Abstract
The receptor for activated C-kinase 1 (RACK1) is a highly conserved WD40 repeat scaffold protein found in a wide range of eukaryotic species from Chlamydymonas to plants and humans. In tissues of higher mammals, RACK1 is ubiquitously expressed and has been implicated in diverse signaling pathways involving neuropathology, cellular stress, protein translation, and developmental processes. RACK1 has established itself as a scaffold protein through physical interaction with a myriad of signaling proteins ranging from kinases, phosphatases, ion channels, membrane receptors, G proteins, IP3 receptor, and with widely conserved structural proteins associated with the ribosome. In the plant Arabidopsis thaliana, RACK1A is implicated in diverse developmental and environmental stress pathways. Despite the functional conservation of RACK1-mediated protein–protein interaction-regulated signaling modes, the structural basis of such interactions is largely unknown. Here we present the first crystal structure of a RACK1 protein, RACK1 isoform A from Arabidobsis thaliana, at 2.4 Å resolution, as a C-terminal fusion of the maltose binding protein. The structure implicates highly conserved surface residues that could play critical roles in protein–protein interactions and reveals the surface location of proposed post-transcriptionally modified residues. The availability of this structure provides a structural basis for dissecting RACK1-mediated cellular signaling mechanisms in both plants and animals.
Keywords: RACK1, protein signaling, scaffolding protein, WD40, propeller, protein–protein interaction, drought
Scaffold proteins are uniquely poised to integrate signals from multiple pathways. Such proteins generate considerable functional diversity by mediating concomitant and/or promiscuous interactions with a vast array of protein partners. The receptor for activated C-kinase 1 (RACK1) is a member of the WD40 repeat family of β-propeller proteins. It was discovered through its ability to function as a scaffold protein, stabilizing signaling complexes involving protein kinase C (Ron and Mochly-Rosen 1994; Ron et al. 1994). In metazoans, RACK1 is now postulated to coordinate many different signal transduction pathways, ranging from cell division to ion channel regulation, by interacting with more than 80 different proteins (McCahill et al. 2002; Alfarano et al. 2005; Sklan et al. 2006). Global control of gene transcription, translation, and ribosome assembly and activation are also emerging as important cellular functions of RACK1 (Ceci et al. 2003; Nilsson et al. 2004). Definitive molecular tests, however, are lacking for most of RACK1's many postulated roles in signal transduction, partly because the structural basis for its interaction with other signaling molecules has not been elucidated.
RACK1 orthologs have also been discovered in primitive eukaryotes, such as Chlamydymonas (Schloss 1990), and are highly conserved in plants (van Nocker and Ludwig 2003), which do not express canonical protein kinase C enzymes (Herold et al. 2002). The sequence of human RACK1 is identical to the orthologs in chicken and mouse, and shares 66% sequence identity to RACK1A from Arabidopsis thaliana. This high degree of sequence conservation likely correlates to functional conservation between these diverse species.
Recently, it has been reported that RACK1A mediates multiple hormonal and developmental signaling pathways in A. thaliana (Chen et al. 2006). Preliminary results indicate that RACK1A plays an important molecular role in regulating diverse environmental stresses including drought response. As opposed to a single gene in metazoans, the A. thaliana genome maintains three different RACK1 genes termed RACK1A, RACK1B, and RACK1C. RACK1B and 1C share 87% and 93% sequence identity to RACK1A.
The first member of the WD repeat family of proteins for which a definitive structure has been determined is the β subunit of the mammalian heterotrimeric (αβγ) G proteins (Wall et al. 1995; Lambright et al. 1996; Sondek et al. 1996). RACK1 orthologs exhibit ∼25% sequence identity to the β subunit of the G proteins. Due to a lack of any reported RACK1 crystal structure, the RACK1 molecule has been modeled based on the Gβ subunit. These models have provided insight into the global fold of RACK1 but lack the fine structural details of diverse loop regions and divergent surface side chain conformations that are likely responsible for differences in binding interactions with protein partners.
To improve our understanding of RACK1 in signaling pathways, we have solved the crystal structure of RACK1A from A. thaliana at 2.4 Å resolution. This structure confirms RACK1A to be a seven-bladed β-propeller protein similar to the Gβ subunit, while demonstrating differences that may explain why RACK1 does not function as a canonical Gβ protein. Two large conserved surface regions have been identified that may serve as potential protein binding points. The surfaces of this molecule may provide a stable platform for multiple simultaneous protein–protein interactions, suggesting a mechanism for the regulation of inherently complex signaling pathways by RACK1. Since the early 1990s when RACK1 was identified as a Gβ subunit homolog in plants and animals, a great deal of functional characterization, mainly in animals, has established RACK1 as a scaffold protein competent to mediate diverse signaling pathways. Apart from generalized phenotypic characterization of the A. thaliana rack1a loss-of-function mutants (Chen et al. 2006), precise biological roles of RACK1A in plants have remained largely unexplored. The availability of structural data will aid in elucidating RACK1A-mediated developmental and environmental stress response pathways in plants.
Results and Discussion
Overall structure
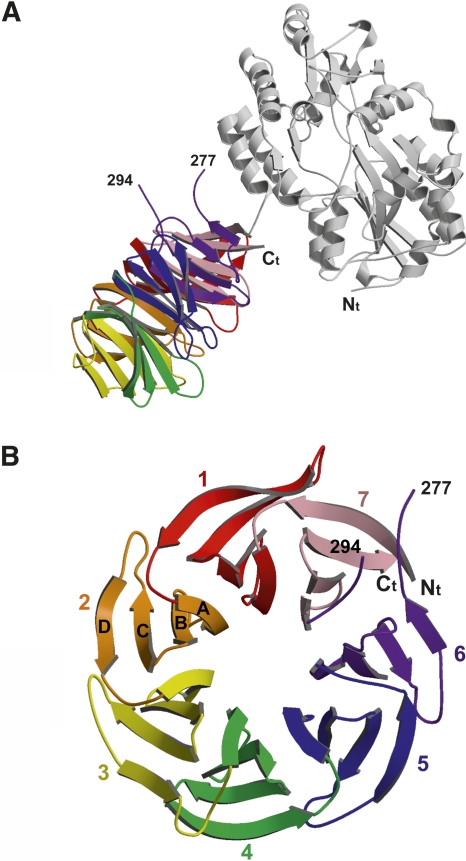
Initial attempts to crystallize native A. thaliana RACK1A failed to produce crystals. Therefore, we fused RACK1A to the C-terminal end of a mutant form of the maltose binding protein (MBP), producing a protein with different surfaces to promote crystallization. The MBP fusion protein used in this study utilized an identical linker to that used previously for MBP-fusion crystallization experiments (Kobe et al. 1999). In addition, MBP residues Asp82 and Lys83 from one loop and Lys239 from another were mutated to alanines in order to reduce surface entropy and potentially facilitate better crystal lattice contacts (Derewenda 2004). Based on secondary structure prediction, residue Gly4 of RACK1A was used as the starting residue to reduce the length and flexibility of the linker between the MBP protein and RACK1A. The fusion protein crystallizes with only one residue in the linker, Gly4 from RACK1A, not involved in secondary structure in either protein (Fig. 1A). Surprisingly, there are no interactions in the crystal structure between MBP and RACK1A of the same molecule. Residues lining the interface between the two domains are ∼5 Å apart. In addition, there are no RACK1A–RACK1A crystal lattice contacts, suggesting a reason why attempts to crystallize RACK1A alone have failed. Because MBP is used only as a vehicle for crystallization, the rest of the discussion will focus on RACK1A.
Figure 1.
(A) Ribbon diagram of the MBP–RACK1A fusion protein. The MBP molecule is colored gray and the RACK1A molecule is colored based on the different blades in the β-propeller. (B) Top view of the β-propeller of RACK1A. Each blade of the propeller is made up of four antiparallel β-strands with β-strand A lining the inside of the open pore and β-strand D on the outside of the propeller.
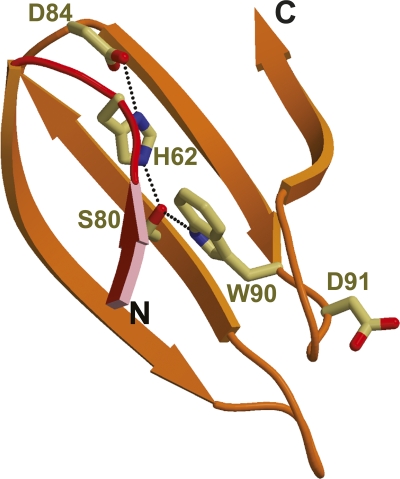
The structure of RACK1A is a canonical seven-bladed β-propeller similar to the heterotrimeric G protein Gβ. This fold was predicted from the presence of WD40 repeats in the sequence (Wall et al. 1995; Sondek et al. 1996; Sondek and Siderovski 2001). Each propeller blade consists of a four-stranded antiparallel β-sheet (Fig. 1B). The blades are arranged in sequential order around a pseudo-sevenfold axis that creates a water accessible channel in the middle of the propeller with a width of ∼9 Å. β-strand A of each blade lines the channel of the propeller while β-strand D is located along the outer surface of the protein. Loops connecting β-strand D from one propeller to β-strand A of the next and β-strand B to C form the top of the propeller, while loops connecting β-strand A to B and C to D line the bottom of the propeller. The sequentially defined WD repeat spans two blades and consists of strand D from one blade and A, B, and C of the following. The last blade therefore consists of β-strands from both the C terminus (β-strands A, B, and C) and N terminus (β-strand D). The WD residues, for which the repeat is named, are located at the end of strand C on the bottom side of the propeller and are conserved in blades 2, 5, and 6. The aspartate residue, of the WD motif, has been substituted with a Lys in blade 1, an asparagine in blades 3 and 4, and a glycine in blade 7. Also conserved within the WD40 repeat is a Asp–His–Ser hydrogen-bonding network that holds the blade together, while connecting it to the adjacent blades (Fig. 2). The major difference between the WD repeats is in the loops connecting the β-strands. It is the structural organization of these loops that provides the distinct features of each member of the WD family and distinguishes RACK1 interactions from those of other WD proteins (Garcia-Higuera et al. 1996; Sklan et al. 2006).
Figure 2.
Ribbon diagram of Asp–His–Ser hydrogen-bonding network responsible for maintaining the integrity of the β-sheet within blade 2 of the β-propeller. His62 lies on the loop connecting blade 1 with blade 2. Trp90 and Asp91 represent the WD repeat of blade 2.
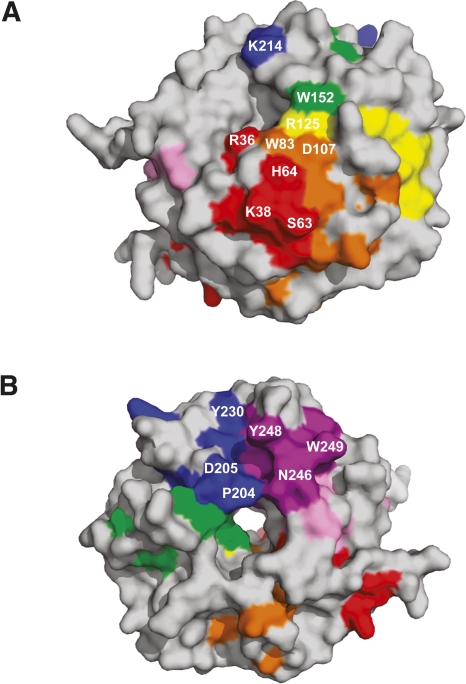
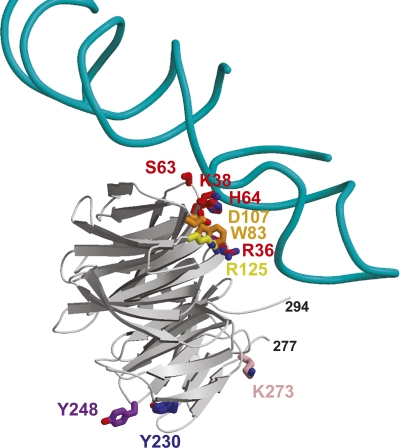
The high degree of sequence identity among different orthologs of RACK1 suggests conservation of biological functions that evolved prior to separation of the plant and animal kingdoms (Neer et al. 1994; Sklan et al. 2006). RACK1 is proposed to be a scaffolding protein, based on the array of proteins that can form interactions with it (Sklan et al. 2006), yet little is known about the specific interactions between RACK1 and its binding partners. Knowledge of specific residues involved in these interactions is particularly lacking. To better understand what residues may be important in complex formation for functional conservation across the plant/animal kingdoms, conserved residues from Figure 3 were mapped to the surface of the protein, revealing two conserved surface regions (Fig. 4A,B). The first conserved region (region 1) is located on the top rim of the propeller involving side chains from residues Arg36, Lys38, Ser63, His64 from blade 1; Trp83, Asp107 from blade 2; Arg125 from blade 3; and Trp152 from blade 4 (Fig. 4A). The second large conserved surface region (region 2) of RACK1 is located on the bottom of the propeller and is mainly comprised of conserved residues Pro204, Asp205, Tyr230 from blade 5 and Asn246, Tyr248, and Trp249 from blade 6 (Fig. 4B). These regions represent potential protein–protein interaction sites. Earlier mutational studies have implicated blade 5 and 6 as major docking stations for interaction with RACK1 supporting region 2 as a protein–protein interaction site (Chang et al. 2001; Steele et al. 2001; Sengupta et al. 2004).
Figure 3.
Sequence alignment of RACK1A from A. thaliana (GI:30685669) versus RACK1 from Homo sapiens (GI:83641897), Drosophila melanogaster (GI:24582618), and S. cerevisiae (GI:6323763), as well as a structural alignment with respect to the crystal structure of Gβ in complex with Gα and Gγ (PDB ID code 1GP2). Secondary structural elements in RACK1A are shown above the alignment and colored according to the ribbon diagram in Figure 1B. Identity between RACK1A from A. thaliana and human RACK1 versus the other proteins is listed at the end of the alignment. Residues conserved in all RACK proteins shown are marked with an asterisk below the residue. Residues in Gβ that are structurally similar to residues in RACK1A are capitalized. Residues of Gβ that line the interface with Gγ interface are colored red. Residues lining the interface with Gα are colored green. Tyrosine RACK1 residues predicted to be phosphorylated are colored purple and the predicted sumoylation site of human RACK1 is underlined.
Figure 4.
Surface diagrams of conserved regions within the RACK1 proteins. (A) Conserved region 1. Conserved residues are colored according to the blade to which they belong based on the ribbon diagram in Figure 1B. Residues Arg36, Trp83, Arg125, Trp152, and Lys214 line the upper rim of the β-propeller pore. (B) Conserved region 2. Tyr230 and Tyr248 on RACK1A are proposed phosphorylation sites. Residue Pro204 is located on the lower rim of the pore of the β-propeller. Molecular surfaces were created using PyMOL (DeLano Scientific).
Comparison to Gβ
When RACK1 was originally cloned from tobacco and from a rat brain cDNA library, it was reported to be a homolog of the Gβ subunit owing to high sequence similarity and the presence of WD40 repeats (Ishida et al. 1993; Ron et al. 1994). Crystal structures of the Gβ subunit and other WD40 repeat proteins revealed that the β-propeller structure is a common feature of WD40 repeat proteins (Wall et al. 1995; Sondek et al. 1996; Sprague et al. 2000; Pickles et al. 2002). The crystal structure of the bovine Gβ subunit has been widely used as a template to build homology models of RACK1 protein. Although WD repeat proteins, including RACK1 (Sondek and Siderovski 2001; Steele et al. 2001), can be modeled onto the Gβ structure, there are both structural and functional reasons for hypothesizing that they serve different functions in most cells (Smith et al. 1999; McCahill et al. 2002). In A. thaliana for example, it has been demonstrated that Gβ and RACK1A have distinct biological functions (Ullah et al. 2003; Chen et al. 2006).
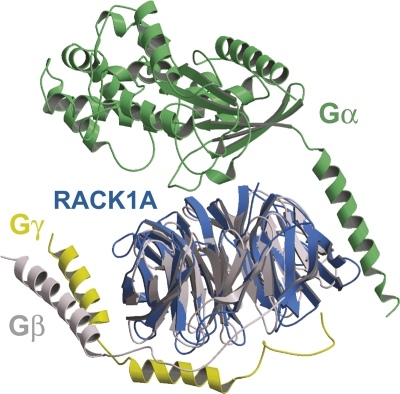
A comparison of the structures of RACK1A and Gβ reveals some possible reasons for the functional differences of these proteins (Fig. 5). Based on structural alignments with bovine Gβ, the overall root-mean-squared deviation for 278 structurally conserved Cα atoms is 1.5 Å, with an overall sequence identity of 23% (PDB ID code 1GP2). The greatest structural deviations between the two proteins are a long helix on the N terminus of Gβ (residues 1–44) and a loop insertion in plant RACK1A between propeller blades 6 and 7 that is unique to the majority of plant RACK1 proteins (Fig. 3). Despite the remaining similarities between the two proteins, it is unlikely that RACK1 binds the Gγ subunit in the same fashion as does Gβ. Gβ and Gγ are tightly associated with the N termini of both proteins engaged in a coiled-coil interaction. RACK1A lacks this helix as well as Asp258 of Gβ within the propeller that forms the interactions with Arg22 of Gβ and Arg27 of Gγ, properly positioning the coiled coil with respect to the propeller domain. In addition, the hydrophobic cleft that exists in Gβ, between β-sheets 1 and 7 that accommodates Phe61 of Gγ, is occupied in RACK1A by larger side chains, so that penetration by a phenylalanine side chains is unlikely. The current position of Tyr248 in the RACK1A structure would also cause significant steric clashes with the Gγ subunit.
Figure 5.
Superposition of the trimeric Gαβγ protein complex with RACK1A. Gα is colored green; Gβ, gray; and Gγ, yellow (PDB ID code 1GP2). RACK1A is colored blue.
The Gα subunit alone also does not bind RACK1. However, the Gβγ dimer and the Gαβγ trimer are both capable of forming complexes with RACK1, with the affinity for the Gαβγ trimer being threefold higher than for the Gβγ dimer (Dell et al. 2002). Dell et al. (2002) also demonstrated that a GST–RACK1 C-terminal truncation consisting of RACK1 residues 1–207 was required for trimeric G protein binding whereas residues 207–317 would not bind the G protein complex. This suggests that the conserved region 1 of RACK1 may be involved in trimeric G protein binding. So, although RACK1 is not a canonical substitute for the Gβ subunit, it can form interactions with the entire trimeric complex.
Despite the structural differences between RACK1A and Gβ, there is evidence to suggest that RACK1 homologs in some species might retain Gβ-like function. Zeller et al. (2007) recently established Asc1, a yeast homolog of RACK1, as a bona fide Gβ subunit since Asc1 has been shown to interact directly with the Gα subunit in a guanine nucleotide-dependent manner. In addition, the Asc1 ortholog in Cryptococcus neoformans, Gib2, has been identified as a Gβ-like subunit that functions as a positive regulator of cAMP, as opposed to a negative regulator in yeast where it regulates melanization and capsule formation associated with virulence (Palmer et al. 2006). This Gβ-like function occurs in the absence of interaction with a Gγ subunit. These proteins are thus described as “non-canonical Gβ subunits.” However, the structural basis of the reported interaction of RACK1 homologs in yeast and in Cryptococcus with the Gα subunit remains to be identified.
RACK1 ribosome interactions
RACK1 has been shown to regulate translation initiation by recruiting PKC to the ribosome (Ceci et al. 2003; Nilsson et al. 2004). Despite the lack of any known PKC ortholog in plants, a recent genome-wide screen for A. thaliana RACK1A interacting proteins has identified at least four ribosomal proteins (data not shown). Based on cryo-EM studies, a model of Saccharomyces cerevisiae RACK1 (derived from Gβ) binding to the 80S ribosomal subunit has been created. This model places RACK1 on the head region of the 40S ribosomal subunit near the mRNA exit channel (Sengupta et al. 2004). In addition, the authors demonstrated that mutations of S. cerevisiae RACK1 at Arg38 and Lys40 (Arg36 and Lys38 of RACK1A, in conserved region 1) disrupt the RACK1 interaction with the 40S subunit (Sengupta et al. 2004). These results are consistent with the crystal structure of RACK1A. A superposition of our RACK1A structure onto the RACK1-40S ribosome model (PDB ID code 1ARI) positions RACK1A so that the top rim of RACK1A is in contact with the ribosome, exposing the bottom rim to solvent (Fig. 6). Thus, the conserved region 1 may lie against the ribosome, exposing conserved region 2 to the solvent. This arrangement would allow RACK1 to associate with both the ribosome and other proteins simultaneously.
Figure 6.
Ribbon diagram of the model of RACK1 binding to the 80S ribosome complex, based on the cryo-EM structure (Sengupta et al. 2004). Shown are the H39 and H40 of rRNA from the 40S subunit and RACK1A. To make this figure, RACK1A was superimposed on the model of RACK1 created from Gβ (PDB ID code 1TRJ). Residues colored red, orange, and yellow are conserved residues from conserved site 1 of RACK1A. The purple tyrosine residues are the sites of putative Src phosphorylation at the second conserved site. The pink lysine is residue 273, the proposed sumoylation site. This figure suggests conserved region 1 could interact with the ribosomal RNA and conserved region 2 could recruit Src to the ribosome.
Proposed post-translational modification of RACK1A
Tyrosine phosphorylation and protein sumoylation have been implicated in regulation of RACK1 function (Chang et al. 2001; Yang and Gregoire 2006). Src-tyrosine kinase-mediated RACK1 phosphorylation has been implicated in regulating cell migration and cell cycle checkpoints in human colon cancer cells (Chang et al. 2001; Doan and Huttenlocher 2007; Mamidipudi et al. 2007). RACK1 has also been found to be a binding partner and inhibitor of Src tyrosine kinase (Chang et al. 1998, 2001). Mutagenesis work by this group identified Tyr228 and Tyr246 as potential phosphorylation sites and suggested a correlation between enhanced tyrosine phosphorylation of RACK1 and binding of RACK1 to Src.
Though no bona fide tyrosine kinase has been identified in plants, tyrosine phosphorylation and dephosphorylation have been documented to regulate growth and development in seeds, hypocotyls, and roots in A. thaliana (Reyes et al. 2006; Hussain et al. 2007; Yemets et al. 2008). It is hypothesized that tyrosine phosphorylation in plants is carried out by dual-specificity serine/threonine/tyrosine (STY) protein kinases (Rudrabhatla et al. 2006). In addition, tyrosine-specific protein kinase-like kinases (TKLs) have also been reported as possible tyrosine kinases (Miranda-Saavedra and Barton 2007). Interestingly, the two proposed tyrosine phosphorylation sites of human RACK1 are conserved in A. thaliana RACK1A (Tyr230 and Tyr248). These residues are located on the bottom side of the propeller in conserved region 2 (Figs. 4B, 6). Tyr230 is located at the start of β-strand D of blade 5 on the surface of the protein, but with the acceptor hydroxyl tucked back into the protein so that it is only slightly accessible. This suggests that some rearrangement might be necessary prior to phosphorylation. However, the side chain of Tyr248 is located at the end of the loop connecting β-strands A and B of blade 6 and is fully exposed to the solvent, with the putative phosphate acceptor hydroxyl pointing away from the protein, easily accessible for modification.
SUMO-1 (Small Ubiquitin-like Modifier) is an ubiquitin-related protein that is covalently conjugated to a diverse assortment of substrate proteins and regulates cellular functions by mediating protein–protein interactions and subcellular compartmentalization, as well as effecting protein stability (Salinas et al. 2004). It has been suggested that the underlying principle of sumoylation is to alter inter- or intramolecular interactions of the modified substrate (Geiss-Friedlander and Melchior 2007). At the molecular level, these functions can be controlled by creating intra-molecular conformational change or by creating or blocking the binding interface (Meulmeester and Melchior 2008). Metazoan RACK1 has a proposed “phospho-sumoyl” switch which is putatively involved in transcriptional repression (Yang and Gregoire 2006). The human RACK1 sequence, 270-LKQEVIST-277, has the consensus LK-x-E/D-x-x-SP found in many transcriptional factors. It has been suggested that phosphorylation of the serine enhances sumoylation of the lysine residue (Yang and Gregoire 2006). In plants, sumoylation is implicated in stress hormone ABA-mediated signaling pathways, attenuating ABA response (Lois et al. 2003). Kurepa and colleagues provided evidence for the potential involvement of sumoylation substrates in mediating elevated temperature stress responses (Kurepa et al. 2003). Miura and colleagues have extensively reviewed the role of sumoylation in plant growth and development (Miura et al. 2007). In A. thaliana RACK1A, there are two predicted sumoylation sites, Lys273 and Lys 277. These sites are located at the end of β-strand D of blade 6, similar to the predicted site for metazoan RACK1. For RACK1A this marks the beginning of a long loop that is not present in other nonplant RACK1 proteins. The presence of Lys273 and Lys277 on an exposed loop is consistent with predicted sumoylation sites (Bernier-Villamor et al. 2002; Macauley et al. 2006). However, RACK1A lacks the consensus Ser required for phosphorylation.
Homo-/heterodimerization of RACK1
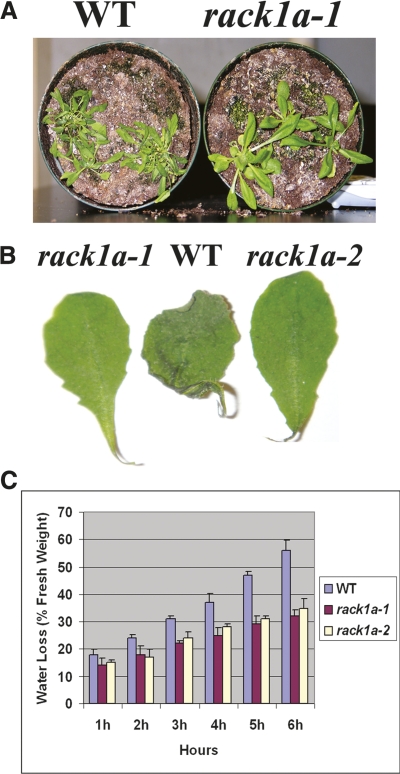
WD40 repeat-mediated protein–protein interactions are well documented. Evidence is accumulating for the homodimeric interactions of the WD40 repeat containing RACK1 proteins (Thornton et al. 2004; Liu et al. 2007). Dimerization events are frequently employed in transducing signals from the cell surface to the nucleus (Klemm et al. 1998). Phosphorylation of mammalian RACK1 at Ser146 promotes dimerization and leads to degradation of HIF-1α, a component of HIF-1, the master regulator of transcriptional response to changes in oxygen levels. Dephosphorylation of Ser146 by calcineurin impairs RACK1 dimerization (Liu et al. 2007). Ser146 is located in one of two loop regions within the propeller domain containing insertion differences between mammalian RACK1 and plant RACK1A (Fig. 3). This loop lacks a serine in RACK1A, instead containing a two-residue insert, GlyGly, which would alter the surface structure of RACK1A compared to mammalian RACK1. In the crystal structure of MBP–RACK1A, there are no RACK1A–RACK1A interactions. The GlyGly insert is located on the opposite face of the propeller from the MBP, suggesting that, if RACK1A were to form a symmetrical homodimer, this interaction could still occur in the fusion protein. Likewise, in the crystal, only the sides of blades 1 and 7 are occluded by the presence of the MBP, suggesting that many other permutations of nonsymmetrical homodimers might be able to form. However, we cannot rule out the possibility of homo- or heterodimerization under specific physiological environments such as water stress conditions in plants. Specially, homo- and heterodimerization between different family members of a protein family can generate considerable functional diversity. In this regard, as opposed to metazoan RACK1 genes, the plant RACK1 protein family has more than one family member, including three members in A. thaliana. As can be seen in Figure 7, the A. thaliana rack1a loss-of-function mutants show significant resistance to water stress conditions, limiting water loss through the guard cells. Presence of this conditional phenotype indicates a physiological and/or structural basis of RACK1A function in A. thaliana. In addition, the availability of more than one family member certainly could provide considerable opportunity for dimerization-mediated regulation of different pathways.
Figure 7.
rack1a mutant plants are resistant to water stress. (A) WT Arabidopsis and rack1a mutant plants were grown with normal water regimen for 3 wk, and then water was withheld for 12 d. Compared to the WT plants, reduced wilting was observed for rack1a mutant plants during water stress. (B) Water loss in detached leaves from 4-wk-old plants placed on lab bench for 12 h. (C) Comparison of rates of water loss from detached leaves of the WT and rack1a mutant plants. Water loss is expressed as the proportion of initial fresh weight. Values are means ± SE from 10 leaves for each of three independent experiments.
Scaffolding proteins, such as RACK1, are uniquely poised to integrate signals from multiple pathways by bringing interacting signaling components into close proximity (Hall and Lefkowitz 2002; Dard and Peter 2006). The repertoire of RACK1 interacting proteins is growing steadily and includes factors that mediate vital cellular processes. The deduced molecular structure of RACK1A will be helpful in exploring its promiscuous and dynamic capacity to interact with multiple proteins, facilitating the integration of diverse signal transduction pathways. This structure lays the foundation for future work involving surface mutagenesis and structural characterization of protein complexes to obtain greater detail of these interactions.
Materials and Methods
Cloning, expression, and purification
The MBP–RACK1A fusion protein was created using pMAL-c2x (New England Biolabs) as the starting vector. The amino acid sequence of MBP contained the mutation E359A and was truncated at Asn367. The linker region encodes three alanine residues and contained a NotI site for cloning. In addition, residues Asp82, Lys83, and Lys239 of MBP were also mutated to alanines. RACK1A containing residues Gly4–Tyr327 from A. thaliana was cloned into this vector using the NotI and BamHI restriction sites. The MBP–RACK1A fusion protein was expressed in BL21(DE3)CodonPlusRIL cells (Stratagene). Cells were grown at 37°C in LB media containing 100 μg/mL ampicillin and 35 μg/mL chloramphincol, shaking at 275 rpm. When the OD600 reached 1.0, the temperature was decreased to 18°C, ITPG was added to a final concentration of 500 μM, and cells were allowed to shake overnight at 275 rpm. Cells were pelleted by centrifugation, resuspended in the sonication buffer (25 mM Tris pH 7.5 and 500 mM NaCl), and sonicated three times for 20 sec each. The insoluble fraction was pelleted by centrifugation and the soluble fraction was loaded in batch onto amylose resin (New England Biolabs). Samples were placed on a rocker at 4°C for 1 h under gentle agitation. Resin was then washed with sonication buffer, followed by elution from the resin in 25 mM Tris pH 7.6, 500 mM NaCl, 1 mM DTT, and 40 mM maltose. Eluted protein was concentrated to 10 mg/mL then loaded onto a 16/60 Superdex200 size exclusion column (GE Healthcare) pre-equilibrated in the same buffer. The purified protein was then dialyzed against 25 mM Tris pH 7.6, 100 mM NaCl, 1 mM DTT, and 5 mM maltose and concentrated to 16.7 mg/mL for crystallization trials.
Crystallization, data collection, and refinement
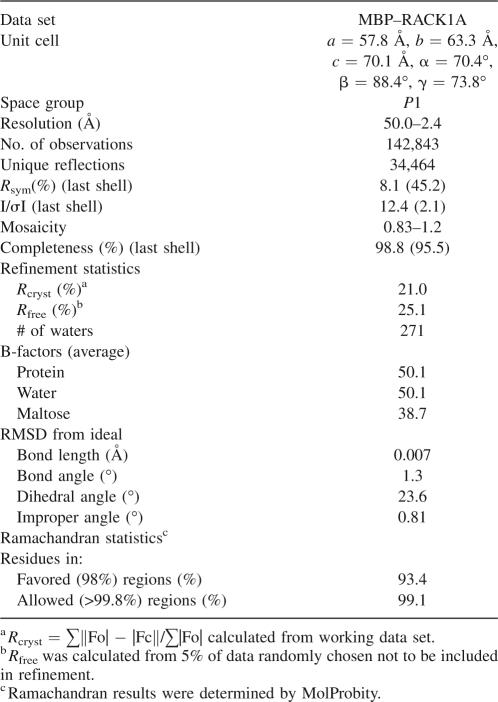
Crystals of MBP–RACK1A formed using the sitting-drop vapor diffusion technique, mixing 1 μL of protein solution with 1 μL of well solution consisting of 100 mM HEPES pH 7.5 and 20% w/v Polyethylene glycol 10,000. To obtain cryo-conditions, 0.66 μL of ethylene glycol was added to the crystallization drop, and the ethylene glycol was allowed to diffuse across the drop for approximately 1 min. A crystal measuring 0.1 mm × 0.05 mm × 0.02 mm was captured in a loop and flash-frozen in liquid nitrogen. The crystal was placed in a stream of nitrogen gas cooled to −180°C. Diffraction data were collected on a Rigaku 007HF generator equipped with VariMaxHF mirrors and a Saturn92CCD detector. Original diffraction was observed to 3.6 Å. To improve the resolution, the crystal was briefly thawed for 8 sec at room temperature, followed by refreezing in the nitrogen gas stream. A data set was collected to 2.4 Å resolution. Data were processed using HKL2000 (Otwinowski and Minor 1997). Molrep from the CCP4 suite was used for molecular replacement to obtain phases (Collaborative Computational Project 1994; Vagin and Teplyakov 1997). The position of the MBP molecule was obtained using residues 1–370 of molecule A from the Protein Data Bank coordinates 1HSJ (Liu et al. 2001). Clear density was visible at this point for the β-propeller domain of RACK1A. RACK1A was positioned by molecular replacement using residues 101–408 of platelet-activating factor acetylhydrolase IB β subunit from coordinates 1VYH (Tarricone et al. 2004). This resulted in correct positioning of the model into the density, but rotated by one WD domain with respect to the sequence alignment. This was corrected by a manual superposition of the starting coordinates of RACK1 onto the molecular replacement solution using O (Jones et al. 1991). The final structure was obtained by iterative cycles of model building and refinement in O (Jones et al. 1991) and CNS (Brunger et al. 1998). Geometry of the structure was assessed using Molprobity (Lovell et al. 2003). Data statistics are shown in Table 1. All structural figures were created using Molscript (Kraulis 1991) and Raster3D (Merritt and Murphy 1994).
Table 1.
Crystallographic data statistics
Plants, growth conditions, and stress treatments
A. thaliana ecotype Col-0 and mutant RACK1A plants were grown in 4-inch round pots filled with a mixture of metromix 510 soil and vermiculite (3:1) in a growth chamber at 22°C, with light intensity of 100 μmol m−2 s−1 and 70% relative humidity under long-day conditions (16-h light/8-h dark cycle). The isolation and characterization of RACK1A plants have been described previously (Chen et al. 2006). To induce water stress, wild-type (WT) and mutant plants were grown with sufficient water for 4 wk, and then water was withheld for 12 d to induce stress conditions.
Water loss measurements
Rosette leaves of mutant and WT plants growing under normal conditions for 4 wk were detached and weighed immediately on a piece of weighing paper, then placed on a laboratory bench and weighed at designated time intervals. Three replicates were done for each line. The percentage of loss of fresh weight was calculated based on the initial weight of the plants.
Protein Data Bank deposition
The coordinates have been deposited in the PDB with ID code 3DM0.
Acknowledgments
We thank Drs. L.G. Pedersen and N. Martin for critical reading of the manuscript. This work is supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 number 1Z01ES080043-20) and by the National Science Foundation RIG grant (H.U.).
Footnotes
Reprint requests to: Lars C. Pedersen, Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, 111 TW Alexander Drive, MD F3-09, Research Triangle Park, NC 27709, USA; e-mail: pederse2@niehs.nih.gov; fax: (919) 541-7880.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035121.108.
References
- Alfarano, C., Andrade, C.E., Anthony, K., Bahroos, N., Bajec, M., Bantoft, K., Betel, D., Bobechko, B., Boutilier, K., Burgess, E., et al. The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor, V., Sampson, D.A., Matunis, M.J., Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Ceci, M., Gaviraghi, C., Gorrini, C., Sala, L.A., Offenhauser, N., Marchisio, P.C., Biffo, S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- Chang, B.Y., Conroy, K.B., Machleder, E.M., Cartwright, C.A. RACK1, a receptor for activated C kinase and a homolog of the β subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol. Cell. Biol. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, B.Y., Chiang, M., Cartwright, C.A. The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J. Biol. Chem. 2001;276:20346–20356. doi: 10.1074/jbc.M101375200. [DOI] [PubMed] [Google Scholar]
- Chen, J.G., Ullah, H., Temple, B., Liang, J., Guo, J., Alonso, J.M., Ecker, J.R., Jones, A.M. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis . J. Exp. Bot. 2006;57:2697–2708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dard, N., Peter, M. Scaffold proteins in MAP kinase signaling: More than simple passive activating platforms. Bioessays. 2006;28:146–156. doi: 10.1002/bies.20351. [DOI] [PubMed] [Google Scholar]
- Dell, E.J., Connor, J., Chen, S., Stebbins, E.G., Skiba, N.P., Mochly-Rosen, D., Hamm, H.E. The βγ subunit of heterotrimeric G proteins interacts with RACK1 and two other WD repeat proteins. J. Biol. Chem. 2002;277:49888–49895. doi: 10.1074/jbc.M202755200. [DOI] [PubMed] [Google Scholar]
- Derewenda, Z.S. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Doan, A.T., Huttenlocher, A. RACK1 regulates Src activity and modulates paxillin dynamics during cell migration. Exp. Cell Res. 2007;313:2667–2679. doi: 10.1016/j.yexcr.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera, I., Fenoglio, J., Li, Y., Lewis, C., Panchenko, M.P., Reiner, O., Smith, T.F., Neer, E.J. Folding of proteins with WD-repeats: Comparison of six members of the WD-repeat superfamily to the G protein β subunit. Biochem. 1996;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander, R., Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., Lefkowitz, R.J. Regulation of G protein-coupled receptor signaling by scaffold proteins. Circ. Res. 2002;91:672–680. doi: 10.1161/01.res.0000037000.74258.03. [DOI] [PubMed] [Google Scholar]
- Herold, M., Cikala, M., MacWilliams, H., David, C.N., Bottger, A. Cloning and characterisation of PKB and PRK homologs from Hydra and the evolution of the protein kinase family. Dev. Genes Evol. 2002;212:513–519. doi: 10.1007/s00427-002-0267-7. [DOI] [PubMed] [Google Scholar]
- Hussain, A., Cao, D., Peng, J. Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein. Planta. 2007;226:475–483. doi: 10.1007/s00425-007-0497-z. [DOI] [PubMed] [Google Scholar]
- Ishida, S., Takahashi, Y., Nagata, T. Isolation of cDNA of an auxin-regulated gene encoding a G protein β subunit-like protein from tobacco BY-2 cells. Proc. Natl. Acad. Sci. 1993;90:11152–11156. doi: 10.1073/pnas.90.23.11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Klemm, J.D., Schreiber, S.L., Crabtree, G.R. Dimerization as a regulatory mechanism in signal transduction. Annu. Rev. Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- Kobe, B., Center, R.J., Kemp, B.E., Poumbourios, P. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P. MOLSCRIPT: A program to produce both detailed and schematic plots of proteins. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Kurepa, J., Walker, J.M., Smalle, J., Gosink, M.M., Davis, S.J., Durham, T.L., Sung, D.Y., Vierstra, R.D. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- Lambright, D.G., Sondek, J., Bohm, A., Skiba, N.P., Hamm, H.E., Sigler, P.B. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Manna, A., Li, R., Martin, W.E., Murphy, R.C., Cheung, A.L., Zhang, G. Crystal structure of the SarR protein from Staphylococcus aureus . Proc. Natl. Acad. Sci. 2001;98:6877–6882. doi: 10.1073/pnas.121013398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.V., Hubbi, M.E., Pan, F., McDonald, K.R., Mansharamani, M., Cole, R.N., Liu, J.O., Semenza, G.L. Calcineurin promotes HIF-1α expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J. Biol. Chem. 2007;282:37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois, L.M., Lima, C.D., Chua, N.H. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis . Plant Cell. 2003;15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell, S.C., Davis, I.W., Arendall W.B., III, de Bakker, P.I., Word, J.M., Prisant, M.G., Richardson, J.S., Richardson, D.C. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Macauley, M.S., Errington, W.J., Scharpf, M., Mackereth, C.D., Blaszczak, A.G., Graves, B.J., McIntosh, L.P. Beads-on-a-string, characterization of ETS-1 sumoylated within its flexible N-terminal sequence. J. Biol. Chem. 2006;281:4164–4172. doi: 10.1074/jbc.M510488200. [DOI] [PubMed] [Google Scholar]
- Mamidipudi, V., Miller, L.D., Mochly-Rosen, D., Cartwright, C.A. Peptide modulators of Src activity in G1 regulate entry into S phase and proliferation of NIH 3T3 cells. Biochem. Biophys. Res. Commun. 2007;352:423–430. doi: 10.1016/j.bbrc.2006.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCahill, A., Warwicker, J., Bolger, G.B., Houslay, M.D., Yarwood, S.J. The RACK1 scaffold protein: A dynamic cog in cell response mechanisms. Mol. Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., Murphy, M.E. Raster3D Version 2.0. Å program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Meulmeester, E., Melchior, F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- Miranda-Saavedra, D., Barton, G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- Miura, K., Jin, J.B., Hasegawa, P.M. Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Nilsson, J., Sengupta, J., Frank, J., Nissen, P. Regulation of eukaryotic translation by the RACK1 protein: A platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palmer, D.A., Thompson, J.K., Li, L., Prat, A., Wang, P. Gib2, a novel Gβ-like/RACK1 homolog, functions as a Gβ subunit in cAMP signaling and is essential in Cryptococcus neoformans . J. Biol. Chem. 2006;281:32596–32605. doi: 10.1074/jbc.M602768200. [DOI] [PubMed] [Google Scholar]
- Pickles, L.M., Roe, S.M., Hemingway, E.J., Stifani, S., Pearl, L.H. Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure. 2002;10:751–761. doi: 10.1016/s0969-2126(02)00768-2. [DOI] [PubMed] [Google Scholar]
- Reyes, D., Rodriguez, D., Nicolas, G., Nicolas, C. Evidence of a role for tyrosine dephosphorylation in the control of postgermination arrest of development by abscisic acid in Arabidopsis thaliana L. Planta. 2006;223:381–385. doi: 10.1007/s00425-005-0135-6. [DOI] [PubMed] [Google Scholar]
- Ron, D., Mochly-Rosen, D. Agonists and antagonists of protein kinase C function, derived from its binding proteins. J. Biol. Chem. 1994;269:21395–21398. [PubMed] [Google Scholar]
- Ron, D., Chen, C.H., Caldwell, J., Jamieson, L., Orr, E., Mochly-Rosen, D. Cloning of an intracellular receptor for protein kinase C: A homolog of the β subunit of G proteins. Proc. Natl. Acad. Sci. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrabhatla, P., Reddy, M.M., Rajasekharan, R. Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 2006;60:293–319. doi: 10.1007/s11103-005-4109-7. [DOI] [PubMed] [Google Scholar]
- Salinas, S., Briancon-Marjollet, A., Bossis, G., Lopez, M.A., Piechaczyk, M., Jariel-Encontre, I., Debant, A., Hipskind, R.A. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol. 2004;165:767–773. doi: 10.1083/jcb.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, J.A. A Chlamydomonas gene encodes a G protein β subunit-like polypeptide. Mol. Gen. Genet. 1990;221:443–452. doi: 10.1007/BF00259410. [DOI] [PubMed] [Google Scholar]
- Sengupta, J., Nilsson, J., Gursky, R., Spahn, C.M., Nissen, P., Frank, J. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- Sklan, E.H., Podoly, E., Soreq, H. RACK1 has the nerve to act: Structure meets function in the nervous system. Prog. Neurobiol. 2006;78:117–134. doi: 10.1016/j.pneurobio.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Siderovski, D.P. γ-Like (GGL) domains: New frontiers in G-protein signaling and β-propeller scaffolding. Biochem. Pharmacol. 2001;61:1329–1337. doi: 10.1016/s0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E., Sigler, P.B. Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Sprague, E.R., Redd, M.J., Johnson, A.D., Wolberger, C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 2000;19:3016–3027. doi: 10.1093/emboj/19.12.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, M.R., McCahill, A., Thompson, D.S., MacKenzie, C., Isaacs, N.W., Houslay, M.D., Bolger, G.B. Identification of a surface on the β-propeller protein RACK1 that interacts with the cAMP-specific phosphodiesterase PDE4D5. Cell. Signal. 2001;13:507–513. doi: 10.1016/s0898-6568(01)00167-x. [DOI] [PubMed] [Google Scholar]
- Tarricone, C., Perrina, F., Monzani, S., Massimiliano, L., Kim, M.H., Derewenda, Z.S., Knapp, S., Tsai, L.H., Musacchio, A. Coupling PAF signaling to dynein regulation: Structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–821. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Thornton, C., Tang, K.C., Phamluong, K., Luong, K., Vagts, A., Nikanjam, D., Yaka, R., Ron, D. Spatial and temporal regulation of RACK1 function and N-methyl-D-aspartate receptor activity through WD40 motif-mediated dimerization. J. Biol. Chem. 2004;279:31357–31364. doi: 10.1074/jbc.M402316200. [DOI] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Temple, B., Boyes, D.C., Alonso, J.M., Davis, K.R., Ecker, J.R., Jones, A.M. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A., Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- van Nocker, S., Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics. 2003;4:50. doi: 10.1186/1471-2164-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, M.A., Coleman, D.E., Lee, E., Iniguez-Lluhi, J.A., Posner, B.A., Gilman, A.G., Sprang, S.R. The structure of the G protein heterotrimer Gi*1 β1 γ2 . Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Yang, X.J., Gregoire, S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Yemets, A., Sheremet, Y., Vissenberg, K., Van Orden, J., Verbelen, J.P., Blume, Y.B. Effects of tyrosine kinase and phosphatase inhibitors on microtubules in Arabidopsis root cells. Cell Biol. Int. 2008;32:630–637. doi: 10.1016/j.cellbi.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Zeller, C.E., Parnell, S.C., Dohlman, H.G. The RACK1 ortholog Asc1 functions as a G-protein β subunit coupled to glucose responsiveness in yeast. J. Biol. Chem. 2007;282:25168–25176. doi: 10.1074/jbc.M702569200. [DOI] [PubMed] [Google Scholar]