Abstract
Two-dimensional (2D) fractionation is a commonly used tool to increase dynamic range and proteome coverage for bottom-up, shotgun proteomics. However, there are few reports comparing the relative separation efficiencies of 2D methodologies using low microgram sample quantities. In order to systematically evaluate 2D separation techniques, we fractionated microgram quantities of E. coli protein extract by seven different methods. The first dimension of separation was performed with either reverse phase high pressure liquid chromatography (RP-HPLC), gel electrophoresis (SDS-PAGE), or strong cation exchange (SCX-HPLC). The second dimension consisted of a standard reverse phase capillary HPLC coupled to an electrospray ionization quadrupole time-of-flight mass spectrometer (ESI-QTOF MS) for tandem mass spectrometric analysis. The overall performance and relative fractionation efficiencies of each technique were assessed by comparing the total number of proteins identified by each method. The protein-level RP-HPLC and the high pH RP-HPLC peptide-level separations performed the best, identifying 281 and 266 proteins, respectively. The on-line pH variance SCX and the SDS-PAGE returned modest performances with 178 and 139 proteins identified, respectively. The off-line SCX had the worst performance with 81 proteins identified. We also examined various chromatographic factors which contribute to separation efficiency, including resolving power, orthogonality, and sample loss.
Keywords: Two-dimensional (2D) LC, MudPIT, shotgun proteomics, strong cation-exchange (SCX) HPLC, fractionation
INTRODUCTION
The identification of proteins from complex biological matrices has traditionally been performed using two-dimensional gel electrophoresis (2-DE). 2-DE separates proteins both by their isoelectric point and molecular weight, producing a high resolution protein map from which individual protein spots are picked, excised, and sequenced[1]. While 2-DE remains a powerful tool, shotgun proteomics utilizing LC-MS/MS has emerged as the technique of choice for large scale protein studies due to its superior throughput and sensitivity[2, 3].
In a typical shotgun proteomics experiment, a complex protein sample is enzymatically digested into peptides which are then separated by HPLC, introduced into a mass spectrometer, fragmented, sequenced and used to identify the parent protein via database searching. Each peptide introduced into the mass spectrometer must be fragmented individually and the rate at which a mass spectrometer can perform this fragmentation is called MS/MS duty cycle. Duty cycle is an important determinant of sampling depth and dynamic range; state-of-the-art, high duty cycle instruments can fragment hundreds of peptides in a single 1D LC-MS/MS experiment. However, a typical biological sample contains thousands of tryptic peptides, resulting in multiple co-eluting peptide species at a given LC elution time which quickly overwhelm the MS/MS acquisition speed of even the highest duty cycle instruments. This duty cycle overload results in a preferential fragmentation of high abundance peptides, introducing an overall sequencing bias towards more highly expressed proteins at the expense of less abundant proteins[4].
In contrast to LC-MS/MS shotgun proteomics, 2-DE employs a divide-and-conquer strategy by resolving proteins into individual spots which can then be selectively excised and sequenced. The high resolution of 2-DE allows the researcher to “hand-pick” the proteins of interest, including those that are not highly expressed, while by-passing the more abundant or less interesting proteins. So while shotgun proteomics generates a much larger number of protein identifications via high throughput sequencing, 2-DE enables the selective sequencing of differentially expressed proteins. In order to couple both the high throughput sequencing of LC-MS/MS and the high resolving power of 2-DE, researchers have coupled multiple fractionation methods in tandem with mass spectrometry, including various HPLC techniques, gel electrophoresis, and capillary electrophoresis (see Washburn and co-workers for an extensive review[5]). However, only a few reports have compared separation efficiencies between different fractionation modes both at the protein and peptide level[6, 7].
All proteins/peptides exhibit different physicochemical attributes based on their amino acid sequences and by exploiting these differences proteins/peptides can be fractionated. The extent to which the proteins/peptides are fractionated depends on the resolving power of the separation technique and the orthogonality of the two separation modes. For example, in 2-DE proteins are first separated by isoelectric point and then by size. These two separation modes have very different selectivities and each exhibits high resolving power, resulting in a very efficient separation. Given that proteins generally exhibit much different physicochemical characteristics from their constitutive peptides, an obvious way of gaining orthogonality in a 2D shotgun experiment is by first fractionating on the protein-level, digesting the fractions, and then fractionating the resulting peptides in the second dimension. A variety of separation modes have been employed to achieve protein level separation, including size exclusion chromatography (SEC)[8, 9], ion exchange chromatography (IEC)[2, 10], isoelectric focusing (IEF)[11–15], 1D gel separation (SDS-PAGE)[16, 17], and reverse phase chromatography (RP)[18–25].
In contrast to a protein-level separation followed by a peptide separation, a two-dimensional peptide-level separation can be performed by employing two or more methods with different separation selectivities. A number of separation modes have been implemented to this end, including strong cation exchange (SCX)[26–31], isoelectric focusing (IEF)[32, 33], capillary electrophoresis (CE)[34, 35], capillary isoelectric focusing (CIEF)[36, 37], and mixed mode pH reverse phase (RP-RP)[38–41].
The protein level and peptide level separations have relative advantages and disadvantages. Proteins are sensitive to precipitation upon exposure to high salt concentrations, pH extremes, and organic solvents. Peptides, on the other hand, are relatively well-behaved in solution and generally do not exhibit solubility issues. In addition, peptides are amenable to stable isotope labeling while whole proteins are not. However, peptide-level separations also have limitations, including the scattering of tryptic peptides from a single parent protein into multiple fractions which can potentially reduce protein identification scores.
The goal of this work is to systematically compare the factors that impact separation efficiencies, such as peak capacity and orthogonality, as evidenced by the total number of protein identifications of the following separation techniques: 1D SDS-PAGE, offline SCX, online SCX, mixed mode pH RP-RP, and protein RP.
EXPERIMENTAL SECTION
Escherichia coli Whole Cell Lysate
50mL of CircleGrow media from MP Biomedicals (Solon, OH) was inoculated with E. coli (DH5-alpha) from Invitrogen (Carlsbad, CA) and grown overnight to saturation. The cells were pelleted by centrifugation at 15,000g for 5 minutes and frozen at −80°C. For protein extraction, the cells were thawed, re-suspended in 1mL of cold water, placed on ice, and extracted with five, 10 second bursts at 30% power with an ultrasonic dismembrator from Microson (Farmingdale, NY). Cell debris was removed by centrifugation at 15,000g for 15 minutes at 4°C. The supernatant was decanted and subjected to protein determination by bichoninic acid (BCA).
Protein Solubility
150µL of E. coli protein extract was diluted to 0.3, 0.6, 0.9 and 1.2µg/µL in the following buffers: 6M urea with 0.1% and 0.05% trifluoroacetic acid (TFA), 0.1% and 0.05% TFA without urea, and 1% acetic acid without urea. For all conditions containing urea, samples were pre-treated with urea before acidification. After treatment, samples were allowed to incubate for 15 minutes at room temperature and then centrifuged at 15,000g for 15 minutes at 4°C. The Eppendorf tubes were then visually inspected for precipitate. Solubility was judged on degree of precipitation and called either soluble (S, no precipitation), partially soluble (S/I, slight precipitation), or insoluble (I, total precipitation). Results are summarized in Table 1.
Table 1.
Protein Solubility in Various Sample Buffers: Soluble (S), Partially Soluble (S/I), and Insoluble (I)
| Protein mg/mL | ||||
|---|---|---|---|---|
| Sample buffer | 0.3 | 0.6 | 0.9 | 1.2 |
| Urea + 0.05% TFA | S | S/I | S/I | I |
| Urea + 0.1 % TFA | S | S/I | S/I | I |
| 0.05 % TFA | S/I | S/I | I | I |
| 0.1 % TFA | S/I | I | I | I |
| 1% acetic acid | I | I | I | I |
Solvent-Assisted Protein Digestion
Proteins were enzymatically digested as described[42]. Protein extracts were diluted into 80% acetonitrile (ACN)/ 20% 200mM ammonium bicarbonate at pH 8. Cysteinyl disulfides were reduced by the addition of 2mM Tris[2-carboxyethyl] phosphine (TCEP) for 30 minutes at 37°C. Reduced disulfides were then alkylated by the addition of 10mM iodoacetamide (IAA) for 30 minutes in the dark. The pH was checked with pH paper and adjusted to pH 8. Trypsin from Promega (Madison, WI) was added at a 1:20 weight-to-weight ratio and incubated for 12 hours at 37°C. After digestion, solvent was removed by vacuum centrifugation.
Off-line Strong Cation Exchange Chromatography (Off-SCX)
100µg of protein digest was reconstituted in 100µL of 25% acetonitrile, 5mM K2HPO4 at pH 3 (HPLC Buffer A). A Rainin Dynamax HPLC equipped with a 100µL sample loop, binary pump, UV detector, and fraction collector was used to deliver the reconstituted digest to a PolySulfoethyl A SCX column (2.1 × 250mm, 5µm, 300Å) from PolyLC (Columbia, MD). Peptides were eluted with a linear gradient of 25% ACN, 500mM K2HPO4 at pH 3 (HPLC Buffer B) from 5–100% over 60 minutes. Fractions were collected every 5 minutes for a total of 12 fractions. Each fraction was vacuum-centrifuged to dryness and reconstituted in 20µL of 0.1% formic acid (FA); 2µL of each reconstituted fraction was analyzed by LC/MS/MS (see below for details).
Off-line High pH Reverse Phase Chromatography (pH-RP)
100µg of protein digest was reconstituted in 100µL of 200mM HCOONH4 (ammonium formate) at pH 10 (HPLC Buffer A). A Rainin HPLC (see Off-SCX for description) was used to deliver the reconstituted digest to a Gemini C18 RP column (2 × 150mm, 3µm, 110Å) from Phenomenex (Torrance, CA). Peptides were eluted with a linear gradient of 100% acetonitrile (HPLC Buffer B) from 5–35% over 60 minutes. Fractions were collected every 5 minutes for a total of 12 fractions. Each fraction was vacuum-centrifuged to dryness and reconstituted in 20µL of 0.1% FA; 2µL of each reconstituted fraction was analyzed by LC/MS/MS (see below for details).
On-line pH Variance Strong Cation Exchange (On-SCX)
10µg of protein digest was reconstituted in 20µL of citric acid buffer at pH 3 from Column Technology (Fremont, CA). The reconstituted peptides were delivered to a SCX trapping column (0.32 × 100mm, 5µm) from Column Technology via a Waters (Milford, MA) capillary HPLC (see LC-MS/MS for description). Ten discrete buffers from Column Technology were used to elute peptides from the trap column via autosampler injection. The buffers consisted of 10mM citric acid adjusted to pH 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, and 8.0 by ammonium hydroxide (see Dai et al. for details[30]). After each injection, the eluted peptides were delivered directly to a capillary reverse phase C18 column and analyzed by MS/MS (see LC-MS/MS section for description).
Off-line Conventional Protein Reverse Phase (cProt)
40µg of undigested protein extract in 100µL of water was delivered to a Macrosphere C18 RP column (2.1 × 250mm, 5µm, 300Å) from Alltech (Lexington, KY) using a Rainin HPLC (see Off-SCX for description). To increase recovery and decrease protein adsorption, the column was heated to 60°C throughout the separation. Proteins were eluted with a linear gradient of 0.1% TFA in acetonitrile (HPLC Buffer B) from 15–55% over 60 minutes (HPLC Buffer A was 0.1% TFA in water). Fractions were collected every 5 minutes for a total of 12 fractions. Each fraction was vacuum-centrifuged to ~50µL and then diluted to 80% acetonitrile/20% 200mM ammonium bicarbonate and digested according to the Solvent-Assisted Protein Digestion procedure. The digested fractions were reconstituted in 20µL of 0.1% FA; 5µL of each reconstituted fraction was analyzed by LC/MS/MS.
Off-line High Recovery Protein Reverse Phase (hrProt)
The procedure for cProt was followed exactly, except an Agilent (Santa Clara, CA) macroporous mRP-C18 column (2.1 × 75mm, 5µm) heated to 80°C was employed instead of the conventional C18 column from Alltech.
Off-line High Recovery Protein Reverse Phase + Urea (hrProt+Urea)
The procedure for hrProt was followed exactly, except urea was added to the protein extract to a concentration of 6M, followed by the addition of TFA to a concentration of 0.1%.
SDS-PAGE (Gel) Separation
40µg of undigested protein extract in 15µL of water was denatured and reduced via the addition of 2.5µL NuPAGE loading buffer and 1µL of NuPAGE reducing agent followed by heating at 70°C for 10 minutes (all NuPAGE products are from Invitrogen). The sample was loaded onto a 12-well pre-cast NuPage Novex 10% Bis-Tris gel. The gel was run at 200V for approximately 45 minutes with NuPAGE MOPS SDS running buffer. The gel was rinsed with water and then stained for 1 hour with Simple Blue SafeStain from Invitrogen (Carlsbad, CA). The gel was then washed with water overnight. After washing, the entire gel lane was excised with a razor blade and then cut into 12 equal size pieces. The individual pieces were then cut into smaller 1mm3 cubes and placed in an Eppendorf tube and subjected to the In-Gel Digestion protocol (see below). After the in-gel digestion, the peptides were reconstituted in 20µL of 0.1% FA; 5µL of each reconstituted fraction was analyzed by LC/MS/MS.
In-Gel Digestion
Gel pieces were destained twice with 250µL of 50% methanol/ 50% 100mM ammonium bicarbonate and then dehydrated with 200µL of 50% acetonitrile/ 50% 100mM ammonium bicarbonate followed by vacuum centrifugation. Proteins were reduced by the addition of 25mM dithiothretiol (DTT) for 30 minutes at 56°C. After the removal of DTT, 25mM iodoacetamide was added for 30 minutes in the dark. The gel pieces were then dehydrated as before and then rehydrated with 25µL of 20ng/µL of trypsin in 25mM ammonium bicarbonate. After digesting overnight at 37°C, the digest solution was transferred to an Eppendorf tube. The gel bound peptides were extracted with 50µL 0.1% FA followed by two additional extractions with 70% acetonitrile/0.1% FA. The digest solution and extract buffers were then combined and dried-down by vacuum centrifugation.
LC-MS/MS
Using a capillary HPLC from Waters, tryptic peptides were delivered to a trap column (PepMap C18, 0.3 × 50mm) from LC Packings (Sunnyvale, CA) via an isocratic flow of 0.1% formic acid in water (LC Buffer A) at a rate of 30µL/min for 3 min. The flow rate was then reduced to 250nL/min, and the peptides were flushed onto an in-house packed capillary column (C18, 75µm × 150mm) and eluted via a 5–45% linear gradient of 0.1% formic acid in acetonitrile (LC Buffer B) over 40 minutes into a nanoelectrospray ionization (nESI) quadrupole time-of-flight (QTOF) mass spectrometer (QTOF Micro) from Waters. Data was collected in positive ion mode from m/z 400 to 2000, followed by data-dependent MS/MS acquisitions from m/z 50 to 2000. The intensity threshold for switching from the survey scan to MS/MS was set at 15 ion counts. The scan time was 0.9 s; inter-scan time, 0.1 s; capillary voltage, 3200 V; and cone voltage, 35 V.
Database Search
Micromass ProteinLynx 2.1 was used to convert the .raw files into .pkl text files for database searching. The .pkl files from individual fractions were combined into one file and searched against the Swiss-Prot database using both Mascot and X!Tandem. The following parameters were used for both search engines: taxonomy was limited to E. coli, parent mass tolerance was 800ppm, fragment mass tolerance was 0.8Da, a maximum of two missed cleavages was allowed, and carbamidomethylation was set as a fixed modification. For the Mascot search results, the significance threshold was set at P<0.05 for protein scores (i.e. scores >25 indicate identity or extensive homology). For the X!Tandem search results, protein expectation values (log E) were required to be <−0.9. Total peptides, unique peptides, protein sequence coverage, and the false discovery rates (FDR) were all derived from the X!Tandem search engine.
RESULTS AND DISCUSSION
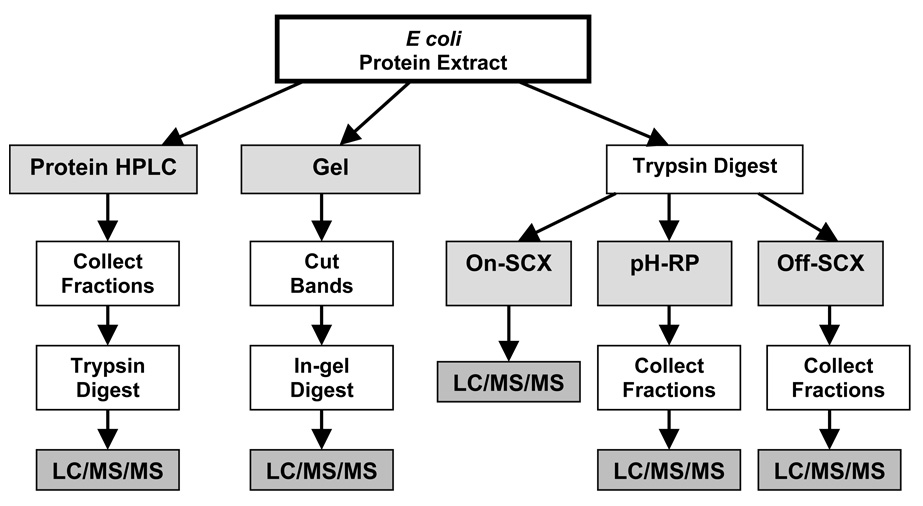
In order to systematically compare the relative efficiencies of 2D separations for low microgram quantities of protein extract, we employed a consistent protein load in the second dimension (10µg), consistent number of collected fractions (10–12), consistent LC-MS/MS conditions, and consistent database search parameters (see Figure 1 for a workflow overview and Table 2 for a summary of method conditions). The total number of proteins identified from each 2D separation scheme was used as the prime indicator of separation efficiency (see Table 3). We also examined the various reasons for the observed disparities between the separation efficiencies, including resolving power, orthogonality, and sample loss, as well as the physicochemical characteristics of the proteins identified by each method.
Figure 1.
Overview of Gel, On-SCX, pH-RP, Off-SCX, and Protein HPLC (includes cProt, hrProt, and hrProt+Urea) workflows.
Table 2.
Summary of 2D Separation Methods
| Type of Separation | 1st Dimension Column Type | 1st Dimension Mobile Phase A: | 1st Dimension Mobile Phase B: | Gradient Percent B: | |
|---|---|---|---|---|---|
| hrProt | Off-line Protein | High Protein Recovery C18 | 0.1% TFA H2O | 0.1% TFA ACN | 15–55 |
| hrProt + Urea | Off-line Protein | High Protein Recovery C18 | 0.1% TFA H2O | 0.1% TFA ACN | 15–55 |
| cProt | Off-line Protein | Conventional 300Å C18 | 0.1% TFA H2O | 0.1% TFA ACN | 15–55 |
| pH-RP | Off-line Peptide | High pH Stable C18 | 200mM NH4Formate pH 10 | ACN | 5–35 |
| On-SCX | On-line Peptide | Strong Cation Exchange | Ammonium Citrate 10mM | Ammonium Citrate 10mM | pH Buffers |
| Off-SCX | Off-line Peptide | Strong Cation Exchange | 25% ACN, 5mM K2HPO4 | 25% ACN, 500mM K2HPO4 | 5–100 |
| Gel | Protein | NA | NA | NA | NA |
Table 3.
Comparison of Separation Methods According to the Number of Total Proteins, Unique Peptides, Total Peptides, Average Percent Sequence Coverage, Average Protein Scores (log E), and False Discovery Rate (FDR)
| Total Proteins |
||||||||
|---|---|---|---|---|---|---|---|---|
| Mascot | X!Tandem | Unique Peptides | Total Peptides | % Unique/Total | Avg % Sequence Coverage | Avg Protein Scores (log E) | %FDR | |
| pH-RP | 266 | 299 | 749 | 870 | 86.1 | 13.1 | −16.3 | 1.16 |
| cProt | 241 | 252 | 731 | 1036 | 70.6 | 19.4 | −19.3 | 1.03 |
| hrProt+Urea | 255 | 287 | 977 | 1509 | 64.7 | 19.3 | −24.9 | 1.05 |
| hrProt | 281 | 271 | 1110 | 1848 | 60.1 | 19.8 | −27.3 | 1.07 |
| Offline SCX | 81 | 105 | 172 | 232 | 74.1 | 8.3 | −8.1 | 1.35 |
| Online SCX | 178 | 199 | 369 | 857 | 43.1 | 9.4 | −9.7 | 1.17 |
| Gel | 139 | 157 | 354 | 476 | 74.4 | 9.5 | −12.2 | 1.35 |
Table 3 summarizes the total proteins identified by Mascot and X!Tandem for each workflow. As can be seen from the table, high pH reverse phase (pH-RP) and high recovery protein reverse phase (hrProt) gave the highest number of proteins identified, followed closely by high recovery protein reverse phase with urea (hrProt+Urea), and conventional protein reverse phase (cProt). The gel electrophoresis (Gel) and on-line strong cation exchange (On-SCX) gave average results while the off-line strong cation exchange (Off-SCX) gave the worst results.
In addition to total proteins identified, we examined the total number of peptides identified (including redundant identifications), the total number of unique peptides (only non-redundant identifications), the average protein sequence coverage, and the protein E-value (the probability score). The protein-based HPLC separations consistently yielded the highest number of total peptides and the highest protein scores. In contrast, the pH-RP had a relatively low number of total peptides, yielding less than half as many of hrProt (870 versus 1848), but had an equal or greater number of protein identifications compared to the protein-based separations. At first glance, this seems counterintuitive, since logically a higher number of identified peptides should yield a higher number of identified proteins. However, if one compares the average protein sequence coverage between the two methods, the hrProt method exhibits much better sequence coverage than the pH-RP. In addition, the hrProt gives a higher average protein probability score. Both of these values reflect the higher number of peptides identified per protein in the hrProt fractionation compared to the pH-RP method.
Although the hrProt method gives higher average protein scores and sequence coverage, the pH-RP still yields a similar or greater number of identified proteins. The reason for this can be seen in the percentage of unique/total peptides given in Table 3. The ratio of unique to total peptides is a measure of how often the instrument fragments the same peptide (MS/MS re-sampling). This re-sampling causes a loss in duty cycle (the instrument spends valuable MS/MS time re-fragmenting the same peptides) and results in a lower number of protein identifications. The substantially higher percentage of unique/total peptides given by the pH-RP method is indicative of a low MS/MS re-sampling rate. So while the hrProt method yields more total peptides than the pH-RP method, the pH-RP method has a higher ratio of unique/total peptides. This is due to the hrProt method having a higher rate of re-sampling events which results in a lower number of protein hits per peptide sequenced. But why do certain workflows exhibit more re-sampling events? Re-sampling is partly dependent upon LC-MS/MS instrument parameters, such as dynamic exclusion, however, in this study the LC-MS/MS parameters have been held constant and thus the re-sampling differences are the result of distinct characteristics of the first dimension separations.
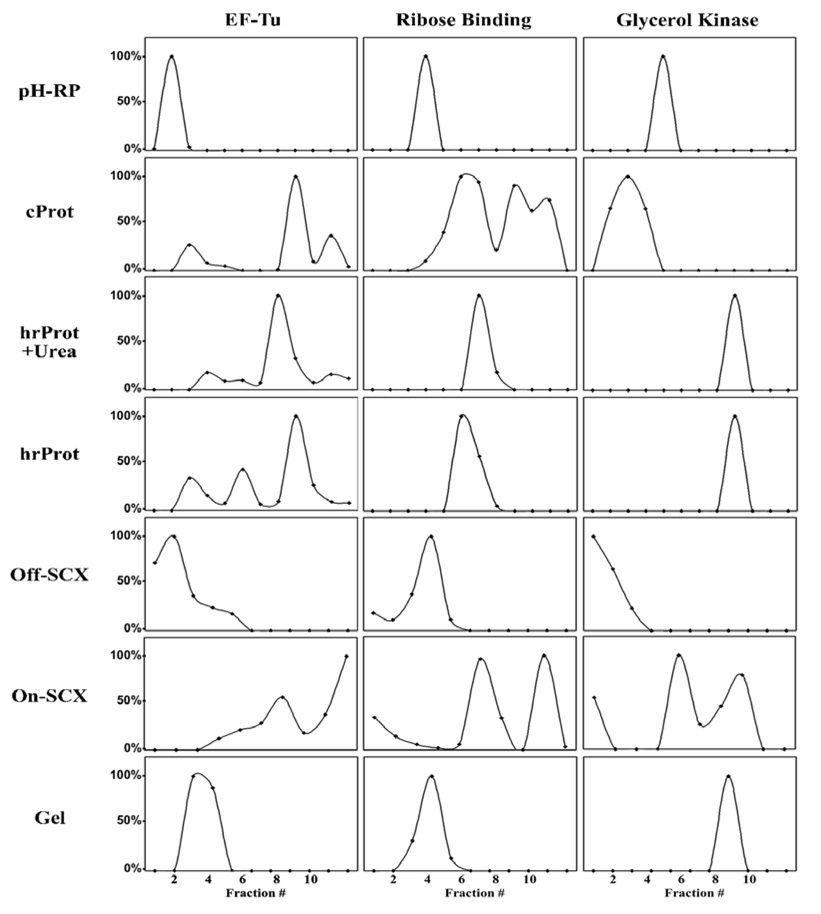
In order to explore the nature of these re-sampling events, we examined the resolution of the seven methods by comparing the elution profiles of three “signature peptides” from three different proteins (see Figure 2). These proteins were picked based on their high abundance in all seven methodologies. Extracted ion chromatograms (XICs) were created across all fractions of every workflow. Using the height of the XICs in each fraction, we were able to generate a virtual elution profile of the protein/peptides in the first dimension of separation. From these XIC chromatograms we compared the relative resolution of each method. As can be seen in Figure 2, the XICs reveal two separate events—peak broadening and peak multiplicity (elution of the same analyte in multiple fractions). Both of these events contribute to re-sampling by introducing the same peptide back to the LC-MS/MS in multiple fractions. The pH-RP workflow exhibits very high resolving power, as evidenced by the discrete distribution of the signature peptides into singular fractions, and thus a low rate of re-sampling and a higher percentage of unique/total peptides.
Figure 2.
Resolution comparion: Elution profiles of three different proteins (Elongation Factor Tu (P02290), Ribose Binding Protein (P02925), and Glycerol Kinase (P08859)) were reconstructed by taking the signal intensity for signature peptides across all collected fractions. The signal intensity is the height of the extracted ion chromatograms (XICs) normalized to 100%.
While pH-RP yields a high resolution separation, SCX yields a much lower resolution separation. Both the On-SCX and the Off-SCX exhibit fairly extreme peak broadening. In addition, the On-SCX exhibits peak multiplicity which results in a high rate of peptide re-sampling. It is not known specifically why this occurs and why it appears to be more pronounced in the On-SCX, which is a pH elution method, relative to the Off-SCX, which is a salt elution method. According to Dai et al., the elution of peptides in the pH mode occurs according to the isoelectric point (pI) of the peptide[30]. However, a peptide possessing multiple functional groups with different pKa’s could interact with the SCX resin in a more complex manner, resulting in a peak elution profile which is not strictly dictated by isoelectric point. If this were the case, then peptides containing fewer ionizable functional groups would elute in a single peak while more complex peptides with multiple ionizable groups would elute in multiple peaks. However, the interaction between individual peptides and the SCX resin is complex and depends on both the pH and ionic strength of the elution buffer.
In regards to the protein based HPLC workflows, both the peak broadening and peak multiplicity are fairly pronounced. These phenomenon have been documented in the literature and have contributed to the general reluctance of researchers to use protein reverse phase in 2D shotgun proteomics[21, 43, 44]. Many factors seem to be involved in the production of peak broadening and multiplicity, including alkyl chain length in the stationary phase, column temperature, mobile phase composition, and the conformation of the protein itself, i.e. native or denatured[23]. For example, it was previously shown that separation efficiency and resolution of intact proteins improved considerably with increasing alkyl chain length. However, potential sample loss due to irreversible protein adsorption onto the C18 stationary phase may counteract the benefits of better separation performance. Thus, the use of C8 column may prove to be beneficial for the separation and identification of low abundance proteins. Due to the inherent complexity of their structures, proteins exhibit very unpredictable interactions with the hydrophobic stationary phase in reverse phase HPLC. In contrast to peptide-based reverse phase HPLC separation, these interactions seem to be a non-partitioning type of separation in which proteins do not actually adsorb into the stationary phase but instead “stick” to the stationary phase due to hydrophobic interactions. As can be imagined, the degree to which a protein’s hydrophobic core is exposed dictates how well it “sticks” to the stationary phase. One of the most important factors in determining hydrophobic core exposure is the extent to which the protein is denatured. Using protein standards, Cohen et al. demonstrated an elution dependence on protein conformation—proteins in their native state exhibit different elution times and peak broadening compared to the same protein in its denatured state[44]. In order to examine the effects of protein denaturation, we used a combination of 6M urea and 0.1% TFA as a sample buffer to denature the whole protein extract before fractionation (hrProt+Urea). However, we observed precipitation at higher protein concentrations (1µg/µL) and deemed it necessary to systematically examine the effects of different acid concentrations on protein solubility. As can be seen from Table 1, the addition of acid to our protein extract causes protein precipitation at all tested concentrations. The addition of 6M urea as a denaturant/solubilizing agent alleviates some of the solubility issues but protein precipitation still occurs at a concentration of 0.6µg/µL. From these experiments, we concluded the maximum concentration we could use with denaturing conditions (6M urea + 0.1% TFA) was 0.4µg/µL. Using a 100µL loop, this concentration yielded a load of 40µg which is 2.5-times lower than that employed for the Off-SCX and pH-RP. In order to adjust for this disparity, we injected 5µL of the digested hrProt and cProt fractions in second dimension instead of the 2µL employed in the Off-SCX and pH-RP workflows. As can be seen from Figure 2, the addition of urea and TFA as denaturants did not significantly diminish peak broadening or multiplicity, nor did it yield more protein identifications and in fact, produced a lower number of protein identifications due possibly to protein precipitation. While we examined some aspects of protein denaturation on separation efficiencies, we did not include disulfide bond reduction or sample pre-heating, both of which enhance denaturation, however, Jorgensen and co-workers report that the reduction of disulfide bonds does not mitigate peak broadening or multiplicity[45].
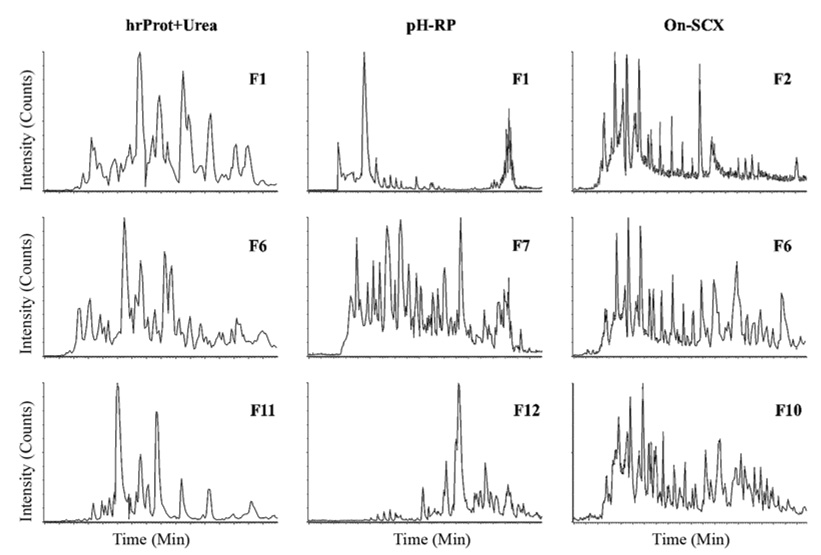
In addition to resolution, the other factor affecting separation efficiency in 2D experiments is the orthogonality of the separations. As mentioned in the introduction, orthogonality is a measure of the relative difference between the separation modes in the first and second dimensions, or the difference in the selectivities of the two modes[39]. Figure 3 exhibits the orthogonality of three workflows by comparing the distribution of peptides in the second dimension RP-HPLC. The chromatograms are base peak intensity (BPI) traces of eluting peptides in three discrete fractions from the beginning, middle, and end of the first dimension separation. As can be seen from all three of the hrProt-Urea BPI chromatograms, the peptides are evenly distributed throughout, indicating good orthogonality. A protein separation followed by digestion and a peptide level separation is inherently orthogonal since the parent proteins exhibit completely different physicochemical properties than their constituent tryptic peptides. All of the protein-based workflows, including the Gel workflow, exhibit good orthogonality (data not shown). The consideration of orthogonality is more important when separating the same species in subsequent fractionations, as with the separation of peptides first by ion exchange and then by reverse phase. Ion exchange and reverse phase selectivities are very different—ion exchange separates by charge while reverse phase separates by hydrophobicity—and thus they exhibit a large degree of orthogonality and a constant elution profile throughout fractions (see Figure 3). In contrast, the pH-RP exhibits a staggered elution profile when examined from the early fraction to the late fraction. The first fraction (F1) exhibits an obvious retention time bias towards early elution; the middle fraction exhibits no bias; and the last fraction (F12) exhibits a bias towards late elution. The pH-RP method employs the same reverse phase stationary phase in the first and second dimensions; the selectivity is changed from the first to second dimensions by simply varying the mobile phase pH. While the pH difference does change the charge on certain functional groups, such as primary amines and carboxylic acids, this change does not constitute a monumental change in hydrophobicity, especially for peptides lacking a large number of these functional groups, and thus the selectivity of the high pH first dimension is not grossly different from the selectivity of the conventional acidic second dimension, resulting in a semi-orthogonal separation.
Figure 3.
Comparison of orthogonality: Base peak intensity (BPI) chromatograms for fractions from the beginning (F1, F2), the middle (F6, F7) and the end (F10–F12) of hrProt+Urea, pH-RP, and On-SCX.
The last factor affecting protein identifications via 2D shotgun proteomics is sample loss. As mentioned above, the Off-SCX mode exhibited good orthogonality and decent resolution, however, due to extreme sample loss this method resulted in the lowest number of protein identifications, total peptides, and unique peptides. Most of the previous studies employing 2D LC-MS/MS with SCX-RP utilized a large amount of protein (1–2mg)[26–30]. The use of such a large amount of proteins makes it difficult to assess sample loss and the relative sensitivity of these 2D methods. In the current study, we did not assess protein loss/recovery directly but instead inferred recoveries based on total proteins and peptides identified. As can be seen from Table 3, Off-SCX produced a pronounced amount of sample loss compared to the other workflows. In contrast, the On-SCX, pH-RP, and protein-based separation methods exhibited a fairly low amount of sample loss. The Gel separation exhibited a fair degree of sample loss as a result of the in-gel digestion which is less efficient than in-solution digestion.
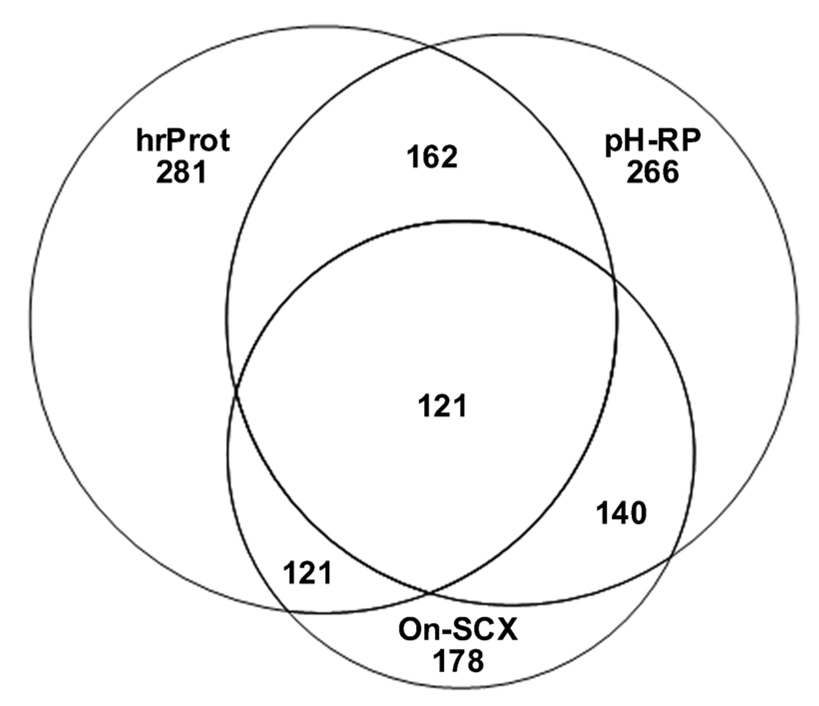
To examine the proteome coverage of different techniques, we compared the number of unique protein identifications in the On-SCX, hrProt, and pH-RP workflows. As shown in Figure 4, hrProt and pH-RP yield a large number of unique protein identifications and On-SCX a significantly lower number. Both the hrProt and pH-RP cover the majority of the On-SCX identified proteins but demonstrate a significant independence from each other. Thus the hrProt and pH-RP are highly complementary to each other, each yielding a high number of unique proteins, while the majority of the proteins identified by On-SCX are encompassed by the other two methods. Employing both hrProt and pH-RP yields a greater proteome coverage compared to either workflow taken independently.
Figure 4.
Venn diagram representing the overlap of Mascot identified proteins in the pH-RP, hrProt, and On-SCX workflows.
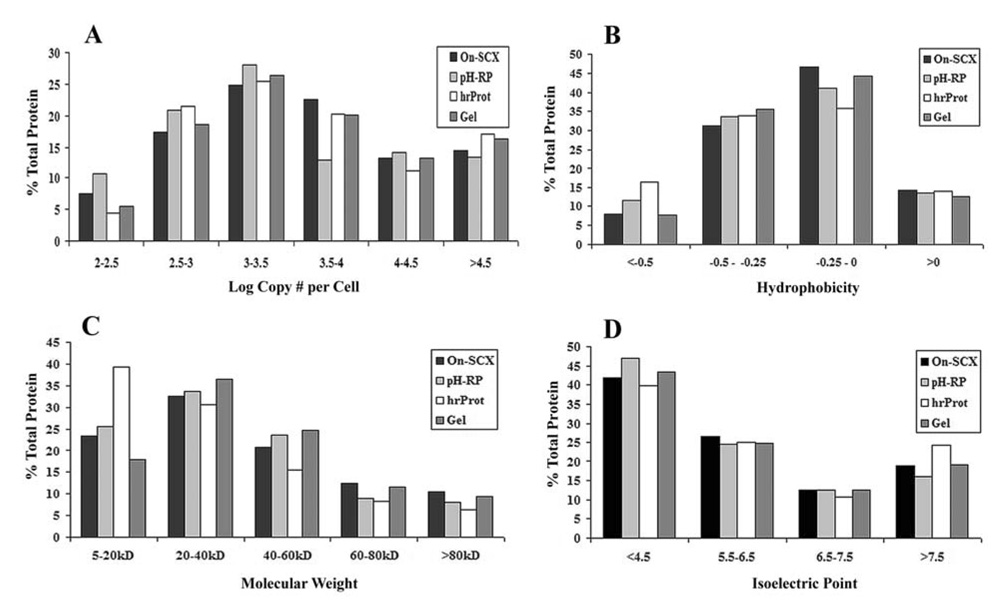
In addition to proteome coverage, resolution, and orthogonality, we also examined the physicochemical characteristics of the identified proteins, including protein abundance, hydrophobicity, molecular weight, and isoelectric point. As can be seen in Figure 5, there seems to be no significant bias for any one method for proteins of a specific hydrophobicity or isoelectric point. In addition, no significant bias is observed in protein abundance, except perhaps a small bias exhibited by the pH-RP workflow in determining low copy number proteins (log copy number below 2.5). However, as can be seen in the molecular weight comparison, the hrProt exhibited a fairly pronounced advantage in identifying low molecular weight species (under 20kD) in comparison with the other methods. It is unknown why this occurs but could be important in regards to experimental design for studies specifically targeting low molecular weight proteins.
Figure 5.
Comparison of protein abundance (A), hydrophobicity (B), molecular weight (C), and isoelectric point (D) between On-SCX, pH-RP, hrProt, and Gel workflows. Protein abundance is reported as the log copy number per cell as determined by Frishman and co-workers[46]. Hydrophobicity was determined using the Kyte-Doolittle scale[47]. The molecular weight and isoelectric point were calculated using the ExPASy Server tool: Compute MW/pI.
CONCLUSIONS
In conclusion, all of the techniques examined in the current study are suitable for 2D shotgun proteomics but each has its advantages and limitations depending on specific experimental design, available equipment, expertise, and monetary resources. The simplest and most cost effective method is the gel-based workflow—this method is cheap and can be performed using equipment commonly available in most biology labs. The off-line HPLC-based methods are more costly and require specialized equipment—an HPLC and columns. However, the off-line HPLC methods are more effective, flexible, and higher throughput than the gel-based method. Within the off-line methods, the peptide separations are easier than the protein-based methods owing to simple sample processing, i.e. en masse digestion versus digestion of individual fractionations. In addition, protein level separations generally preclude the use of stable isotope labeling for quantitative shotgun proteomics. However, protein level separations are superior for spectral counting due to the high number of total peptides identified. Also, protein level separations seem to be superior for smaller proteins and are generally more effective at removing residual abundant proteins after immunodepletion of serum than peptide HPLC[25]. The on-line methods are the most technically challenging of all of the methods and require the most complicated and expensive equipment. However, sample loss is mitigated when comparing the same separation mode, i.e. SCX. Finally, the coupling of two highly resolving and completely orthogonal separation techniques such as CIEF/nano-RPLC may offer an attractive alternative for enhanced proteome coverage using minute quantities of proteins[37,48].
ACKNOWLEDGEMENTS
This work was supported in part by the School of Pharmacy and the Wisconsin Alumni Research Foundation at the University of Wisconsin-Madison, and the National Institutes of Health through grant AI0272588. J.A.D. acknowledges an American Foundation for Pharmaceutical Education (AFPE) pre-doctoral fellowship. L.L. acknowledges an Alfred P. Sloan Research Fellowship. We would also like to thank Agilent for providing the mRP-C18 column free of charge.
REFERENCES
- 1.Klose J, Kobalz U. Electrophoresis. 1995;16:1034–1059. doi: 10.1002/elps.11501601175. [DOI] [PubMed] [Google Scholar]
- 2.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Nat.Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 3.Yates JR, 3rd, Carmack E, Hays L, Link AJ, Eng JK. Methods Mol.Biol. 1999;112:553–569. doi: 10.1385/1-59259-584-7:553. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Sadygov RG, Yates JR., 3rd Anal.Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 5.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP. Chem.Rev. 2007;107:3654–3686. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 6.Gan CS, Reardon KF, Wright PC. Proteomics. 2005;5:2468–2478. doi: 10.1002/pmic.200401266. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Gong Y, Wang Y, Wu S, Cai Y, He P, Lu Z, Ying W, Zhang Y, Jiao L, He H, Zhang Z, He F, Zhao X, Qian X. Proteomics. 2005;5:3423–3441. doi: 10.1002/pmic.200401226. [DOI] [PubMed] [Google Scholar]
- 8.Opiteck GJ, Jorgenson JW. Anal.Chem. 1997;69:2283–2291. doi: 10.1021/ac961156d. [DOI] [PubMed] [Google Scholar]
- 9.Gao J, Opiteck GJ, Friedrichs MS, Dongre AR, Hefta SA. J.Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- 10.Butt A, Davison MD, Smith GJ, Young JA, Gaskell SJ, Oliver SG, Beynon RJ. Proteomics. 2001;1:42–53. doi: 10.1002/1615-9861(200101)1:1<42::AID-PROT42>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Giorgianni F, Desiderio DM, Beranova-Giorgianni S. Electrophoresis. 2003;24:253–259. doi: 10.1002/elps.200390021. [DOI] [PubMed] [Google Scholar]
- 12.Walker AK, Rymar G, Andrews PC. Electrophoresis. 2001;22:933–945. doi: 10.1002/1522-2683()22:5<933::AID-ELPS933>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Wall DB, Kachman MT, Gong S, Hinderer R, Parus S, Misek DE, Hanash SM, Lubman DM. Anal.Chem. 2000;72:1099–1111. doi: 10.1021/ac991332t. [DOI] [PubMed] [Google Scholar]
- 14.Kachman MT, Wang H, Schwartz DR, Cho KR, Lubman DM. Anal.Chem. 2002;74:1779–1791. doi: 10.1021/ac011159c. [DOI] [PubMed] [Google Scholar]
- 15.Yan F, Subramanian B, Nakeff A, Barder TJ, Parus SJ, Lubman DM. Anal.Chem. 2003;75:2299–2308. doi: 10.1021/ac020678s. [DOI] [PubMed] [Google Scholar]
- 16.Lee CL, Hsiao HH, Lin CW, Wu SP, Huang SY, Wu CY, Wang AH, Khoo KH. Proteomics. 2003;3:2472–2486. doi: 10.1002/pmic.200300586. [DOI] [PubMed] [Google Scholar]
- 17.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc.Natl.Acad.Sci.U.S.A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans CR, Jorgenson JW. Anal.Bioanal Chem. 2004;378:1952–1961. doi: 10.1007/s00216-004-2516-2. [DOI] [PubMed] [Google Scholar]
- 19.Zolla L, Bianchetti M. J.Chromatogr.A. 2001;912:269–279. doi: 10.1016/s0021-9673(01)00532-5. [DOI] [PubMed] [Google Scholar]
- 20.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. Proc.Natl.Acad.Sci.U.S.A. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson RV, Glazer AN. Anal.Biochem. 1990;188:295–299. doi: 10.1016/0003-2697(90)90609-d. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Hendrickson CL, Emmett MR, Marshall AG. Anal.Chem. 1999;71:4397–4402. doi: 10.1021/ac990011e. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Balgley BM, Rudnick PA, Lee CS. J. Chromatogr.A. 2005;1073:35–41. doi: 10.1016/j.chroma.2004.08.140. [DOI] [PubMed] [Google Scholar]
- 24.Marshall J, Jankowski A, Furesz S, Kireeva I, Barker L, Dombrovsky M, Zhu W, Jacks K, Ingratta L, Bruin J, Kristensen E, Zhang R, Stanton E, Takahashi M, Jackowski G. J.Proteome Res. 2004;3:364–382. doi: 10.1021/pr034039p. [DOI] [PubMed] [Google Scholar]
- 25.Martosella J, Zolotarjova N, Liu H, Nicol G, Boyes BE. J.Proteome Res. 2005;4:1522–1537. doi: 10.1021/pr050088l. [DOI] [PubMed] [Google Scholar]
- 26.Washburn MP, Wolters D, Yates JR., 3rd Nat.Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 27.Wolters DA, Washburn MP, Yates JR., 3rd Anal.Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 28.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J.Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 29.Vollmer M, Horth P, Nagele E. Anal.Chem. 2004;76:5180–5185. doi: 10.1021/ac040022u. [DOI] [PubMed] [Google Scholar]
- 30.Dai J, Shieh CH, Sheng QH, Zhou H, Zeng R. Anal.Chem. 2005;77:5793–5799. doi: 10.1021/ac050251w. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Jacobs JM, Camp DG, 2nd, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Anal.Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 32.Cargile BJ, Talley DL, Stephenson JL., Jr Electrophoresis. 2004;25:936–945. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 33.Xiao Z, Conrads TP, Lucas DA, Janini GM, Schaefer CF, Buetow KH, Issaq HJ, Veenstra TD. Electrophoresis. 2004;25:128–133. doi: 10.1002/elps.200305700. [DOI] [PubMed] [Google Scholar]
- 34.Tong W, Link A, Eng JK, Yates JR., 3rd Anal.Chem. 1999;71:2270–2278. doi: 10.1021/ac9901182. [DOI] [PubMed] [Google Scholar]
- 35.Figeys D, van Oostveen I, Ducret A, Aebersold R. Anal.Chem. 1996;68:1822–1828. doi: 10.1021/ac960191h. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Balgley BM, DeVoe DL, Lee CS. Anal.Chem. 2003;75:3145–3152. doi: 10.1021/ac034014+. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Rudnick PA, Evans EL, Li J, Zhuang Z, Devoe DL, Lee CS, Balgley BM. Anal.Chem. 2005;77:6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 38.Gilar M, Olivova P, Daly AE, Gebler JC. J.Sep.Sci. 2005;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 39.Gilar M, Olivova P, Daly AE, Gebler JC. Anal.Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 40.Delmotte N, Lasaosa M, Tholey A, Heinzle E, Huber CG. J.Proteome Res. 2007;6:4363–4373. doi: 10.1021/pr070424t. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura T, Kuromitsu J, Oda Y. J.Proteome Res. 2008;7:1007–1011. doi: 10.1021/pr7005878. [DOI] [PubMed] [Google Scholar]
- 42.Strader MB, Tabb DL, Hervey WJ, Pan C, Hurst GB. Anal.Chem. 2006;78:125–134. doi: 10.1021/ac051348l. [DOI] [PubMed] [Google Scholar]
- 43.Cohen KA, Schellenberg K, Benedek K, Karger BL, Grego B, Hearn MT. Anal.Biochem. 1984;140:223–235. doi: 10.1016/0003-2697(84)90158-1. [DOI] [PubMed] [Google Scholar]
- 44.Cohen SA, Benedek K, Tapuhi Y, Ford JC, Karger BL. Anal.Biochem. 1985;144:275–284. doi: 10.1016/0003-2697(85)90117-4. [DOI] [PubMed] [Google Scholar]
- 45.Eschelbach JW, Jorgenson JW. Anal.Chem. 2006;78:1697–1706. doi: 10.1021/ac0518304. [DOI] [PubMed] [Google Scholar]
- 46.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyte J, Doolittle RF. J.Mol.Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Balgley BM, Rudnick PA, Evans EL, Devoe DL, Lee CS. J. Proteome. Res. 2005;4:36–42. doi: 10.1021/pr049876l. [DOI] [PubMed] [Google Scholar]