Abstract
A covalent dimer of the bacteriophage MS2 coat protein was created by performing genetic fusion of two copies of the gene while removing the stop codon of the first gene. The dimer was crystallized in the cubic F432 space group. The organization of the asymmetric unit together with the F432 symmetry results in an arrangement of subunits that corresponds to T = 3 octahedral particles. The octahedral particles are probably artifacts created by the particular crystal packing. When it is not crystallized in the F cubic crystal form, the coat protein dimer appears to assemble into T = 3 icosahedral particles indistinguishable from the wild-type particles. To form an octahedral particle with closed surface, the dimer subunits interact at sharper angles than in the icosahedral arrangement. The fold of the covalent dimer is almost identical to the wild-type dimer with differences located in loops and in the covalent linker region. The main differences in the subunit packing between the octahedral and icosahedral arrangements are located close to the fourfold and fivefold symmetry axes where different sets of loops mediate the contacts. The volume of the wild-type virions is 7 times bigger than that of the octahedral particles.
Keywords: MS2, virus, icosahedron, octahedron, coat protein, dimer
There are five regular platonic solids—the tetrahedron, octahedron, cube, dodecahedron, and icosahedron. Two pairs of them share a common symmetry and are duals to each other: the octahedron and the cube and the dodecahedron and the icosahedron. Crick and Watson (1956) predicted that virus capsids have cubic symmetry. The assumption was that regular platonic solids allow a single type of asymmetric unit to assemble into a well-defined spherical structure. When structures of several viruses were determined it turned out that the virions have icosahedral symmetry. The prediction was true in the sense that the particles possess cubic symmetry, since it is a subset of the icosahedral symmetry. There are only very few examples of virus-related structures that have lower symmetry than the icosahedral one. The only exception with an experimentally determined structure is the octahedral subviral particle formed by the hepatitis B surface antigen (Gilbert et al. 2005). A model involving tetrahedral and octahedral particles was created for subviral particles assembled in vitro from the coat protein of murine polyomavirus (Salunke et al. 1989).
There are 60 identical elements in structures with icosahedral symmetry. Most viruses have bigger genomes than could be packed into a shell built from 60 subunits of a reasonable molecular size. In these viruses each of the 60 structural elements is composed of several protein chains. If the subunit interactions in the icosahedral packing with more than 60 subunits are to be quasi-similar, only certain multiples of 60 are possible (Caspar and Klug 1962). The allowed multiples of 60 can be expressed by the triangulation number T = h 2 + hk + k 2, where h and k are any integers including zero. When T > 1, a particular subunit must be able to adapt to several distinct sets of contacts depending on its location in the capsid. The concept of the triangulation number can be applied to the other platonic solids as well. The T will then express the multiples of symmetry elements of the given solid—60 for the dodecahedron, 24 for the octahedron and the cube, and 12 for the tetrahedron—a T = 3 octahedron is built from 24 × 3 = 72 subunits.
Structures of several small RNA phages belonging to the Leviviridae family have been determined by crystallography (Valegård et al. 1990). The wild-type capsid of phage MS2 consists of 180 chemically identical copies of the coat protein organized in a T = 3 icosahedral shell. The icosahedral asymmetric unit contains three subunits that are designated A, B, and C. There are strong pairwise interactions between the monomers, and therefore the capsid can be considered as built from 90 dimers. The interactions between the two coat protein monomers in the dimer are mostly hydrophobic. Two C-terminal helices fit into a groove formed by the helices and an N-terminal hairpin of the other monomer. Two five-stranded sheets, one from each monomer, form a 10-stranded sheet in the dimer. Two types of dimers are found in the capsid, one at the quasi-twofold axis formed by subunits A and B and one at the icosahedral twofold axis composed of two C subunits. Five B subunits interact around the icosahedral fivefold axis, and three of each of the A and C subunits participate in the contacts around the quasi-sixfold axis. The overall shape of the particle is spherical, with a diameter of 280 Å. The structures of the coat proteins of other small RNA phages such as Qβ (Golmohammadi et al. 1996), GA (Tars et al. 1997), and PP7 (Tars et al. 2000) differ mostly in the loop regions mediating the contacts around the five- and quasi-sixfold icosahedral axes.
It is possible to genetically fuse the two MS2 coat protein subunits, and the protein retains the ability to fold normally because the C and N termini of the two subunits in the wild-type dimer are located close to each other. Peabody and Lim (1996) fused two copies of the coat protein gene next to each other by removing the stop codon of the first gene. Besides the removal of the stop codon, the second residue (serine) from the second subunit was removed. The final covalent coat protein dimer (CCPD) gene encoded 257 residues. The CCPD subunits assembled into virion-like particles (Peabody and Lim 1996). The CCPD subunits were more stable than the dimers of the wild-type subunits (Peabody 1997). We have now crystallized CCPD in the space group F432. The unit cell edge is of length 220 Å, which is too small to accommodate the icosahedral T = 3 virus-like particle. The CCPD crystal arrangement corresponds to octahedral particles with T = 3 quasi-symmetry.
Results
Electron microscopy
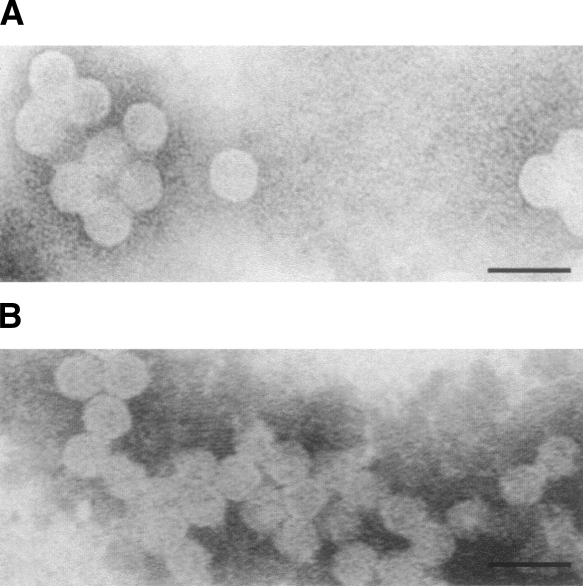
As revealed by electron microscopy the CCPD is able to self-assemble into virus-like particles (Fig. 1A). These particles are indistinguishable from the virus-like particles assembled from the wild-type coat protein at the resolution of the electron microscope (Fig. 1B). Both the wild-type and the CCPD particles appear to have a diameter of ∼280 ± 20 Å.
Figure 1.
EM pictures of wild-type and CCPD particles from solution. Electron microscopy photographs of negatively stained solution samples of (A) CCPD particles and (B) virus-like MS2 particles formed by the wild-type subunits. Scale bar, 50 nm.
Quality of the model
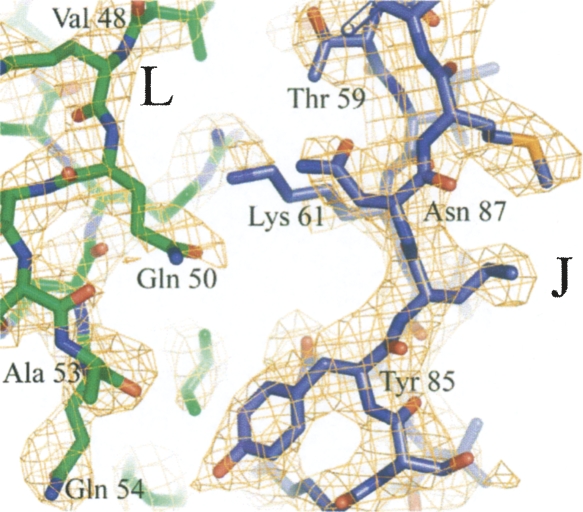
The electron density map at 3.5 Å resolution was continuous throughout most of the main chain of all the three subunits. Discontinuities were observed in the intersubunit linker regions, where there are two alternative conformations for the main chain. Part of the model with electron density is shown in Figure 2. In the octahedral crystal the CCPD occupies positions with twofold symmetry both at true crystallographic twofold axes and at quasi-twofold axes. The CCPD is twofold symmetric except at the N and C termini and at the link between the original subunits (Fig. 3A–C). The orientation of the CCPD is random with respect to these twofold axes. The interpretation of the density was challenging mainly at these regions where the CCPD subunits overlap but adopt different conformations. The model building was greatly facilitated by the similarity between the CCPD structure and the wild-type subunits. The structure of the wild-type subunits used as an initial model was changed only very little with the exception of the loop regions where the contacts differ between the wild-type icosahedral and the octahedral particles.
Figure 2.
Electron density at the interface between J and L CCPD subunits. Part of the interface between the J (blue) and L (green) subunits is shown. Electron density contoured at 1σ is shown in orange.
Figure 3.
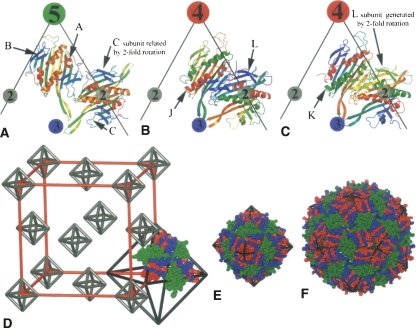
Comparison of the wild-type subunit arrangement in MS2 virions to the CCPD subunits in F432 crystal. (A–C) Comparison of the coat protein dimer arrangement in the wild-type icosahedral particles (A) to the packing of CCPD subunits in the octahedral “particles” (B,C). Edges of the triangular faces of the icosahedron (A) and the octahedron (B,C) are shown as black lines together with the positions of the rotation axes. Individual protein chains are rainbow colored from the N terminus in blue to the C terminus in red. The icosahedral asymmetric unit (A) consists of the three subunits A, B, and C. There are two positions for a subunit dimer in the icosahedral arrangement and a CCPD subunit in the octahedral arrangement (compare A to B and C). Since the CCPD subunits have an approximate twofold symmetry, each dimer position can be occupied by two alternative orientations of the CCPD subunit. The region of the icosahedral AB dimer is in the octahedral arrangement occupied by CCPD subunits J or K where the two halves (corresponding to two wild-type subunits) have different structures. The region of the wild-type CC dimer is occupied by the CCPD subunit L. Since the L subunit is positioned on the true twofold rotation axis, the two halves (corresponding to the wild-type subunits) have identical structures except for the linker region and the N and C termini. The twofold rotation axis positions another copy of the L subunit on top of itself (compare B and C). (D) Scheme of the organization of the CCPD subunits in the F432 unit cell. Subunits J, K, and L from three asymmetric units composing one triangular face of the T = 3 octahedron are shown. The length of the edge of the unit cell shown in red is 220 Å. Parts of the subunits J and K that correspond to the subunit A are colored in blue, parts corresponding to the B subunit in red. Subunit L, which is equivalent to the dimer of C subunits, is shown in green. This coloring was used to make the comparison to the wild-type particles easier. (E) Octahedral assembly of CCPD subunits superimposed on an octahedral wire frame. The 24 asymmetric units from the F432 crystal architecture form the octahedral “particles.” CCPD subunits are colored as in D. (F) Model of a wild-type MS2 T = 3 virion superimposed on an icosahedral wire frame. The A subunits are shown in blue, B in red, and C in green. Panels E and F are in scale to allow size comparison.
A model was built for all residues of the three subunits in the asymmetric unit. The model of the asymmetric unit consists of three molecules of 257 residues each, which have 50% occupancy. The R-factor is 0.273 for the 6079 reflections between 45 and 3.5 Å used in the refinement. The R-free is 0.312 for 308 reflections. The analysis of the overall quality of the structure as judged by the Ramachandran plot showed that 3.8% of the residues were in disallowed regions and 74.9% in favored regions using the criterion of Molprobity (Lovell et al. 2003). Most of the residues that appear as Ramachandran outliers are located in the loop regions. The coordinates were deposited in the Protein Data Bank (entry 2vtu).
Organization of the F432 asymmetric unit and description of CCPD fold
The space group F432 is the most complex one of the space groups allowed for protein crystals. The unit cell consists of 96 asymmetric units that are related by 24 different rotations and four translations. The 24 rotations of the asymmetric units grouped around a common origin correspond to the 24 octahedral symmetry operators (Fig. 3D). Any object that crystallizes in this space group adopts an octahedral organization. Since there are three halves of the CCPD subunits in the crystallographic asymmetric unit, the subunit arrangement corresponds to T = 3 octahedral particles (Fig. 3D,E). The three CCPD subunits were named J, K, and L. Each of the CCPD subunits corresponds to a dimer of the wild-type coat protein.
The MS2 wild-type coat protein subunit consists of an N-terminal hairpin with β-strands A and B, which are exposed on the surface of the particle, a five-stranded β-sheet exposed to the particle interior composed of β-strands C to G and a C-terminal helical part containing helices A and B, positioned on the outer surface of the capsid (Fig. 4A). The naming convention of the wild-type subunit was kept for the CCPD. The particular half of the CCPD subunit is always specified in the text. The CCPD subunit J resembles the subunit dimer AB in the wild-type T = 3 icosahedral particle (Fig. 3F). Its N-terminal half corresponds to the A subunit and the C-terminal half corresponds to the B subunit (Fig. 3A,B). There is a covalent linkage at the place of the C terminus of the A subunit and the N terminus of the B subunit. The molecule K resembles the BA dimer and is related to subunit J by an approximate twofold rotation (Fig. 3A,C). The subunits J and K occupy the same space in the crystal, and this is the reason for their 50% occupancy (Fig. 3A,B). Their structures differ only in the linker regions and at the N and C termini. The CCPD subunit L is analogous to the CC dimer in the wild-type T = 3 icosahedral particle (Fig. 3B,C). It is located on the crystallographic twofold axis, which places another copy of the L subunit on top of itself (Fig. 3B,C), and thus the L subunit has 50% occupancy as well. The two halves of the L subunit differ only in the region of the intersubunit linker; otherwise their structures are identical. The molecules J and K participate in the contacts around the fourfold and threefold symmetry axes. The molecule L links two threefold symmetry axes and has the twofold symmetry axis running through its center (Fig. 3B,C).
Figure 4.
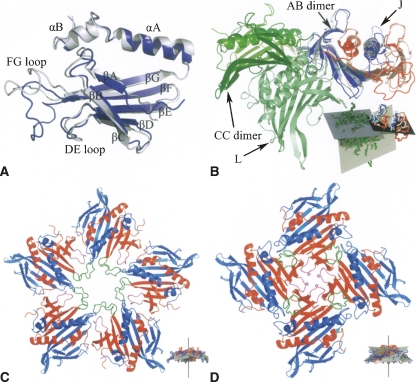
Structure of the coat protein subunit and differences in interactions between the wild-type and CCPD subunits. (A) Comparison of the structure of the wild-type MS2 B subunit (blue) to residues 1–129 of the K CCPD subunit (light blue). Names of secondary structure elements are shown. (B) Comparison of the relative positioning of the AB and CC wild-type dimers in the T = 3 virions to the covalent dimers J and L as observed in the F432 crystal. The J subunit (residues 1–129 in light blue and 130–257 in light red) was superimposed on the AB dimer (A is colored in blue and B in red). The wild-type CC dimer (colored in dark green) points up, while the L subunit (light green) points down. The inset shows the least squares planes of the AB dimer in black (the least squares plane of the CCPD subunits J and K is almost identical with that of AB dimer since the subunits were superimposed), of the CC dimer in blue, and of the L subunit in gray. Subunit coloring is consistent with the main figure. (C) Decamer formed by five dimers of A (blue) and B (red) wild-type MS2 subunits as observed in the T = 3 virions. The FG loops of the B subunits are shown in green and the DE loops in purple. The inset shows a side view of the decamer with the least squares planes of each dimer displayed in gray. The fivefold rotation axis relating the subunits is shown in black. Subunits are colored as in the main figure except for the loop regions. (D) Contacts of four J CCPD subunits around the crystallographic fourfold symmetry axis. Residues 1–129 corresponding to the wild-type A subunit are colored in blue, residues 130–257 corresponding to the B subunit in red. The regions corresponding to the FG loop of the B subunit are shown in green and the DE loop in purple. The inset shows a side view of the CCPD J subunit tetramer, using the same colors as in the main figure, except for the loop regions. The least squares planes of each subunit are shown in gray. The fourfold rotation axis is shown in black. Subunits are colored consistently with the main figure except for the loop regions.
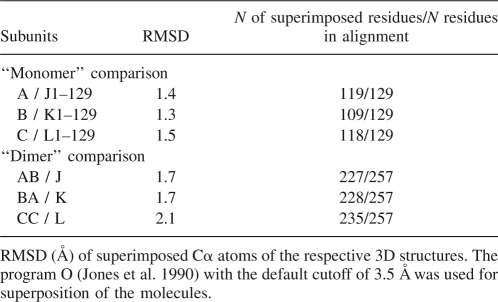
Table 1 shows the rms deviations of the Cα atoms between the wild-type subunits and the corresponding parts of the CCPD subunits after superposition. Although the rms deviations for superimposed residues are similar, the biggest differences are between the wild-type subunit B and the N-terminal half of the CCPD subunit K, where it was possible to relate only 109 of the 129 residues used in the superposition. The main differences are located in the FG and DE loop regions (Fig. 4A). A comparison of the CCPD subunits to the AB and CC dimers shows that the CC dimer is more different from the L subunit than the J and K subunits from the AB dimer. The differences between the individual CCPD subunits are relatively small. The subunits J and K are almost identical with an rms deviation of 0.2 for 252 residues. The rms deviation between subunits J (or K) and L is 2.2 for 211 residues.
Table 1.
Structural similarity of wild-type and covalent CC dimer MS2 subunits
Discussion
Structural differences of the CCPD to wild-type coat protein subunits that allow octahedral packing
Protein molecules of identical amino acid sequence can adopt different conformations. The coat protein subunits of viruses with a triangulation number of T > 1 are well-characterized examples of this structural heterogeneity since several of the variants are present within the structure of a single capsid. In spite of the structural variation, the assembly of the coat proteins into viral capsids is tightly regulated and rarely results in formation of aberrant structures during the virus life cycle. Some examples of coat proteins forming structures other than the native ones are known. Brome mosaic virus coat protein can assemble into two types of particles consisting of 180 or 120 subunits (Krol et al. 1999). Occasionally, rod-like particles were observed within the icosahedral virion preparations of viruses from the Papovaviridae family (Kiselev and Klug 1969). Rod-shaped virus-like particles were also observed in preparations of mutant coat protein of the phage Qβ (Cielens et al. 2000). Finally, particles of different sizes together with tubular structures are produced during hepatitis B virus infection (Seeger et al. 2007).
The CCPD subunits can form both octahedral and icosahedral particles. The main difference between the octahedral and icosahedral arrangement is that there are fourfold instead of fivefold contacts, which results in a more bent contact interface (Fig. 4C,D). The angle between the least squares plane of the wild-type AB dimer and the fivefold rotation axis is 64° (Fig. 4C), while it is 58° between the least squares plane of J and K subunits and the fourfold rotation axis (Fig. 4D). Since the least squares planes of the dimers are tangential to the particle surface, the angle would be 90° if the interactions around the axis were completely flat; the smaller the angle is, the more bent the interface is. A second difference is in the interaction of AB and CC dimers to the J (or K) and L molecules in the octahedral assembly. The binding interfaces are completely distinct (Fig. 4B). In the octahedral arrangement the L molecule is under the molecules J and K while the AB and CC contacts are flat (Fig. 4B). The angle between the least squares planes of the dimers AB and CC is 14° while it is 57° for the molecules J (or K) and L (Fig. 4B). The more open angle of interaction results in bigger curvature of the particle surface and thus allows formation of an octahedral instead of an icosahedral particle.
There are several differences in the loop regions between the wild-type AB and CC dimers and the J, K, and L subunits that explain the different packing. The main differences are in the FG loop of the part of J and K subunits that corresponds to the B subunit and participates in the tetramer contacts (Fig. 4A). The FG loop is folded up (Fig. 4D) and does not make any contacts to the part of the CCPD corresponding to the A molecule. In the icosahedral capsid the FG loop of the B subunit, which participates in the pentamer contacts, interacts with the A subunit from the neighboring dimer (Fig. 4C). The contacts around the fourfold axis are instead mediated by the DE loops (Fig. 4D). Other differences are located in the loop regions forming the J and L interface. A comparison of the residues participating in the contacts between the wild-type AB and CC dimers and CCPD J and L subunits is shown in Figure 5. The interactions of the EF loop of the N-terminal part of subunit L with the strand F of the N-terminal part of the subunit J is shown in Figure 2.
Figure 5.
Comparison of residues mediating the interaction between the wild-type AB and C1C6 dimers and between the J and L CCPD subunits. The amino acid sequence of the wild-type MS2 subunits is shown as a part of the CCPD subunit sequence—numbers of the first and last residues of the respective wild-type and CCPD subunit in each line are shown. Secondary structure elements following the wild-type protein naming convention are shown above the amino acid sequence. Regions of β-strands are shown as bars and α-helices as stars. The wild-type AB dimer is superimposed on the J CCPD subunit and the C1C6 dimer on the L CCPD subunit. The serines B-2 and C6-2 shown as small bold letters are not present in the CCPD sequences. Residues with gray background participate in the contacts of both CCPD and wild-type subunits. Residues shown as underlined and bold letters participate only in the contacts between the CCPD subunits. Residues highlighted in italics and bold participate only in the contacts of the wild-type subunits. The interacting residues were identified using the protein interfaces, surfaces, and assemblies service PISA at European Bioinformatics Institute, http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html (Krissinel and Henrick 2007).
Comparison of the subunit interactions in the icosahedral particle to the F432 crystal and its consequences for particle stability
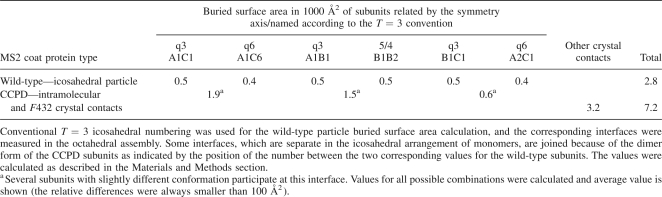
Upon crystallization in the F432 space group the CCPD subunits rearrange from icosahedral particles into the octahedral organization. The process requires initial disassembly of the icosahedral particles. A reason for the CCPD subunit reassembly may be the conditions in the phase separation from which the crystals were growing. The phase separations usually contain high concentrations of protein and polyethylene glycol (Sica et al. 1996), which could destabilize the icosahedral particles and favor formation of the cubic crystals. Wild-type MS2 was crystallized in 1.5% PEG 6000, 0.2 M sodium phosphate pH 7.4 (Valegård et al. 1986). The concentration of salt and PEG was lower than in the CCPD crystallization conditions, and the phase separation was not observed. The crystal packing of the CCPD subunits is very dense, with a 53% protein fraction of the crystal volume. A comparison of the buried surface area of the wild-type T = 3 particles and of the octahedral assembly of CCPD subunits is shown in Table 2.
Table 2.
Buried surface area comparison
Most of the crystal contacts between the octahedral “particles” are formed by the α-helices of the L subunits. The helices from the two halves of the CCPD L subunit are ∼1 Å further from each other, 1 Å closer to the β-sheet, and 2 Å shifted along the helix axis toward the C terminus of the helix relative to the position of the helices in the wild-type CC dimer.
The octahedral variants of the interfaces A1C1 and A1C6 are relatively enlarged because the J (or K) and L subunits interact at a sharper angle, which allows more residues to come into contact (Fig. 4B). Even though the orientation of the interacting subunits is different, 39 out of 119 residues involved in the binding are common to both interfaces (Fig. 5). The buried surface area cannot be directly converted into association energy, but we believe that it indicates the relative importance of the individual interactions. The possible reason for the crystallization of the CCPD in the F432 space group may be the higher stability of the cubic arrangement over the icosahedral particles, because the average buried area in the F432 crystal per subunit is bigger.
Octahedral packing and particle size comparison
The diameter of the CCPD particles observed in the electron microscopy images is comparable to the diameter of the virus-like wild-type particles (Fig. 1A,B). Crystals of the CCPD icosahedral particles diffracting to low resolution were obtained (0.1 M Bicine pH 9.0, 20% PEG 5000). The size and shape of the CCPD icosahedral particles appears to be indentical to the wild-type virions except for the areas around the linker region (data not shown). From the size comparison of the icosahedral and octahedral particles based on the X-ray models (Fig. 3E,F), the icosahedral particles have a 1.6 times bigger diameter. This difference should be clearly visible on the electron microscopy images. We conclude that the CCPD subunits assembled into icosahedral T = 3 particles (Fig. 1A), but under the crystallization conditions these particles disassembled and the dimer formed T = 3 octahedra. Even though the biological relevance of our observation is limited, it clearly demonstrates that the shift from icosahedral to octahedral packing is possible by simply rearranging the building blocks while retaining their overall orientation.
There are only 24 asymmetric units in the octahedral compared to 60 in the icosahedral arrangement. The curvature of the surface of the octahedral particle is bigger and with the same size of asymmetric unit the octahedral particle is smaller than the icosahedral. The inner volume of the T = 3 wild-type MS2 particle is 5.4 × 106 Å3 while the volume of the T = 3 octahedral particle is only 0.75 × 106 Å3. Not only is the inner volume of the icosahedral particle 7 times bigger, but the whole octahedral particle with a maximum outer radius of 90 Å could fit into the icosahedral with minimal inner radius 103 Å. The size comparison clearly demonstrates that the icosahedral packing is more convenient in terms of efficiency evaluated as the ratio between the subunit size and particle volume available for the genome storage.
Materials and Methods
Cloning, protein production, purification, and sequence verification
The CCPD gene corresponding to the dl-13 version of the coat protein single-chain dimer as described in Peabody and Lim (1996), was cloned into the expression plasmid pBAD under the control of the arabinose promotor. The sequence was verified (data not shown). The plasmid was transformed into BL21-AI cells. Bacteria were grown to OD = 0.5, induced with 0.2% arabinose, and grown for 4 more hours. Cells were collected by centrifugation and broken by sonication. Cell debris were removed by centrifugation at 35,000g. The resulting supernatant was applied to a Sepharose CL4B size exclusion column. The resulting fractions containing virus particles were pooled and concentrated. Purity of the protein was checked on a SDS polyacrylamide gel.
Crystallization and data collection
Crystals of CCPD were obtained at 25°C using the hanging-drop technique with a bottom solution containing 0.32 M Na2HPO4, 0.08 M NaH2PO4, and 5% PEG 8000. The drops were prepared by mixing 5 μL of bottom solution with an equal volume of CCPD solution (10–20 mg/mL). Octahedron-shaped crystals formed within 5 months. For data collection, crystals of a size ∼0.1 mm were soaked for 2–3 min in mother liquor containing 30% glycerol and immediately frozen in liquid nitrogen. Data were collected from a single crystal at 100 K on the ADSC Quantum3 CCD detector at beamline ID 14-4 at the ESRF synchrotron radiation source in Grenoble, France. An oscillation range of 1° was used during data collection. The crystal diffracted to 3.5 Å resolution. Data were processed and scaled using the HKL2000 package (Otwinowski and Minor 1996). The crystals were decaying during the data collection, probably because of radiation damage. As a result, the intensities of symmetry-related reflections and Friedel pairs changed continuously. To minimize the errors in the data, only images 7–32 from the beginning of the data collection were used to calculate the final data set. The statistics from data collection and scaling is shown in Table 3.
Table 3.
Scaling statistics
Structure determination
The crystals were of space group F432. The crystallographic asymmetric unit contained three CCPD subunits with 50% occupancy. The wild-type MS2 subunit (PDB entry 2MS2) with trimmed loop regions was used as a search model for the molecular replacement (Golmohammadi et al. 1993). The position of the L subunit was identified with the program CNS (Brünger et al. 1998). The position of the J and K subunits was located using the program Beast (Read 2001).
The model was built using the program O (Jones et al. 1990), starting from the MS2 wild-type coordinates. Initially, a model corresponding to the three wild-type subunits with 100% occupancy was built and refined, then the linker region was added and the CCPD subunits with 50% occupancy were created. The model was refined by manual rebuilding alternating with coordinate refinement in the program CNS. Van der Waals and electrostatic interactions between the subunits J and K, not related by crystallographic symmetry and interactions of the subunit L with its symmetry-related copy occupying the same space, were switched off using the “igroupstatement” command during the refinement in CNS (Brünger et al. 1998). Other calculations were done using Collaborative Computational Project, Number 4 (1994). Because of the low resolution no water molecules were added to the model.
Calculation of the particle radius and volume
The maximum particle radius was calculated as a distance of the furthermost atom of the asymmetric unit from the center of the particle. The minimal radius was calculated accordingly. For the particle volume calculations PDB files containing coordinates for the whole icosahedral and octahedral particles were generated. The volume of the particle was then calculated with the use of the program MAMA (Kleywegt and Jones 1999). Two masks were generated: the first covering the protein shell and the second one covering the whole particle including the internal cavity. The difference of the two volumes defined the size of the particle cavity.
Buried surface area calculation
The surface area of individual protein subunits and their dimers was calculated using the program Areaimol (Lee and Richards 1971). The buried surface area was calculated as [(surface area of the dimer AB) − (surface area of molecule A) − (surface area of molecule B)]/2.
Least squares plane and intersubunit angle calculation
The least squares planes were calculated with the program Moleman2 using the command ONo LS_plane_odl. Angles between the planes were calculated with command ONo ANgle_ls_planes (Kleywegt and Jones 1996). Angles between the planes and rotation axes were calculated manually. The planes were displayed in O (Jones et al. 1990), and the final figures were generated with Molray (Harris and Jones 2001).
Electron microscopy
The purified wild-type and CCPD particles were applied to the nitrocellulose-covered copper grids and stained with 2.5% aqueous uranyl acetate. The grids were examined with a Hitachi H-7100 electron microscope operated at 100 kV.
Acknowledgments
We thank the staff at the ESRF France. P.P. acknowledges very helpful advice on various calculations and imaging from Gerard Kleywegt, Alwyn Jones, and Mark Harris. This work was supported by the Swedish Research Council.
Footnotes
Reprint requests to: Pavel Plevka, Uppsala University, Department of Cell and Molecular Biology, Box 596, S751 24 Uppsala, Sweden; e-mail address: pavel@xray.bmc.uu.se; fax: 46-18-53-69-71.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036905.108.
References
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Caspar, D.L.D., Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cielens, I., Ose, V., Petrovskis, I., Strelnikova, A., Renhofa, R., Kozlovska, T., Pumpens, P. Mutilation of RNA phage qβ virus-like particles: From icosahedrons to rods. FEBS Lett. 2000;482:261–264. doi: 10.1016/s0014-5793(00)02061-5. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Crick, F.H.C., Watson, J.D. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- Gilbert, R.J., Beales, L., Blond, D., Simon, M.N., Lin, B.Y., Chisari, F.V., Stuart, D.I., Rowlands, D.J. Hepatitis B small surface antigen particles are octahedral. Proc. Natl. Acad. Sci. 2005;102:14783–14788. doi: 10.1073/pnas.0505062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golmohammadi, R., Valegård, K., Fridborg, K., Liljas, L. The refined structure of bacteriophage MS2 at 2.8 Å resolution. J. Mol. Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- Golmohammadi, R., Fridborg, K., Bundule, M., Valegård, K., Liljas, L. The crystal structure of bacteriophage Qb at 3.5 Å resolution. Structure. 1996;4:543–554. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- Harris, M., Jones, T.A. Molray—a web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1201–1203. doi: 10.1107/s0907444901007697. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Bergdoll, M., Kjeldgaard, M. O: A macromolecule modeling environment. In: Bugg C., Ealick S., editors. Crystallographic and modeling methods in molecular design. Springer-Verlag; New York: 1990. pp. 189–199. [Google Scholar]
- Kiselev, N.A., Klug, A. The structure of viruses of the papilloma-polyoma type V. tubular variants built of pentamers. J. Mol. Biol. 1969;40:155–171. doi: 10.1016/0022-2836(69)90465-3. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., Jones, T.A. Phi/psi-chology: Ramachandran revisited. Structure. 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., Jones, T.A. Software for handling macromolecular envelopes. Acta Crystallogr. D Biol. Crystallogr. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- Krissinel, E., Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krol, M.A., Olson, N.H., Tate, J., Johnson, J.E., Baker, T.S., Ahlquist, P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B., Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lovell, S.C., Davis, I.W., Arendall W.B., III, de Bakker, P.I., Word, J.M., Prisant, M.G., Richardson, J.S., Richardson, D.C. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W. Jr, Sweet R.M., editors. Methods in enzymology. Academic Press; New York: 1996. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Peabody, D.S. Subunit fusion confers tolerance to peptide insertions in a virus coat protein. Arch. Biochem. Biophys. 1997;347:85–92. doi: 10.1006/abbi.1997.0312. [DOI] [PubMed] [Google Scholar]
- Peabody, D.S., Lim, F. Complementation of RNA binding site mutations in MS2 coat protein heterodimers. Nucleic Acids Res. 1996;24:2352–2359. doi: 10.1093/nar/24.12.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Salunke, D.M., Caspar, D.L., Garcea, R.L. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger, C., Zoulim, F., Mason, W.S. Fields virology. 5th ed. Lippincott; Philadelphia, PA: 2007. pp. 2977–3029. [Google Scholar]
- Sica, F., Adinolfi, S., Vitagliano, L., Zagari, A., Capasso, S., Mazzarella, L. Cosolute effect on crystallization of two dinucleotide complexes of bovine seminal ribonuclease from concentrated salt solutions. J. Cryst. Growth. 1996;168:192–197. [Google Scholar]
- Tars, K., Bundule, M., Fridborg, K., Liljas, L. The crystal structure of bacteriophage GA and a comparison of phages belonging to the major groups of Escherichia coli leviviruses. J. Mol. Biol. 1997;271:759–773. doi: 10.1006/jmbi.1997.1214. [DOI] [PubMed] [Google Scholar]
- Tars, K., Fridborg, K., Bundule, M., Liljas, L. Crystal structure of phage PP7 from Pseudomonas aeruginosa at 3.7 Å resolution. Virology. 2000;272:331–337. doi: 10.1006/viro.2000.0373. [DOI] [PubMed] [Google Scholar]
- Valegård, K., Unge, T., Montelius, I., Strandberg, B., Fiers, W. Purification, crystallization and preliminary X-ray data of the bacteriophage MS2. J. Mol. Biol. 1986;190:587–591. doi: 10.1016/0022-2836(86)90244-5. [DOI] [PubMed] [Google Scholar]
- Valegård, K., Liljas, L., Fridborg, K., Unge, T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]