Abstract
Islet amyloid polypeptide (IAPP, also known as amylin) is the major protein component of pancreatic amyloid fibers in type II diabetes and is normally cosecreted with insulin from the β-cells of the pancreas. IAPP forms amyloid fibrils rapidly at concentrations well below those found in vivo, yet progression of type II diabetes occurs over many years. Insulin, a known inhibitor of IAPP fibrillogenesis, exists as a dense crystalline or near-crystalline core in the secretory vesicle, while IAPP localizes to the region between the crystal and the secretory vesicle membrane. In vitro, IAPP fibrillogenesis is both accelerated by lipid membranes and inhibited by monomeric insulin. In this work, we investigate insulin–IAPP–lipid interactions in vitro under conditions chosen to approximate native secretory vesicle physiology and the amyloid disease state. The effect of insulin on IAPP fibrillogenesis is investigated using fluorescence spectrometry. Additionally, interactions of IAPP and lipids with crystalline insulin are studied using fluorescence microscopy. We find that, while soluble states of insulin and IAPP do not interact significantly, large assemblies of either insulin (crystals) or IAPP (fibers) can lead to stable IAPP–insulin interactions. The results raise the possibility of multiple physiological interactions between these two β-cell hormones.
Keywords: islet amyloid polypeptide, insulin, amyloid inhibition, protein-membrane interactions, peptide–crystal interactions, amylin, protein aggregation
Islet amyloid polypeptide (IAPP) and insulin are hormones cosecreted by the β-cells of the pancreas. In late stages of type II diabetes (T2D), pancreatic β-cell death is accompanied by deposition of IAPP amyloid fibers in ∼90% of patients (Westermark and Grimelius 1973). In an analogous manner, deposition of amyloid by the Alzheimer's β-peptide (Aβ) is associated with neuronal death in Alzheimer's disease (Caughey and Lansbury 2003). In general, the association of amyloid with cell death is supported by the observation that soluble oligomeric states of amyloid precursors, including IAPP and Aβ, induce disease-like toxicity when added to cultured cells (Janson et al. 1999) or injected into model animals (Lesne et al. 2006). Association of IAPP with T2D is further supported by a number of recent transgenic rodent models engineered to express human IAPP (Fox et al. 1993; Hoppener et al. 1993; D'Alessio et al. 1994; Soeller et al. 1998; Butler et al. 2004). For example, the human islet amyloid polypeptide transgenic (HIP) rat develops diabetes between 5 and 10 mo of age, with a progressive pathology strikingly similar to human T2D (Butler et al. 2004; Matveyenko and Butler 2006).
In vitro, IAPP forms amyloid fibers in ∼30 h timescale or faster at protein concentrations orders of magnitude below those present in vivo. This process is further accelerated to a timescale of ≤100 min in the presence of anionic phospholipid vesicles (Knight and Miranker 2004; Jayasinghe and Langen 2005). In contrast, progression of T2D takes many years. Some factor(s), such as chaperones, must act to prevent and/or clear amyloid deposits in vivo. One surprising candidate is insulin, which has been demonstrated to inhibit IAPP fiber formation in vitro (Westermark et al. 1996; Jaikaran et al. 2003; Larson and Miranker 2004; Gilead et al. 2006).
Insulin and IAPP are co-regulated at the expression level and have complementary hormone activities. Expression of both genes is controlled by common promoter elements (German et al. 1992). Protein levels are maintained at an ∼1:100 ratio of IAPP to insulin in healthy β-cells, and the highest reported ratio in disease is 1:20 (Gedulin et al. 1991; Hull et al. 2004). Both molecules are packaged into secretory granules as proproteins, which are cleaved by the same prohormone convertases, PC1 and PC2 (Docherty and Steiner 2003; Young 2005). As a putative paracrine hormone, IAPP has effects complementary to insulin; for example, both suppress glucagon secretion by islet α-cells (Kruger et al. 2006). IAPP may also provide feedback on the secretion of insulin (D'Alessio et al. 1994; Gebre-Medhin et al. 1998). Additionally, the insulin-degrading enzyme (IDE), an important component of metabolism, has also been shown to degrade IAPP and prevent accumulation of amyloid fibers in cell culture (Bennett et al. 2003). Recent crystal structures of IDE bound to either IAPP or insulin reveal intriguing similarities in the interaction with the two peptides (Shen et al. 2006). Both peptides can self-interact; insulin forms crystals in vivo and IAPP forms amyloid fibers. Since IAPP and insulin are cosecreted, the potential for interaction exists throughout the secretory pathway. Production of both peptides increases during the early development of T2D, leading to stresses to the secretory pathway that have been implicated in β-cell death (Huang et al. 2007; Laybutt et al. 2007). Finally, as both insulin and IAPP are localized at high concentrations in the secretory granule, it is important to understand the molecular interactions between these peptides under such conditions.
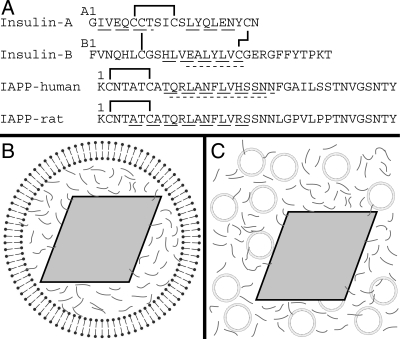
IAPP is known to interact with insulin when one component is in an immobilized or insoluble state. Notably, a kinetic study of insulin inhibition of IAPP fiber formation revealed strong interactions of insulin with IAPP fibers, but no significant interaction with monomeric IAPP was apparent (Larson and Miranker 2004). Inhibition of fiber formation appeared to be mediated by the binding of insulin to fiber ends. Insulin has also been observed to bind both to the sides of IAPP fibers and to IAPP immobilized on a surface plasmon resonance chip (Jaikaran et al. 2003). Finally, a study of IAPP and insulin sub-peptides suggested a potential for interaction between residues 7 and 19 on IAPP and the B chain of insulin (Gilead et al. 2006). This interaction site corresponds to a region of homology between the two peptides that, in part, mediates binding to IDE (Fig. 1A; Shen et al. 2006). However, interactions between fully solubilized forms of insulin and IAPP have not been reported.
Figure 1.
Schematic of peptide sequences and experimental conditions. (A) Sequences of insulin A and B chains, as well as human and rat versions of IAPP. Solid lines indicate disulfide bonds. Wide dashed underlines indicate regions identified as α-helical either in the T6 crystal structure of insulin (1MSO) (Smith et al. 2003), by EPR for membrane-bound human IAPP (Apostolidou et al. 2008), or by NMR for rat IAPP in solution (Williamson and Miranker 2007). Regions that bind the catalytic cleft of IDE are underlined with short dashes (Shen et al. 2006). (B) Schematic of an insulin secretory granule. A dense crystalline granule core of insulin (gray rhombus) is surrounded by a lipid bilayer and by IAPP (gray lines) in the halo region. (C) Schematic of experimental conditions for insulin crystal binding experiments. Synthetic liposomes (circles) and IAPP are added to preparations of insulin crystals. Illustrations are not to scale.
Within the secretory granule, insulin is packaged into an insoluble crystalline state. Conditions approximating the secretory granule (10 mM Zn++, pH ∼6.0) lead to rapid crystallization of insulin into a hexameric conformation known as T6 (Smith et al. 2003). In vivo, the effective concentration of insulin in the granule is ∼40 mM, and crystalline lattices consistent with the T6 crystal spacing are observable (Blundell et al. 1972). These crystals occupy ∼50%–90% of the secretory granule volume, and the remaining granule contents occupy the “halo” region between the crystal and the vesicle membrane (Fig. 1B). IAPP is generally found within this region, although one study reports IAPP localized to the crystal surface as well (Lukinius et al. 1989). The 1:100 molar ratio of IAPP to insulin corresponds to 0.8–4 mM IAPP in the halo region. Thus, IAPP exists at very high concentrations in close proximity to both the lipid membrane and the insulin crystal.
In vitro, IAPP fibrillogenesis is strongly inhibited by insulin but strongly accelerated by lipid membranes (Larson and Miranker 2004; Knight et al. 2006). Here, we investigate the effect of insulin on lipid-catalyzed fiber formation in order to understand the mechanism of insulin–IAPP–lipid interactions under conditions relevant to physiology. Further, we examine the interactions of soluble IAPP and lipids with crystalline insulin at secretory granule pH. We find that the presence of insulin as an ordered array (crystals) is sufficient to produce a strong interaction between the two peptides.
Results
The goal of this study is to characterize ternary interactions between IAPP, lipid membranes, and insulin. Our investigation measures two independent phenomena: (1) IAPP fiber formation and (2) binding of IAPP to the surface of insulin crystals. Here, the combined effects of insulin and lipid on fiber formation are characterized by established techniques in order to clarify the mechanism of insulin inhibition. The interactions of IAPP with crystalline insulin are investigated using microscopy with fluorescent tagged proteins and lipids (Fig. 1C). The insulin crystals used are prepared at secretory vesicle pH and Zn++ concentrations, with assays conducted at insulin:IAPP ratios of 50:1. Lipid-catalyzed fiber formation kinetics are measured as a function of the insulin:IAPP ratio at both vesicular and extracellular pH. These conditions were chosen to approximate the normal secretory granule environment and the conditions of extracellular IAPP fibrillogenesis.
Effects of insulin on lipid-catalyzed fiber formation
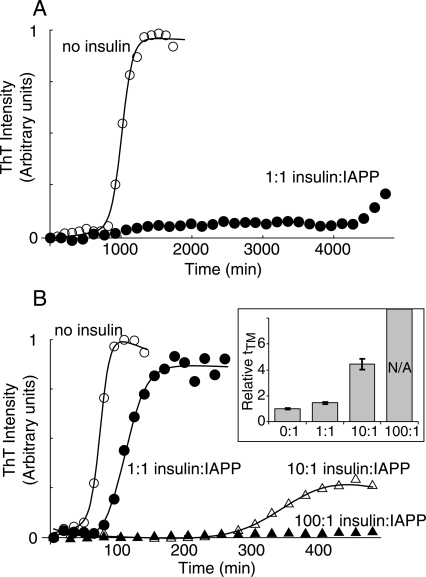
IAPP fiber formation is dramatically inhibited by insulin in the absence of lipid (Fig. 2A). Insulin has previously been shown to strongly inhibit IAPP fibrillogenesis reactions accelerated by the cosolvent (1,1,1,3,3,3)-hexafluoro-2-propanol (HFIP) (Larson and Miranker 2004). In order to test whether this inhibition similarly occurs in the absence of HFIP, fiber formation was measured with 10 μM IAPP in 50 mM sodium citrate, pH 6.0, with 10 mM Zn++ and ∼1% dimethyl sulfoxide (DMSO). IAPP forms fibers under these conditions in 800 ± 300 min, but requires >4000 min in the presence of equimolar insulin (Fig. 2A). The use of pH 6.0 and 10 mM Zn++ serves to approximate the conditions of the secretory granule (Hutton 1982; Hutton et al. 1983). Similarly, we approximate the extracellular environment by conducting reactions at pH 7.4 and without Zn++. Under these conditions, equimolar insulin also strongly inhibits fiber formation, with no conversion observed in reactions monitored for 2000 min (data not shown). In the absence of Zn++, insulin exists as a mixture of soluble monomer, dimer, and hexamer states (Pekar and Frank 1972). In both cases, insulin inhibition is qualitatively comparable to that observed in HFIP-catalyzed reactions.
Figure 2.
Inhibition of IAPP fiber formation by insulin, monitored by Thioflavin T (ThT) fluorescence. (A) Reaction kinetics of 10 μM IAPP with (filled) and without (open) 10 μM insulin at pH 6.0 with 10 mM Zn++ in the absence of liposomes. (B) Fiber formation of 10 μM IAPP at pH 7.4 with 1.3 mM DOPG. Data in A and B are representative of experiments performed in triplicate, in parallel with controls lacking IAPP (Supplemental Fig. 2). The transition midpoint time (tTM) was extracted from sigmoid fits (solid lines). (Inset) Mean values of tTM normalized to zero insulin, for reactions performed in triplicate. No transition was observed for 100:1 insulin:IAPP. Error bars are ±1 SD.
Insulin is a much less effective inhibitor of bilayer-catalyzed IAPP fiber formation. Fibrillogenesis of 10 μM IAPP catalyzed by 1.3 mM 1,2-dioleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)] (DOPG) liposomes occurs with a transition midpoint time (tTM) of 74 ± 5 min in the absence of insulin but is undetectable after 500 min in the presence of 1 mM insulin (Fig. 2B). Intermediate inhibition is observed at lower insulin:IAPP ratios; for example, an approximate fourfold increase in tTM is induced by 100 μM insulin. Notably, only a 1.4-fold increase in tTM is induced by equimolar insulin (Fig. 2B, closed circles). This is in marked contrast to the fivefold or greater change in tTM observed with equimolar insulin in the absence of lipid (Fig. 2A). Clearly, insulin is a less potent inhibitor of lipid-catalyzed fiber formation. However, lipid-catalyzed IAPP fiber formation is still strongly inhibited by insulin at the 100-fold excess concentration found in the β-cell secretory granule (Fig. 2B).
Binding to insulin crystals
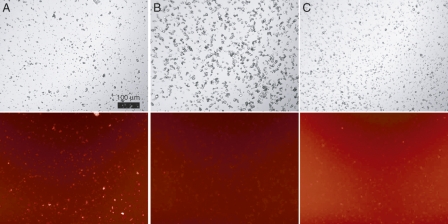
IAPP readily binds arrays of insulin in the form of crystals. Insulin crystals are the major component of secretory granules in vivo, surrounded by both soluble IAPP and a lipid bilayer (Fig. 1B). In order to determine whether IAPP interacts with insulin crystals under physiologically relevant conditions, we prepared insulin microcrystals in vitro using an adaptation of standard crystallization conditions (Baker et al. 1988; Martin and Zilm 2003). The conditions for crystallization (pH ∼6, high [Zn++]) are closely similar to the environment of the maturing secretory granule (Hutton 1982; Hutton et al. 1983). This results in a distribution of crystal sizes on the order of 1–10 μm in diameter (Fig. 3A, top). Addition of rhodamine-labeled IAPP (rhodamine-IAPP) to a final concentration of 2 μM results in the crystal surface becoming fluorescent (Fig. 3A, bottom). In contrast, no localized fluorescence is observed when rhodamine-IAPP is added to lysozyme crystals under matched buffer conditions (Fig. 3B). Additionally, 2 μM rhodamine-labeled bovine serum albumin (rhodamine-BSA) does not render insulin crystals fluorescent (Fig. 3C). These controls suggest the existence of specificity in IAPP–insulin crystal interactions. Using our observed average crystal size, the published unit cell dimensions (Baker et al. 1988), and assuming smooth crystal surfaces with one IAPP binding site per insulin monomer, we estimate the effective concentration of binding sites to be <1 μM in these experiments. Thus, as the concentrations of both binding partners are 2 μM or less, the easily visible fluorescence at the crystal surface suggests an affinity of low micromolar range or stronger. Clearly, IAPP can interact with insulin crystal surfaces at concentrations well below that found in vivo.
Figure 3.
Rhodamine-IAPP binding to insulin crystals. Microcrystals were prepared of either (A,C) human insulin or (B) hen egg white lysozyme, and washed into CZ6 buffer. To these were added (A,B) 2 μM rhodamine-IAPP or (C) 2 μM rhodamine-BSA. Crystals are clearly visible in phase contrast mode (top). Protein binding is evidenced by bright crystal surfaces when viewed with a Texas Red fluorescence filter (bottom). Note that rhodamine-BSA contains multiple fluorophores per molecule thus, the background fluorescence appears brighter. Scale bar (100 μm) applies to all panels.
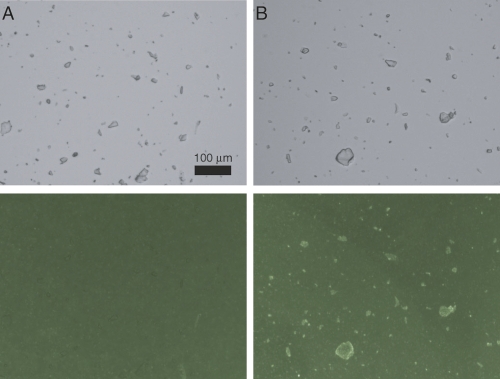
IAPP can simultaneously bind insulin crystal surfaces and lipid bilayers. DOPG liposomes were prepared that contained 0.25% fluorescent lipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-DOPE). Addition of these liposomes to insulin crystals yielded only uniform background fluorescence (Fig. 4A). To test for IAPP-mediated interactions between lipids and insulin crystals, we used a variant of IAPP from rat that binds DOPG membranes but does not form fibers (Jayasinghe and Langen 2005). Addition of 7 μM rat IAPP to the liposome/crystal mixture results in marked localization of fluorescence to the crystal surface (Fig. 4B). Human IAPP could not be used under these conditions, as it aggregates and gives punctate fluorescence not localized to crystal surfaces (data not shown). Neutralization of the liposome negative charge by rat IAPP is likely important for the observed colocalization. Wholly zwitterionic liposomes formed from 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), for example, can localize to the crystal surface in the absence of peptide (data not shown). Taken together, these results suggest that insulin crystals can interact with both IAPP and lipids under solution conditions comparable to those in the β-cell secretory granule.
Figure 4.
Fluorescent lipid binding to insulin crystals. Microcrystals of human insulin were incubated in CZ6 buffer with 0.13 mM DOPG liposomes containing 0.25% NBD-DOPE in the absence (A) or presence (B) of 7 μM rat IAPP. Crystals are clearly visible in phase contrast mode (top). Lipid binding is evidenced by bright crystal surfaces when viewed with a FITC fluorescence filter (bottom). Scale bar (100 μm) applies to all panels.
Discussion
Our investigation of IAPP–lipid–insulin interactions revealed three significant observations: (1) Insulin inhibition of IAPP fiber formation is strongly diminished when fibrillogenesis is lipid-catalyzed; (2) IAPP readily and specifically interacts with insulin crystal surfaces; (3) IAPP can mediate interactions between lipid bilayers and condensed insulin in vitro.
Insulin inhibition of IAPP fiber formation was previously studied using the co-solvent HFIP as a means to accelerate fiber formation to timescales appropriate for the laboratory (Larson and Miranker 2004). Fiber formation takes place in ∼1 h in 2% HFIP but ∼10–20 h in buffer alone (Fig. 2A). In that study, 2 μM insulin was sufficient to inhibit 25 μM IAPP fiber formation ∼10-fold. The capacity of insulin to act substoichiometrically was accounted for by modeling the binding of IAPP to fiber ends. The concentration of fiber ends is small relative to total protein, and binding can be expected to reduce the elongation rate. Since initiation of new fibers (primary nucleation) is comparatively rare, a reduction of the elongation rate strongly affects the overall rate of fiber formation. This mechanism is consistent with our measurements of inhibition performed here for reactions not catalyzed by HFIP.
In contrast, catalysis by lipid bilayers is not readily inhibited by insulin. In a lipid-catalyzed reaction, fiber elongation occurs predominantly at the bilayer surface (Knight and Miranker 2004; Lopes et al. 2007). The local concentration of IAPP at the surface is therefore greatly elevated. Importantly, insulin is not a membrane binding protein. Thus, the ratio of IAPP to insulin near fiber ends may be greatly increased at the membrane relative to bulk solution. A competitive inhibitor's efficacy is dependent on its relative local concentration at the site of action. Therefore, the results presented here are consistent with inhibition due to binding fiber ends. Our observations are also consistent with a recent report of IAPP fiber-induced membrane disruption, in which a 10-fold excess of insulin was necessary to observe significant reduction in vesicle leakage (Engel et al. 2008). Overall, insulin inhibition of fiber elongation is much less effective when IAPP and fiber ends, but not insulin, are concentrated at the liposome surface.
IAPP and insulin more readily interact when one component is present as an array. Our findings are consistent not only with insulin binding to fibrillar IAPP, but we correspondingly observe that soluble IAPP can bind to the surface of insulin crystals (Fig. 3). Under conditions similar to those used here, 100 μM IAPP does not appear to interact with 1 mM insulin in solution (Supplemental Fig. 1). This suggests the binding affinity is in the millimolar range or weaker. This observation further supports the conclusion that insulin inhibition of fiber formation occurs through interaction at fiber ends. In addition, insulin has been directly observed to bind IAPP fibers or IAPP immobilized on a surface plasmon resonance chip (Jaikaran et al. 2003). Here we have made the complementary observation that IAPP binds insulin crystals. Taken together, this suggests that insulin:IAPP interactions are mediated through condensed states.
Binding of IAPP to insulin crystals and inhibition of IAPP fiber formation by soluble insulin may involve different interfaces. In a recent study, fragments of either IAPP or insulin B chain were conjugated to a cellulose membrane and assayed for binding to full-length versions of the opposing peptide (Gilead et al. 2006). IAPP was observed to interact with fragments of insulin B chain corresponding to either residues B9–20 or B23–30. However, only the B9–20 segment was observed to inhibit IAPP fiber formation. In the T6 insulin crystal structure, residues B9–20 are located in the interior of the hexamer and participate in interface contacts (Smith et al. 2003). In contrast, residues B23–30 are exposed on the surface of the hexamer. Thus, it is plausible that the two modes of IAPP–insulin binding observed here correspond to interactions involving distinct residues.
IAPP and lipids bind to insulin crystals in vitro under conditions that approximate the secretory granule lumen. In this physicochemical study of IAPP surface interactions, practical considerations result in conditions that differ from physiological in two significant ways. First, the 100% DOPG membranes used in this study contain more negative surface charge than physiological membranes. This composition was chosen for fiber formation kinetic experiments, since its binding to IAPP and catalysis of amyloid formation have been well documented (Knight and Miranker 2004; Knight et al. 2006). IAPP in diabetic amyloid deposits is wild type, suggesting that alterations in local environment, such as lipids, are responsible for catalysis of amyloid formation in disease (Hull et al. 2004). Under non-diabetic physiological conditions, lipid-bound protein states that lead to amyloid fibers are unlikely to be highly populated. The second major difference is that concentrations of IAPP within the secretory granule halo region are ∼2 orders of magnitude higher than the concentrations used in our study (Hull et al. 2004). Thus, the interactions observed here, especially between IAPP and insulin, may be even more favored at the high IAPP concentrations found in vivo.
The structure and oligomeric state of IAPP upon binding to insulin crystals are not known. The in vivo roles, if any, of ternary interactions involving IAPP, insulin, and lipids in packaging and secretion are also unknown. It is intriguing to note, however, that IAPP knockout mice exhibit elevated glucose-stimulated insulin secretion (Gebre-Medhin et al. 1998). Plainly, the inhibition of IAPP fiber formation is not the evolved function of insulin. Rather, this phenomenon suggests that specific molecular interactions have the potential to take place in the unusual environment of a secretory pathway. Perturbation of these interactions may then result in amyloid assembly and related toxicity.
Materials and Methods
Materials
DOPG was from Avanti and dissolved in chloroform; DMSO was from J.T. Baker; Thioflavin T (ThT) was from Acros; human insulin was from Roche; tetramethylrhodamine-BSA was from Molecular Probes; human IAPP was synthesized using standard t-Boc methods and purified by the W.M. Keck facility (New Haven, CT); rat IAPP and N-terminally labeled rhodamine-IAPP were synthesized by the W.M. Keck facility, and cyclized and purified in house.
Peptide stocks
IAPP stocks were prepared in DMSO (human IAPP) or water (rat IAPP) as described previously (Knight and Miranker 2004). DOPG liposomes (100-nm diameter) were prepared by extrusion as described previously (Knight and Miranker 2004). Insulin stocks were prepared by dissolution into 20 mM HCl for crystallization, or dilute NaOH for use in fiber formation reactions. All experiments were either in KP7.4 buffer (50 mM potassium phosphate, 100 mM KCl, pH 7.4) or CZ6 buffer (50 mM sodium citrate, 10 mM ZnSO4, pH 6.0) as indicated. These buffers have ionic strengths of 0.23 and 0.26, respectively.
IAPP fiber formation kinetics
IAPP fiber formation was monitored by 50 μM ThT fluorescence in real time as described previously (Knight and Miranker 2004). Kinetic data were fit to sigmoid curves, from which the inflection point of the curve was extracted as the tTM. The tTM of IAPP fiber formation can vary by a factor of ∼2 between different IAPP stocks and as a function of the age of the peptide stock. Therefore, all reactions that are directly compared here were initiated and monitored simultaneously.
Insulin crystal preparation
Crystallization conditions were 0.86 mM insulin, 10 mM ZnSO4, 50 mM sodium citrate, 10% (v/v) acetone. These conditions generate the T6 form of the insulin crystal (Baker et al. 1988). Microcrystallization was initiated by centrifugation of this solution in a speed-vac until the volume had decreased to ∼20% of the original (Martin and Zilm 2003). Crystallization was then allowed to complete overnight at 4°C. The crystals were washed into CZ6 buffer at a total concentration of ∼1 mM insulin. We measured the residual concentration of soluble insulin under these conditions to be ∼15 μM (data not shown), which is similar to previously reported values for insulin crystals (Georgiou and Vekilov 2006). Notably, this concentration of soluble insulin is too low to significantly inhibit bilayer-catalyzed IAPP fiber formation (Fig. 2). The presence of crystals also does not significantly inhibit fiber formation (data not shown).
Fluorescence microscopy
IAPP and/or lipids were added to insulin crystals (total insulin concentration ∼100 μM) within 4-μL wells on glass slides, and were incubated 10 min at room temperature prior to visualization with a Zeiss Axioskop microscope. Images were acquired in phase mode, or in fluorescence mode using filters for Texas Red (Fig. 3) or fluorescein isothiocyanate (FITC) (Fig. 4). Acquired images were colorized and contrast was not adjusted. All images shown together were acquired with the same exposure times and were processed identically.
Electronic supplemental material
Electronic supplemental material includes HSQC NMR spectra of 0.1 mM IAPP with and without 1 mM insulin (Supplemental Fig. 1); Thioflavin T fluorescence data for kinetic control measurements lacking IAPP (Supplemental Fig. 2); and the corresponding experimental methods, figure legends, and references.
Acknowledgments
We thank Dr. O. Kerscher for assistance with fluorescence microscopy. This work was supported by NIH grant DK54899 to A.D.M.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Andrew D. Miranker, Department of Molecular Biophysics and Biochemistry, Yale University, 260 Whitney Avenue, New Haven, CT 06520-8114, USA; e-mail: andrew.miranker@yale.edu; fax: (203) 432-5175.
Abbreviations: IAPP, islet amyloid polypeptide; T2D, type II diabetes; Aβ, Alzheimer's β-peptide; IDE, insulin-degrading enzyme; DMSO, dimethyl sulfoxide; HFIP, (1,1,1,3,3,3)-hexafluoro-2-propanol; tTM, transition midpoint time (at which fibrillogenesis is 50% complete); ThT, Thioflavin T; DOPG, 1,2-dioleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)]; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; BSA, bovine serum albumin; NBD-DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl).
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036350.108.
References
- Apostolidou, M., Jayasinghe, S.A., Langen, R. Structure of α-helical membrane-bound hIAPP and its implications for membrane-mediated misfolding. J. Biol. Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, E.N., Blundell, T.L., Cutfield, J.F., Cutfield, S.M., Dodson, E.J., Dodson, G.G., Hodgkin, D.M., Hubbard, R.E., Isaacs, N.W., Reynolds, C.D., et al. The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- Bennett, R.G., Hamel, F.G., Duckworth, W.C. An insulin-degrading enzyme inhibitor decreases amylin degradation, increases amylin-induced cytotoxicity, and increases amyloid formation in insulinoma cell cultures. Diabetes. 2003;52:2315–2320. doi: 10.2337/diabetes.52.9.2315. [DOI] [PubMed] [Google Scholar]
- Blundell, T.L., Dodson, G., Hodgkin, D.M., Mercola, D. Insulin: The structure in the crystal and its reflection in chemistry and biology. Adv. Protein Chem. 1972;26:279–402. [Google Scholar]
- Butler, A.E., Jang, J., Gurlo, T., Carty, M.D., Soeller, W.C., Butler, P.C. Diabetes due to a progressive defect in β-cell mass in rats transgenic for human islet amyloid polypeptide (HIP rat): A new model for type 2 diabetes. Diabetes. 2004;53:1509–1516. doi: 10.2337/diabetes.53.6.1509. [DOI] [PubMed] [Google Scholar]
- Caughey, B., Lansbury, P.T. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- D'Alessio, D.A., Verchere, C.B., Kahn, S.E., Hoagland, V., Baskin, D.G., Palmiter, R.D., Ensinck, J.W. Pancreatic expression and secretion of human islet amyloid polypeptide in a transgenic mouse. Diabetes. 1994;43:1457–1461. doi: 10.2337/diab.43.12.1457. [DOI] [PubMed] [Google Scholar]
- Docherty, K., Steiner, D.F. The molecular and cell biology of the β cell. In: Porte D., et al., editors. Ellenberg and Rifkin's diabetes mellitus. 6th ed. McGraw-Hill; New York: 2003. pp. 23–41. [Google Scholar]
- Engel, M.F., Khemtemourian, L., Kleijer, C.C., Meeldijk, H.J., Jacobs, J., Verkleij, A.J., de Kruijff, B., Killian, J.A., Hoppener, J.W. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc. Natl. Acad. Sci. 2008;105:6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, N., Schrementi, J., Nishi, M., Ohagi, S., Chan, S.J., Heisserman, J.A., Westermark, G.T., Leckstrom, A., Westermark, P., Steiner, D.F. Human islet amyloid polypeptide transgenic mice as a model of non-insulin-dependent diabetes mellitus (NIDDM) FEBS Lett. 1993;323:40–44. doi: 10.1016/0014-5793(93)81444-5. [DOI] [PubMed] [Google Scholar]
- Gebre-Medhin, S., Mulder, H., Pekny, M., Westermark, G., Tornell, J., Westermark, P., Sundler, F., Ahren, B., Betsholtz, C. Increased insulin secretion and glucose tolerance in mice lacking islet amyloid polypeptide (amylin) Biochem. Biophys. Res. Commun. 1998;250:271–277. doi: 10.1006/bbrc.1998.9308. [DOI] [PubMed] [Google Scholar]
- Gedulin, B., Cooper, G.J., Young, A.A. Amylin secretion from the perfused pancreas: Dissociation from insulin and abnormal elevation in insulin-resistant diabetic rats. Biochem. Biophys. Res. Commun. 1991;180:782–789. doi: 10.1016/s0006-291x(05)81133-7. [DOI] [PubMed] [Google Scholar]
- Georgiou, D.K., Vekilov, P.G. A fast response mechanism for insulin storage in crystals may involve kink generation by association of 2D clusters. Proc. Natl. Acad. Sci. 2006;103:1681–1686. doi: 10.1073/pnas.0506526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German, M.S., Moss, L.G., Wang, J., Rutter, W.J. The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical β-cell nuclear complexes. Mol. Cell. Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilead, S., Wolfenson, H., Gazit, E. Molecular mapping of the recognition interface between the islet amyloid polypeptide and insulin. Angew. Chem. Int. Ed. Engl. 2006;45:6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- Hoppener, J.W., Verbeek, J.S., de Koning, E.J., Oosterwijk, C., van Hulst, K.L., Visser-Vernooy, H.J., Hofhuis, F.M., van Gaalen, S., Berends, M.J., Hackeng, W.H., et al. Chronic overproduction of islet amyloid polypeptide/amylin in transgenic mice: Lysosomal localization of human islet amyloid polypeptide and lack of marked hyperglycaemia or hyperinsulinaemia. Diabetologia. 1993;36:1258–1265. doi: 10.1007/BF00400803. [DOI] [PubMed] [Google Scholar]
- Huang, C.J., Lin, C.Y., Haataja, L., Gurlo, T., Butler, A.E., Rizza, R.A., Butler, P.C. High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress mediated β-cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes. 2007;56:2016–2027. doi: 10.2337/db07-0197. [DOI] [PubMed] [Google Scholar]
- Hull, R.L., Westermark, G.T., Westermark, P., Kahn, S.E. Islet amyloid: A critical entity in the pathogenesis of type 2 diabetes. J. Clin. Endocrinol. Metab. 2004;89:3629–3643. doi: 10.1210/jc.2004-0405. [DOI] [PubMed] [Google Scholar]
- Hutton, J.C. The internal pH and membrane potential of the insulin-secretory granule. Biochem. J. 1982;204:171–178. doi: 10.1042/bj2040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton, J.C., Penn, E.J., Peshavaria, M. Low-molecular-weight constituents of isolated insulin-secretory granules. Bivalent cations, adenine nucleotides and inorganic phosphate. Biochem. J. 1983;210:297–305. doi: 10.1042/bj2100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaikaran, E.T., Nilsson, M.R., Clark, A. Pancreatic β-cell granule peptides form heteromolecular complexes which inhibit islet amyloid polypeptide fibril formation. Biochem. J. 2003;377:709–716. doi: 10.1042/BJ20030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, J., Ashley, R.H., Harrison, D., McIntyre, S., Butler, P.C. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- Jayasinghe, S.A., Langen, R. Lipid membranes modulate the structure of islet amyloid polypeptide. Biochemistry. 2005;44:12113–12119. doi: 10.1021/bi050840w. [DOI] [PubMed] [Google Scholar]
- Knight, J.D., Miranker, A.D. Phospholipid catalysis of diabetic amyloid assembly. J. Mol. Biol. 2004;341:1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- Knight, J.D., Hebda, J.A., Miranker, A.D. Conserved and cooperative assembly of membrane-bound α-helical states of islet amyloid polypeptide. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- Kruger, D.F., Martin, C.L., Sadler, C.E. New insights into glucose regulation. Diabetes Educ. 2006;32:221–228. doi: 10.1177/0145721706286568. [DOI] [PubMed] [Google Scholar]
- Larson, J.L., Miranker, A.D. The mechanism of insulin action on islet amyloid polypeptide fiber formation. J. Mol. Biol. 2004;335:221–231. doi: 10.1016/j.jmb.2003.10.045. [DOI] [PubMed] [Google Scholar]
- Laybutt, D.R., Preston, A.M., Akerfeldt, M.C., Kench, J.G., Busch, A.K., Biankin, A.V., Biden, T.J. Endoplasmic reticulum stress contributes to β cell apoptosis in type 2 diabetes. Diabetologia. 2007;50:752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- Lesne, S., Koh, M.T., Kotilinek, L., Kayed, R., Glabe, C.G., Yang, A., Gallagher, M., Ashe, K.H. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Lopes, D.H., Meister, A., Gohlke, A., Hauser, A., Blume, A., Winter, R. Mechanism of islet amyloid polypeptide fibrillation at lipid interfaces studied by infrared reflection absorption spectroscopy. Biophys. J. 2007;93:3132–3141. doi: 10.1529/biophysj.107.110635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinius, A., Wilander, E., Westermark, G.T., Engstrom, U., Westermark, P. Co-localization of islet amyloid polypeptide and insulin in the B cell secretory granules of the human pancreatic islets. Diabetologia. 1989;32:240–244. doi: 10.1007/BF00285291. [DOI] [PubMed] [Google Scholar]
- Martin, R.W., Zilm, K.W. Preparation of protein nanocrystals and their characterization by solid state NMR. J. Magn. Reson. 2003;165:162–174. doi: 10.1016/s1090-7807(03)00253-2. [DOI] [PubMed] [Google Scholar]
- Matveyenko, A.V., Butler, P.C. β-cell deficit due to increased apoptosis in the human islet amyloid polypeptide transgenic (HIP) rat recapitulates the metabolic defects present in type 2 diabetes. Diabetes. 2006;55:2106–2114. doi: 10.2337/db05-1672. [DOI] [PubMed] [Google Scholar]
- Pekar, A.H., Frank, B.H. Conformation of proinsulin. A comparison of insulin and proinsulin self-association at neutral pH. Biochemistry. 1972;11:4013–4016. doi: 10.1021/bi00772a001. [DOI] [PubMed] [Google Scholar]
- Shen, Y., Joachimiak, A., Rosner, M.R., Tang, W.J. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G.D., Pangborn, W.A., Blessing, R.H. The structure of T6 human insulin at 1.0 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2003;59:474–482. doi: 10.1107/s0907444902023685. [DOI] [PubMed] [Google Scholar]
- Soeller, W.C., Janson, J., Hart, S.E., Parker, J.C., Carty, M.D., Stevenson, R.W., Kreutter, D.K., Butler, P.C. Islet amyloid-associated diabetes in obese A(vy)/a mice expressing human islet amyloid polypeptide. Diabetes. 1998;47:743–750. doi: 10.2337/diabetes.47.5.743. [DOI] [PubMed] [Google Scholar]
- Westermark, P., Grimelius, L. The pancreatic islet cells in insular amyloidosis in human diabetic and non-diabetic adults. Acta Pathol. Microbiol. Scand. [A] 1973;81:291–300. doi: 10.1111/j.1699-0463.1973.tb03538.x. [DOI] [PubMed] [Google Scholar]
- Westermark, P., Li, Z.C., Westermark, G.T., Leckstrom, A., Steiner, D.F. Effects of β cell granule components on human islet amyloid polypeptide fibril formation. FEBS Lett. 1996;379:203–206. doi: 10.1016/0014-5793(95)01512-4. [DOI] [PubMed] [Google Scholar]
- Williamson, J.A., Miranker, A.D. Direct detection of transient α-helical states in islet amyloid polypeptide. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A. Historical background. Adv. Pharmacol. 2005;52:1–18. doi: 10.1016/S1054-3589(05)52001-5. [DOI] [PubMed] [Google Scholar]