Abstract
Pentachlorophenol (PCP) is a persistent chemical contaminant which has been extensively investigated in terms of its toxicology and metabolism. Similar to PCP, other chlorinated phenol derivatives are also widely present in the environment from various sources. Even though some of the chlorine-substituted phenols, and particularly PCP, are well known inhibitors of phenol sulfotransferases (SULTs), these compounds have been shown to undergo sulfation in humans. In order to investigate the enzymatic basis for sulfation of PCP in humans, we have studied the potential for PCP as well as the mono-, di-, tri-, and tetra-chlorinated phenols to serve as substrates for human hydroxysteroid sulfotransferase, hSULT2A1. Our results show that all of these compounds are substrates for this isoform of sulfotransferase and the highest rates of sulfation are obtained with PCP, trichlorophenols, and tetrachlorophenols. Much lower rates of sulfation were obtained with isomers of monochlorophenol and dichlorophenol as substrates for hSULT2A1. Thus, the sulfation of polychlorinated phenols catalyzed by hSULT2A1 may be a significant component of their metabolism in humans.
Introduction
Pentachlorophenol (PCP) is a synthetic molecule that has been used throughout the world for purposes as varied as antimicrobial agent, detergent and wood preservative. Even though its use has been restricted after the 1980s, it is still widely present in the environment, and it is considered as a chemical pollutant of concern (1). The air and the food chain are main pathways for human exposure to this compound, and it is possible to detect significant concentrations of PCP in the plasma of various age groups including newborns (2–4). Acute toxicity of PCP is usually occupationally related with significant damage to various organs, and chronic exposure also results in severe health disorders (5–8). Concentrations of PCP in the blood of exposed individuals can vary widely, and values ranging from 1.5 µg/L to 90 µg/L have been reported (4,8,9). Furthermore, there is a wide tissue distribution for PCP and a relatively long elimination half-life (9–12). Even though PCP is not directly classified as a human carcinogen, it has been shown to be involved in the initiation and/or promotion of carcinogenesis in animal studies (13–14). Moreover, some of the oxidized metabolites of PCP are reactive electrophiles towards DNA and proteins (15–17).
The biotransformation of PCP has been investigated in a wide range of animal species (9,18–25). In these studies, the unchanged parent molecule and its glucuronide and sulfate conjugates are found, as well as other metabolites. Interestingly, besides the formation of the reactive metabolite, tetrachloro-1,4-benzoquinone, other chlorinated phenols were also identified as metabolites of PCP (21). This was parallel with the identification of PCP as one of the metabolites of hexachlorobenzene (26). In humans, sulfation was found to be a dominant excretion pathway for low concentrations of chlorinated phenols when investigated in saw-mill workers exposed to 2,4,6-tri-, 2,3,4,6-tetra-, and penta-chlorophenol (27).
Even though the chlorinated phenols are subject to sulfation in humans, some of these compounds, particularly PCP, are well known for their characteristic inhibition of the phenol sulfotransferases (i.e., sulfotransferase family 1, or SULT1, enzymes) (28). However, their potential to interact with a human hydroxysteroid sulfotransferase (e.g., the family 2 enzyme, hSULT2A1) has not received attention. hSULT2A1 catalyzes the sulfation of various endogenous molecules (e.g. hydroxysteroids and bile acids) and xenobiotics bearing aliphatic or benzylic alcohol functional groups. Moreover, a few phenols such as 1-hydroxypyrene (29) and certain hydroxylated polychlorinated biphenyls (30) can serve as substrates for this enzyme. Additionally, hSULT2A1 has been reported to catalyze the sulfation of phenols such as estrone (31), raloxifene (32), and 4-hydroxytamoxifen (32), although hSULT1E1 and hSULT1A1 are likely the major SULTs involved for these particular phenols under physiological conditions. While PCP and several related phenols with multiple chlorine atoms clearly inhibit many phenol sulfotransferases (i.e., SULT1 enzymes), we have now investigated the hypothesis that these molecules serve as substrates for hydroxysteroid sulfotransferase hSULT2A1.
Materials and Methods
Chemicals and Biochemicals
2-Mercaptoethanol, dehydroepiandrosterone (DHEA), PAPS, potassium phosphate, methylene blue, 2-chlorophenol, 3-chlorophenol, 4-chlorophenol, 2,3-dichlorophenol, 2,4-dichlorophenol, 2,5-dichlorophenol, 2,6-dichlorophenol, 3,4-dichlorophenol, 3,5-dichlorophenol, 2,4,6-trichlorophenol, 2,3,4-trichlorophenol, 2,4,5-trichlorophenol and pentachlorophenol were purchased from Sigma Chemical Co. (St. Louis, MO). PAPS was further purified by a previously described procedure (33) to a purity greater than 99% as judged by HPLC. 2,3,6-Trichlorophenol, 2,3,5-trichlorophenol, 3,4,5-trichlorophenol, 2,3,4,6-tetrachlorophenol, 2,3,5,6-tetrachlorophenol and 2,3,4,5-tetrachlorophenol were obtained from Supelco (Bellefonte, PA). Each of the chlorinated phenols was of the highest purity commercially available from the manufacturer. Sucrose, chloroform and anhydrous sodium sulfate were from Fisher Scientific (Pittsburgh, PA). Tris-HCl was obtained from RPI (Mt. Prospect, IL). DE52 was purchased from Whatman (Fairfield, IL). Hydroxyapatite (Bio-Gel HT) was from Bio-Rad Laboratories (Hercules, CA), and Tween 20 was obtained from J.T. Baker Chemicals (Philipsburg, NJ).
Expression and Purification of Human SULT2A1
Expression of hSULT2A1 in Escherichia coli cells and the preparation of cell extract were carried out according to the previously described procedure (30). The cell extract obtained (1 g protein) was applied to a DE52 anion exchange column (2.5 × 17 cm) equilibrated with buffer A (50 mM Tris-HCl buffer, pH 7.5, containing 0.25 M sucrose, 1 mM DTT, 10% (v/v) glycerol and 0.05% (v/v) Tween 20). After removing the proteins which did not bind to this column by washing with buffer A, the protein was eluted with a linear gradient formed between 250 mL of buffer A and 250 mL of buffer A containing 0.5 M potassium chloride. The fractions containing hSULT2A1 were combined and concentrated by ultrafiltration (YM10 membrane; Millipore Corporation, Bedford, MA). The concentration of potassium chloride was then reduced through successive dilution and concentration by ultrafiltration, with the dilutions carried out using the same buffer to be employed for the subsequent hydroxyapatite chromatography step (i.e., buffer B: 10 mM potassium phosphate, 0.25 M sucrose, 1 mM DTT and 0.05% (v/v) Tween 20, pH 6.8). The resulting protein (0.4 g) was applied to a column of hydroxyapatite (2.5 × 9.0 cm) equilibrated with buffer B. Buffer B was used to wash the column in order to remove all proteins that did not bind to the column, and elution was then carried out with a linear gradient formed between 200 mL of buffer B and 200 mL of buffer B containing 0.4 M potassium phosphate. The fractions containing hSULT2A1 activity were pooled and concentrated by ultrafiltration. In order to prepare this mixture for the following hydroxyapatite chromatography the phosphate concentration was then elevated to 30 mM by successive dilution/concentration with buffer C (buffer C: 30 mM potassium phosphate, 0.25 M sucrose, 1 mM DTT and 0.05% (v/v) Tween 20, pH 6.8). A new column (2.5 × 9.0 cm) of hydroxyapatite equilibrated with buffer C was prepared and the concentrated protein (0.1 g) from the first hydroxyapatite column was then applied to the second hydroxyapatite column. Proteins that did not bind to the column were removed by washing with buffer C, and elution of SULT2A1 was carried out with a linear gradient formed between 75 mL of buffer C and 75 mL of buffer C containing 0.3 M potassium phosphate. The fractions containing hSULT2A1 with the highest specific activity were analyzed by SDS-PAGE, and 36 mg of purified hSULT2A1 was obtained. The resulting purified enzyme was judged to be homogeneous by SDS-PAGE with Coomassie Blue staining. At each step of purification, protein content was determined by the modified Lowry method (34). The subunit relative molecular mass of the homogeneous protein was found to be 33.7 kDa, and the kinetic behavior of the protein with DHEA as substrate was completely consistent with the previously reported data for the native enzyme isolated from human liver (31). A summary of the purification (Table S1) and figures showing results at each step in the purification (Figure S1–Figure S4) are included in the Supporting Information for this article (available at http://pubs.acs.org).
Assay of Chlorinated Phenols as Substrates of Human SULT2A1
The potential of all chlorinated phenols to serve as substrates for hSULT2A1 was investigated using a standard methylene blue assay (35,36). Rates of sulfation were determined at 10 µM and 50 µM concentrations of each chlorinated phenol using the following procedure. Each 400 µL of reaction mixture contained 0.25 M potassium phosphate buffer at pH 7.0, 0.2 mM PAPS, 7.5 mM 2-mercaptoethanol, chlorinated phenol, and 2.5 % (v/v) ethanol as co-solvent for chlorinated phenols. The reactions were initiated by the addition of 32 µg of purified hSULT2A1 and incubated at 37°C for 10 min. Then, the reactions were terminated by the addition of 0.5 mL methylene blue reagent and 1.5 mL chloroform. The chlorinated phenol-dependent formation of a product organic sulfate that formed a paired ion with methylene blue in the chloroform phase was measured at 651 nm.
In order to make a relative comparison of the degree of sulfation that accounted for any day-to-day experimental variability in enzyme activity, the rate of sulfation of dehydroepiandrosterone (DHEA) at 50 µM concentration was determined using the same procedure described above. For each set of experiments, sulfation of the chlorinated phenol derivative (at 10 µM and 50 µM) was analyzed together with the sulfation of 50 µM DHEA. The overall standard deviation of all hSULT2A1-catalyzed DHEA-sulfation reactions was 13% of the mean.
Although the methylene blue assay has been routinely used for the determination of SULT activities, and it has been successfully applied to various substrates including steroids (35), phenols (37), amines (38), and benzylic alcohols (39), confirmation of the assay methodology was obtained by the comparison of the rates of sulfation of representative chlorinated phenols (i.e., 3,5-dichlorophenol, 3,4,5-trichlorophenol, 2,3,4,6-tetrachlorophenol, and pentachlorophenol) at 50 µM using an HPLC method for the determination of substrate-dependent formation of PAP catalyzed by the SULT (36). These assays were carried out at 37°C and at pH 7.0 with the amounts of enzyme and other components directly scaled to a 30 µL reaction mixture for comparison to the methylene blue assay. At the end of the 10 min incubation time, the reactions were terminated by the addition of 30 µL methanol and the substrate-dependent formation of PAP from PAPS was determined by HPLC as previously described (36). The rates of hSULT2A1-catalyzed sulfation of 3,5-dichlorophenol, 3,4,5-trichlorophenol, 2,3,4,6-tetrachlorophenol, and pentachlorophenol in HPLC assays were found to be 7.8 ± 0.2, 36.2 ± 1.4, 20.2 ± 1.0, and 46.9 ± 0.7 nmoles of product/min/mg hSULT2A1, respectively. These results were consistent with those obtained for the product sulfates using the methylene blue assay for 3,5-dichlorophenol, 3,4,5-trichlorophenol, 2,3,4,6-tetrachlorophenol, and pentachlorophenol (i.e., 7.2 ± 0.2, 38.2 ± 0.7, 21.6 ± 0.2, and 48.0 ± 0.9, respectively).
Control experiments under the assay conditions used for these studies indicated the dependence of the hSULT2A1-catalyzed reaction product formation on both time and protein concentration for both PCP and for 3,5-dichlorophenol as representative substrates. There was no detectable sulfation of the co-solvent (i.e., 2.5% v/v ethanol) in control experiments without DHEA or a chlorinated phenol. Unless otherwise noted, assays were done in duplicate and the means ± standard errors are reported.
Solubility of Chlorinated Phenols in the Assay
In order to assess solubility of compounds under the experimental conditions, a previously described light scattering method was employed at 400 nm using a Perkin-Elmer LS55 Luminescence Spectrometer (40).
Results
Investigation of Chlorinated Phenols as Substrates of hSULT2A1
In analyzing the potential for chlorinated phenols to serve as substrates for hSULT2A1, we first determined their solubility at 10 µM and 50 µM concentrations under the specific conditions of assay (Materials and Methods). All of the compounds were found to be soluble at these concentrations (data not shown). The hSULT2A1-catalyzed formation of sulfates from chlorinated phenols was analyzed by the methylene blue paired ion extraction method, and the rates of sulfation were compared using the rate of sulfation of 50 µM DHEA as reference.
As illustrated in Table 1 and Table 2, all of the chlorinated phenols were substrates with a relative activity range of 0.3–161% when compared with the rate of sulfation of 50 µM DHEA. The velocities at 50 µM were higher than the ones at 10 µM for all tested compounds. The highest relative activity was observed with 2,3,4,5-tetrachlorophenol (161% relative activity) and the monochlorinated phenols were the weakest substrates (around 0.3–4% relative activity). Even though variabilities were seen among the members of each group of compounds, there was a general tendency of increased sulfation with increased chlorination of the phenol.
Table 1.
Monochlorophenols and dichlorophenols as substrates for hSULT2A1.
| Molecule | Structure | Sulfation at 10 µM (nmol/min/mg) | Sulfation at 50 µM (nmol/min/mg) | Relative activity (%)a |
|---|---|---|---|---|
| 2-Chlorophenol | 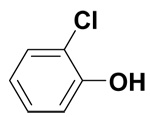 |
0 | 0.2 ± 0.1 | 0.3 |
| 3-Chlorophenol | 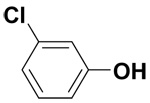 |
0 | 1.4 ± 0.7 | 2.3 |
| 4-Chlorophenol | 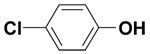 |
0 | 1.6 ± 0.5 | 3.8 |
| 2,3-Dichlorophenol | 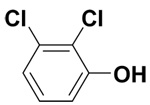 |
0.6 ± 0.2 | 4.2 ± 0.5 | 9.8 |
| 2,4-Dichlorophenol | 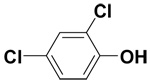 |
1.1 ± 0.3 | 2.7 ± 0.3 | 5.4 |
| 2,5-Dichlorophenol | 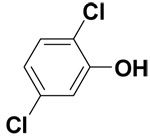 |
1.7 ± 0.1 | 6.9 ± 0.2 | 13.3 |
| 2,6-Dichlorophenol | 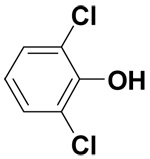 |
0.3 ± 0 | 2.5 ± 0.5 | 5.8 |
| 3,4-Dichlorophenol | 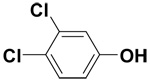 |
0.3 ± 0.2 | 2.9 ± 0.2 | 6.9 |
| 3,5-Dichlorophenol | 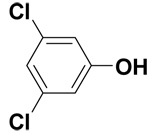 |
2.3 ± 0.4 | 7.2 ± 0.2 | 17.1 |
Relative activity is expressed as a percentage based on the ratio of the rate of sulfation of the chlorinated phenol at 50 µM to the rate of sulfation of 50 µM DHEA under the same assay conditions.
Table 2.
Tri-, tetra-, and pentachlorophenols as substrates for hSULT2A1.
| Molecule | Structure | Sulfation at 10 µM (nmol/min/mg) | Sulfation at 50 µM (nmol/min/mg) | Relative activity (%)a |
|---|---|---|---|---|
| 2,3,4-Trichlorophenol | 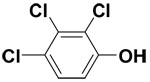 |
10.1 ± 1.0 | 30.9 ± 0.3 | 55.2 |
| 2,3,5-Trichlorophenol | 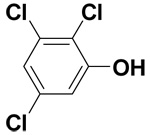 |
14.6 ± 0.8 | 47.7 ± 0.7 | 106.4 |
| 2,3,6-Trichlorophenol | 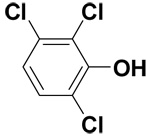 |
10.5 ± 0.7 | 28.4 ± 0.8 | 65.9 |
| 2,4,5-Trichlorophenol | 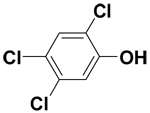 |
14.4 ± 1.3 | 46.0 ± 1.1 | 81.7 |
| 2,4,6-Trichlorophenol | 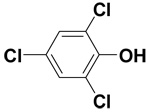 |
9.3 ± 0.1 | 26.2 ± 0.2 | 44.7 |
| 3,4,5-Trichlorophenol | 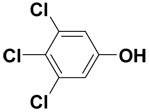 |
12.1 ± 1.0 | 38.2 ± 0.7 | 89.3 |
| 2,3,4,5-Tetrachlorophenol | 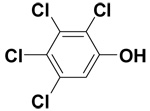 |
16.0 ± 0.5 | 67.1 ± 1.7 | 160.9 |
| 2,3,4,6-Tetrachlorophenol | 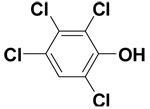 |
10.1 ± 1.5 | 21.6 ± 0.2 | 51.2 |
| 2,3,5,6-Tetrachlorophenol | 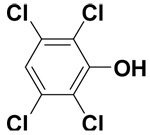 |
4.0 ± 0.3 | 12.9 ± 2.6 | 29.5 |
| Pentachlorophenol | 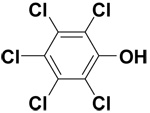 |
13.2 ± 0.2 | 48.0 ± 0.9 | 91.2 |
Relative activity is expressed as a percentage based on the ratio of the rate of sulfation of the chlorinated phenol at 50 µM to the rate of sulfation of 50 µM DHEA under the same assay conditions.
Only the monochlorophenols were not sulfated at 10 µM, but these compounds were sulfated at very low rates when present at 50 µM. The range of relative activities for dichlorophenols was found to be 5–17% of the rate for DHEA, and 3,5-dichlorophenol showed the highest relative ability to serve as a substrate in this group. Trichlorophenols exhibited rates of sulfation 5–10 times greater than dichlorophenols at both 10 and 50 µM levels. Among the trichlorophenols, 2,3,5-trichlorophenol, 2,4,5-trichlorophenol, and 3,4,5-trichlorophenol were sulfated at the highest rates. Rates of sulfation for tetrachlorophenols suggest that steric effects of two chlorine atoms ortho to the phenolic hydroxyl may decrease the rate of catalysis, since 2,3,4,5-tetrachlorophenol showed the highest relative rate of sulfation.
Discussion
It has long been known that PCP and several other chlorinated phenols are inhibitors of phenol sulfotransferases (i.e., SULT1 enzymes) in both in vitro and in vivo studies (28,41–46), and that their effects on these enzymes differ from their interactions with hydroxysteroid sulfotransferases (SULT2 enzymes). For example, studies with female rat hepatic cytosol indicated that PCP inhibited sulfation of 4-nitrophenol (a substrate for SULT1 enzymes), but had little effect on the sulfation of 5-hydroxychrysene and DHEA (substrates for hydroxysteroid sulfotransferases) (41). Likewise, in vivo studies in the rat have indicated that while PCP was an excellent inhibitor of the sulfation of SULT1 substrates such as 1-naphthol (43), acetaminophen (45), and harmol (46), there was no inhibition of the sulfation of DHEA at two different doses administered by i.v. infusion (47). Such differences in the effects of PCP on SULT1 and SULT2 enzymes were even more prominent at a high dose infusion of DHEA (50 mg/kg), where an increase in DHEA-sulfate serum concentrations was observed with PCP-treatment (47). Although in vivo studies on the effects of PCP on the sulfation of 5-hydroxymethylchrysene (41) and DHEA (41,47) in the rat did not address the possible formation of PCP-sulfate, sulfation has been shown to be a significant pathway for excretion of low doses of PCP in humans (27). Thus, when our results on the ability of human hydroxysteroid sulfotransferase hSULT2A1 to catalyze sulfation of PCP are considered along with previous studies on urinary excretion of PCP-sulfate, it is reasonable to conclude that hSULT2A1 contributes to the sulfation of PCP in humans. Human sulfotransferases such as hSULT2B1a and hSULT2B1b might also contribute to this observed sulfation, but the ability of these SULTs to catalyze the formation of PCP-sulfate remains to be investigated.
After the identification of PCP as a substrate for hSULT2A1 we also analyzed the substrate specificity of this enzyme with other chlorinated phenols. Similar to PCP, mono-, di-, tri-, and tetrachlorophenol derivatives are also widely present in the environment. Some of these chlorinated phenols are metabolites of hexachlorobenzene or PCP, and some of them are produced for industrial purposes (48–49). Our studies with two concentrations (i.e., 10 µM and 50 µM) of chlorinated phenols indicated variations in their abilities to serve as substrates for hSULT2A1. Monochlorophenols have been previously shown to be substrates for phenol sulfotransferases (i.e., SULT1 enzymes) (50). Our studies with the hydroxysteroid sulfotransferase hSULT2A1 indicated that these compounds are only very weak substrates of this enzyme. Thus any contribution of hSULT2A1 in the sulfation of these monochlorophenols is expected to be very low. Dichlorophenols were better substrates in comparison to monochlorophenols, since sulfation was observed at concentrations of both 10 µM and 50 µM. However, their relative activities were still low in comparison to DHEA and to other higher chlorinated phenol derivatives. Moreover, it is possible that some lower chlorinated phenols might potentially serve as substrates for SULT1 enzymes, and these structure-activity relationships will be the subject of future studies as well.
As illustrated in Table 2, the rates of sulfation seen for many of the trichloro-, and tetrachlorophenols were comparable to those seen for DHEA. Two compounds, 2,3,5-trichlorophenol and 2,3,4,5-tetrachlorophenol, exhibited higher rates of sulfation in comparison to DHEA under these conditions. As suggested for pentachlorophenol, our results with tri- and tetra-chlorophenols indicated a possible detoxication pathway of these compounds involving the contribution of hSULT2A1. This potential for excretion through metabolic sulfation assumes additional importance, since some of these compounds are also known as inhibitors of phenol sulfotransferases such as those involved in regulation of estrogen concentrations (51).
The overall increase in the rate of sulfation with increasing number of chlorine atoms may be primarily related to the increasing lipophilicity of the compounds. Indeed, according to the crystal structures of this enzyme, the substrate binding pocket of hSULT2A1 is composed of hydrophobic amino acid residues (52). However, this is not sufficient to explain the different rates of sulfation for compounds within each group. Thus, steric interactions between the active site residues of the enzyme and specific chlorine atoms on the phenol must also play a role. In general, the presence of two chlorine atoms meta to the phenolic hydroxyl was found to be important for higher rates of sulfation. Indeed, all of the compounds that indicated the highest relative activities among the members of individual groups (3,5-dichlorophenol, 2,3,5-trichlorophenol and 2,3,4,5-tetrachlorophenol) have this di-meta chlorine substitution. Nevertheless, it should be emphasized that, in addition to these initial observations, more detailed kinetic analyses of each of the 19 chlorinated phenols will be needed to establish firm structure-activity relationships for substrates and to assess the extent to which substrate inhibition may be occurring for individual congeners. While the current studies clearly show the ability of a large number of chlorinated phenols to serve as substrates for hSULT2A1, the presence of substrate inhibition for some, but perhaps not all, substrates of hSULT2A1 would complicate analysis of structure-activity relationships. Additional studies will also be necessary to assess the potential for each of the chlorinated phenols to compete with DHEA as a substrate for hSULT2A1. This will require analysis of multiple concentrations of both DHEA and the chlorinated phenol, since the potential for such interactions would be dependent upon kinetic parameters of both the chlorinated phenol and DHEA.
In summary, we have determined that PCP and related chlorinated phenols, many of which are known as inhibitors of phenol sulfotransferases, are substrates for the human hydroxysteroid sulfotransferase hSULT2A1. The importance of the contribution of this enzyme towards the detoxication of these molecules under in vivo conditions will be the subject of future investigations.
Supplementary Material
All figures and tables related to the method utilized for the purification of recombinant human SULT2A1 are available free of charge via the internet at http://pubs.acs.org.
Acknowledgements
This study was supported by the National Institutes of Health through research grants R01 CA038683 from the National Cancer Institute and P42 ES013661 from the National Institute of Environmental Health Sciences. Support from the NIEHS through P30 ES05605 is also acknowledged. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- DE52
Diethylaminoethyl cellulose anion exchange column
- DHEA
dehydroepiandrosterone (5-androsten-3β-ol-17-one)
- DTT
dithiothreitol
- hSULT2A1
human hydroxysteroid sulfotransferase 2A1
- PAP
adenosine 3’5’-diphosphate
- PAPS
3’-phosphoadenosine 5’-phosphosulfate
- PCP
pentachlorophenol
- SULT
sulfotransferase
- Tris-HCl
[tris(hydroxymethyl)aminomethane] hydrochloride.
References
- 1.ATSDR. Toxicological Profile for Pentachlorophenol (update) vol.xix. Atlanta, GA: US Department of Health and Human Services, Agency for Toxic Substance and Disease Registry; 2001. Public health statement for pentachlorophenol; 269 pp. http://www.atsdr.cdc.gov/toxprofiles/phs51.html. [Google Scholar]
- 2.Wilson NK, Chuang JC, Morgan MK, Lordo RA, Sheldon LS. An observational study of the potential exposures of preschool children to pentachlorophenol, bisphenol-A, and nonylphenol at home and daycare. Environ. Res. 2007;103:9–20. doi: 10.1016/j.envres.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Thompson TS, Treble RG. Preliminary results of a survey of pentachlorophenol levels in human urine. Bull. Environ. Contam. Toxicol. 1994;53:274–279. doi: 10.1007/BF00192044. [DOI] [PubMed] [Google Scholar]
- 4.Carrizo D, Grimalt JO, Fito NR, Torrent M, Sunyer J. Pentachlorobenzene, hexachlorobenzene, and pentachlorophenol in children’s serum from industrial and rural populations after restricted use. Ecotoxicol. Environ. Saf. 2007 doi: 10.1016/j.ecoenv.2007.08.021. “in press”. [DOI] [PubMed] [Google Scholar]
- 5.O’Malley MA, Carpenter AV, Sweeney MH, Fingerhut MA, Marlow DA, Halperin WE, Mathias CG. Chloracne associated with employment in the production of pentachlorophenol. Am. J. Ind. Med. 1990;17:411–421. doi: 10.1002/ajim.4700170401. [DOI] [PubMed] [Google Scholar]
- 6.Jorens PG, Schepens PJ. Human pentachlorophenol poisoning. Hum. Exp. Toxicol. 1993;12:479–495. doi: 10.1177/096032719301200605. [DOI] [PubMed] [Google Scholar]
- 7.Begley J, Reichert EL, Rashad MN, Klemmer HW. Association between renal function tests and pentachlorophenol exposure. Clin. Toxicol. 1977;11:97–106. doi: 10.3109/15563657708989823. [DOI] [PubMed] [Google Scholar]
- 8.Daniel V, Huber W, Bauer K, Suesal C, Mytilineos J, Melk A, Conradt C, Opelz G. Association of elevated blood levels of pentachlorophenol with cellular and humoral immunodeficiencies. Arch. Environ. Health. 2001;56:77–83. doi: 10.1080/00039890109604057. [DOI] [PubMed] [Google Scholar]
- 9.Uhl S, Schmid P, Schlatter C. Pharmacokinetics of pentachlorophenol in man. Arch. Toxicol. 1986;58:182–186. doi: 10.1007/BF00340979. [DOI] [PubMed] [Google Scholar]
- 10.Wagner SL, Durand LR, Inman RD, Kiigemagi U, Deinzer ML. Residues of pentachlorophenol and other chlorinated contaminants in human tissues: Analysis by electron capture gas chromatography and electron capture negative ion mass spectrometry. Arch. Environ. Contam. Toxicol. 1991;21:596–606. doi: 10.1007/BF01183883. [DOI] [PubMed] [Google Scholar]
- 11.Hattamer-Frey HA, Travis CC. Pentachlorophenol: Environmental partitioning and human exposure. Arch. Environ. Contam. Toxicol. 1989;18:482–489. doi: 10.1007/BF01055013. [DOI] [PubMed] [Google Scholar]
- 12.Barbieri F, Colosio C, Schlitt H, Maroni M. Urine excretion of pentachlorophenol (PCP) in occupational exposure. J. Pestic. Sci. 1995;43:259–262. [Google Scholar]
- 13.Chhabra RS, Maronpot RM, Bucher JR, Haseman JK, Toft JD, Hejtmancik MR. Toxicology and carcinogenesis studies of pentachlorophenol in rats. Toxicol. Sci. 1999;48:14–20. doi: 10.1093/toxsci/48.1.14. [DOI] [PubMed] [Google Scholar]
- 14.Umemura T, Kai S, Hasegawa R, Sai K, Kurukawa Y, Williams GM. Pentachlorophenol produces liver oxidative stress and promotes but does not initiate hepatocarcinogenesis in B6C3F1 mice. Carcinogenesis. 1999;20:1115–1120. doi: 10.1093/carcin/20.6.1115. [DOI] [PubMed] [Google Scholar]
- 15.Vaidyanathan VG, Villalta PW, Sturla SJ. Nucleobase dependent reactivity of a quinone metabolite of pentachlorophenol. Chem. Res. Toxicol. 2007;20:913–919. doi: 10.1021/tx600359d. [DOI] [PubMed] [Google Scholar]
- 16.Lin PH, La DK, Upton PB, Swenberg JA. Analysis of DNA adducts in rats exposed to pentachlorophenol. Carcinogenesis. 2002;23:365–369. doi: 10.1093/carcin/23.2.365. [DOI] [PubMed] [Google Scholar]
- 17.Waidyanatha S, Lin PH, Rappaport SM. Characterization of chlorinated adducts of hemoglobin and albumin following administration of pentachlorophenol to rats. Chem. Res. Toxicol. 1996;9:647–653. doi: 10.1021/tx950172n. [DOI] [PubMed] [Google Scholar]
- 18.Braun WH, Blau GE, Chenoweth MB. The metabolism/pharmacokinetics of pentachlorophenol in man, and a comparison with the rat and monkey. In: Deichmann WB, editor. Toxicology and Occupational Medicine: Proceedings of the Tenth Inter-American Conference on Toxicology and Occupational Medicine. North Holland, New York: Elsevier; 1979. pp. 289–296. [Google Scholar]
- 19.Ahlborg UG, Lindgren JE, Mercier M. Metabolism of pentachlorophenol. Arch. Toxicol. 1974;32:271–281. doi: 10.1007/BF00330109. [DOI] [PubMed] [Google Scholar]
- 20.Ahlborg UG, Larsson K, Thunberg T. Metabolism of pentachlorophenol in vivo and in vitro. Arch. Toxicol. 1978;40:45–53. doi: 10.1007/BF00353278. [DOI] [PubMed] [Google Scholar]
- 21.Renner G, Hopfer C. Metabolic studies on pentachlorophenol in rats. Xenobiotica. 1990;20:573–582. doi: 10.3109/00498259009046872. [DOI] [PubMed] [Google Scholar]
- 22.Renner G, Müke W. Transformations of pentachlorophenol Part I: Metabolism in animals and man. Toxicol. Environ. Chem. 1986;11:9–29. [Google Scholar]
- 23.Stehly GR, Hayton WL. Metabolism of pentachlorophenol by fish. Xenobiotica. 1989;19:75–81. doi: 10.3109/00498258909034678. [DOI] [PubMed] [Google Scholar]
- 24.Benner DB, Tjeerdema RS. Toxicokinetics and biotransformation of pentachlorophenol in the topsmelt (Atherinops affinis) J. Biochem. Toxicol. 1993;8:111–117. doi: 10.1002/jbt.2570080302. [DOI] [PubMed] [Google Scholar]
- 25.Sacco JC, James MO. Sulfonation of environmental chemicals and their metabolites in the polar bear (Ursus maritimus) Drug Metab. Dispos. 33:1341–1348. doi: 10.1124/dmd.105.004648. [DOI] [PubMed] [Google Scholar]
- 26.To-Figueras J, Sala M, Otero R, Barrot C, Santiago-Silva M, Rodamilans M, Herrero C, Grimalt J, Sunyer J. Metabolism of hexachlorobenzene in humans: Association between serum levels and urinary metabolites in a highly exposed population. Environ. Health Perspect. 1997;105:78–83. doi: 10.1289/ehp.9710578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekari K, Luotamo M, Lindroos L, Aitio A. Urinary excretion of chlorinated phenols in saw-mill workers. Int. Arch. Occup. Environ. Health. 1991;63:57–62. doi: 10.1007/BF00406199. [DOI] [PubMed] [Google Scholar]
- 28.Mulder GJ, Scholtens E. Phenol sulfotransferase and uridine diphosphate glucuronyltransferase from rat liver in vivo and in vitro. Biochem. J. 1977;165:553–559. doi: 10.1042/bj1650553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma B, Shou M, Schrag ML. Solvent effect on cDNA-expressed human sulfotransferase (SULT) activities in vitro. Drug. Metab. Dispos. 2003;31:1300–1305. doi: 10.1124/dmd.31.11.1300. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Apak TI, Lehmler HJ, Robertson LW, Duffel MW. Hydroxylated polychlorinated biphenyls are substrates and inhibitors of human hydroxysteroid sulfotransferase SULT2A1. Chem. Res. Toxicol. 2006;19:1420–1425. doi: 10.1021/tx060160+. [DOI] [PubMed] [Google Scholar]
- 31.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochem. J. 1989;260:641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falany JL, Pillof DE, Leyh TS, Falany CN. Sulfation of raloxifene and 4-hydroxytamoxifen by human cytosolic sulfotransferases. Drug Metab. Dispos. 2006;34:361–368. doi: 10.1124/dmd.105.006551. [DOI] [PubMed] [Google Scholar]
- 33.Sekura RD. Adenosine 3’-phosphate 5’-phosphosulfate. Methods Enzymol. 1981;77:413–415. [Google Scholar]
- 34.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal. Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 35.Nose Y, Lipman F. Separation of steroid sulfokinases. J. Biol. Chem. 1958;233:1348–1351. [PubMed] [Google Scholar]
- 36.Sheng J, Sharma V, Duffel MW. Current Protocols in Toxicology. Vol.1. New York: John Wiley & Sons; 2001. Measurement of aryl and alcohol sulfotransferase activity; pp. 4.5.1–4.5.9. [DOI] [PubMed] [Google Scholar]
- 37.Sekura RD, Duffel MW, Jakoby WB. Aryl sulfotransferases. Methods Enzymol. 1981;77:197–206. doi: 10.1016/s0076-6879(81)77026-5. [DOI] [PubMed] [Google Scholar]
- 38.Ramaswamy SG, Jakoby WB. Sulfotransferase assays. Methods Enzymol. 1987;143:201–207. doi: 10.1016/0076-6879(87)43038-3. [DOI] [PubMed] [Google Scholar]
- 39.Duffel MW, Janns MN. Aryl sulfotransferase IV catalyzed sulfation of 1-napthalene-methanol. In: Kocsis JJ, Jollow DJ, Witmer CM, Nelson JO, Snyder R, editors. Biological Reactive Intermediates III-Mechanisms of Action in Animal Models and Human Disease. New York, NY: Plenum; 1986. pp. 197pp. 415–422. [Google Scholar]
- 40.Blomquist CH, Kotts CE, Hakanson EY. A simple method for detecting steroid aggregation and estimating solubility in aqueous solutions. Anal. Biochem. 1978;87:631–635. doi: 10.1016/0003-2697(78)90714-5. [DOI] [PubMed] [Google Scholar]
- 41.Okuda H, Nojima H, Watanabe N, Watabe T. Sulphotransferase mediated activation of the carcinogen 5-hydroxymethylchrysene. Species and sex differences in tissue distribution of the enzyme activity and a possible participation of hydroxysteroid sulphotransferases. Biochem. Pharmacol. 1989;38:3003–3009. doi: 10.1016/0006-2952(89)90008-7. [DOI] [PubMed] [Google Scholar]
- 42.Fayz S, Cherry WF, Dawson JR, Mulder GJ, Pang KS. Inhibition of acetaminophen sulfation by 2,6-dichloro-4-nitrophenol in the perfused rat liver preparation. Lack of a compensatory increase of glucuronidation. Drug Metab. Dispos. 1984;12:323–329. [PubMed] [Google Scholar]
- 43.Boles JW, Klaassen CD. Effects of molybdate and pentachlorophenol on the sulfation of α-naphthol. Toxicol.Lett. 1999;106:1–8. doi: 10.1016/s0378-4274(99)00017-x. [DOI] [PubMed] [Google Scholar]
- 44.Daimon H, Sawada S, Asakura S, Sagami F. Inhibition of sulfotransferase affecting in vivo genotoxicity and DNA adducts induced by safrole in rat liver. Teratog. Carcinog. Mutagen. 1998;17:327–337. doi: 10.1002/(sici)1520-6866(1997)17:6<327::aid-tcm3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 45.Boles JW, Klaassen CD. Effects of molybdate and pentachlorophenol on the sulfation of acetaminophen. J. Toxicol. 2000;146:23–25. doi: 10.1016/s0300-483x(00)00159-1. [DOI] [PubMed] [Google Scholar]
- 46.Meerman JHN, Sterenberg HMJ, Mulder GJ. Use of pentachlorophenol as long-term inhibitor of sulfation of phenols and hydroxamic acids in the rat in vivo. Biochem. Pharmacol. 1983;32:1587–1593. doi: 10.1016/0006-2952(83)90332-5. [DOI] [PubMed] [Google Scholar]
- 47.Boles JW, Klaassen CD. Effects of molybdate and pentachlorophenol on the sulfation of dehydroepiandrosterone. Toxicol. Appl. Pharmacol. 1998;151:105–109. doi: 10.1006/taap.1998.8448. [DOI] [PubMed] [Google Scholar]
- 48.Borzelleca JF, Hayes JR, Condie LW, Egle JL. Acute toxicity of monochlorophenols, dichlorophenols and pentachlorophenol in the mouse. Toxicol. Lett. 1985;29:39–42. doi: 10.1016/0378-4274(85)90197-3. [DOI] [PubMed] [Google Scholar]
- 49.Hill RH, Head SL, Baker S, Gregg M, Shealy DB, Bailey SL, Williams CC, Sampson EJ, Needham LL. Pesticide residues in urine of adults living in The United States: Reference range concentrations. Environ. Res. 1995;71:99–108. doi: 10.1006/enrs.1995.1071. [DOI] [PubMed] [Google Scholar]
- 50.Campbell NRC, Van Loon JA, Sundaram RS, Ames MM, Hansch C, Weinshilboum R. Human and rat liver phenol sulfotransferase: Structure-Activity relationships for phenolic substrates. Mol. Pharmacol. 1987;32:813–819. [PubMed] [Google Scholar]
- 51.Harris RM, Kirk CJ, Waring RH. Non-genomic effects of endocrine disrupters: Inhibition of estrogen sulfotransferase by phenols and chlorinated phenols. Mol. Cell. Endocrinol. 2005;244:72–74. doi: 10.1016/j.mce.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Rehse PH, Zhou M, Lin SX. Crystal structure of human dehydroepiandrosterone sulfotransferase in complex with substrate. Biochem. J. 2002;364:165–171. doi: 10.1042/bj3640165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All figures and tables related to the method utilized for the purification of recombinant human SULT2A1 are available free of charge via the internet at http://pubs.acs.org.


