Tanimura S, Kohno M. Efficient suppression of FGF-2-induced ERK activation by the cooperative interaction among mammalian Sprouty isoforms. J Cell Sci. 2005;118:5861–71. doi: 10.1242/jcs.02711. [DOI] [PubMed] [Google Scholar]
- Peters K, Ornitz D, Werner S, Williams L. Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev Biol. 1993;155:423–30. doi: 10.1006/dbio.1993.1040. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–4. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- Rousseau F, el Ghouzzi V, Delezoide AL, Legeai-Mallet L, Le Merrer M, Munnich A, Bonaventure J. Missense FGFR3 mutations create cysteine residues in thanatophoric dwarfism type I (TD1) Hum Mol Genet. 1996;5:509–12. doi: 10.1093/hmg/5.4.509. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Saugier P, Le Merrer M, Munnich A, Delezoide AL, Maroteaux P, Bonaventure J, Narcy F, Sanak M. Stop codon FGFR3 mutations in thanatophoric dwarfism type 1. Nat Genet. 1995;10:11–2. doi: 10.1038/ng0595-11. [DOI] [PubMed] [Google Scholar]
- Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13:1361–6. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni M, Raz R, Coffin JD, Levy D, Basilico C. STAT1 mediates the increased apoptosis and reduced chondrocyte proliferation in mice overexpressing FGF2. Development. 2001;128:2119–29. doi: 10.1242/dev.128.11.2119. [DOI] [PubMed] [Google Scholar]
- Sakai K, Hiripi L, Glumoff V, Brandau O, Eerola R, Vuorio E, Bosze Z, Fassler R, Aszodi A. Stage-and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol. 2001;19:761–7. doi: 10.1016/s0945-053x(00)00122-0. [DOI] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–24. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–64. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, Raiborg C, Stenmark H, Madshus IH. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol Biol Cell. 2004;15:3591–604. doi: 10.1091/mbc.E04-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WC, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton WA, Fu XY. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–92. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- Sunters A, McCluskey J, Grigoriadis AE. Control of cell cycle gene expression in bone development and during c-Fos-induced osteosarcoma formation. Dev Genet. 1998;22:386–97. doi: 10.1002/(SICI)1520-6408(1998)22:4<386::AID-DVG8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Ayada T, Ichiyama K, Kohno R, Yonemitsu Y, Minami Y, Kikuchi A, Maehara Y, Yoshimura A. Sprouty2 and Sprouty4 are essential for embryonic morphogenesis and regulation of FGF signaling. Biochem Biophys Res Commun. 2007;352:896–902. doi: 10.1016/j.bbrc.2006.11.107. [DOI] [PubMed] [Google Scholar]
- Tavormina PL, Shiang R, Thompson LM, Zhu YZ, Wilkin DJ, Lachman RS, Wilcox WR, Rimoin DL, Cohn DH, Wasmuth JJ. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet. 1995;9:321–8. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Wong A, Lamothe B, Lee A, Schlessinger J, Lax I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc Natl Acad Sci U S A. 2002a;99:6684–9. doi: 10.1073/pnas.052138899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. Embo J. 2002b;21:4796–4808. doi: 10.1093/emboj/cdf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Lim J, Low BC, Chen Q, Guy GR. Evidence for direct interaction between Sprouty and Cbl. J Biol Chem. 2001;276:5866–75. doi: 10.1074/jbc.M006945200. [DOI] [PubMed] [Google Scholar]
- Yang X, Webster JB, Kovalenko D, Nadeau RJ, Zubanova O, Chen PY, Friesel R. Sprouty genes are expressed in osteoblasts and inhibit fibroblast growth factor-mediated osteoblast responses. Calcif Tissue Int. 2006;78:233–40. doi: 10.1007/s00223-005-0231-4. [DOI] [PubMed] [Google Scholar]
- Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, Kurihara T, Rogi T, Tanaka S, Suda M, Tamura N, Ogawa Y, Nakao K. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10:80–6. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lin Y, Itaranta P, Yagi A, Vainio S. Expression of Sprouty genes 1, 2 and 4 during mouse organogenesis. Mech Dev. 2001;109:367–70. doi: 10.1016/s0925-4773(01)00526-3. [DOI] [PubMed] [Google Scholar]