Abstract
The Medium-Chain Dehydrogenase/Reductase Engineering Database (MDRED, http://www.mdred.uni-stuttgart.de) has been established to serve as an analysis tool for a systematic investigation of sequence–structure–function relationships. It includes sequence and structure information of 2684 and 42 medium-chain dehydrogenases/reductases (MDRs), respectively. Although MDRs are very diverse in sequence, they have a conserved tertiary structure. MDRs are assigned to 199 homologous families and 29 superfamilies. For each family, annotated multiple sequence alignments are provided, and functionally relevant residues are annotated. Twenty-five superfamilies were classified as zinc-containing MDRs, four as non-zinc-containing MDRs. For the zinc-containing MDRs, three subclasses were identified by systematic analysis of a variable loop region, the quaternary structure determining loop (QSDL): the class of short, medium, and long QSDL, which include 11, 3, and 5 superfamilies, respectively. The length of the QSDL is predictive for tetramer (short QSDL) and dimer (long QSDL) formation. The class of medium QSDL includes both tetrameric and dimeric MDRs. The shape of the substrate-binding site is highly conserved in all zinc-containing MDRs with the exception of two variable regions, the substrate recognition sites (SRS): two residues located on the QSDL (SRS1) and, for the class of long QSDL, one residue located in the catalytic domain (SRS2). The MDRED is the first online-accessible resource of MDRs that integrates information on sequence, structure, and function. Annotation of functionally relevant residues assist the understanding of sequence–structure–function relationships. Thus, the MDRED serves as a valuable tool to identify potential hotspots for engineering properties such as substrate specificity.
Keywords: sequence–structure–function relationship, protein family database, protein family classification, structure analysis
The protein family of medium-chain dehydrogenases/reductases (MDRs) comprises a large enzyme family with a broad range of enzymatic activities. They are found in all kingdoms of life and are involved in metabolism, regulatory processes, and protection against cell damage (Jörnvall et al. 1999; Nordling et al. 2002). Despite their low sequence similarity, they have a similar size of 350 to 400 residues and a conserved overall structure formed by two domains, a cofactor binding domain and a catalytic domain (Jörnvall et al. 1978). While all MDRs use NAD(H) or NADP(H) as cofactor, they can be divided into two classes with a different reaction mechanism: zinc-containing and non-zinc-containing MDRs (Nordling et al. 2002). In addition, many MDRs bind a second, non-catalytic, structural zinc ion (Eklund et al. 1976; Chase Jr. 1999).
Most MDRs are active as dimers or tetramers. Previously, a loop segment in the catalytic domain subsequent to the structural zinc binding site has been suggested to mediate quaternary structure formation (Jörnvall 1977; Persson et al. 1994; Norin et al. 1997). In addition, this loop segment is part of the substrate-binding site (Shafqat et al. 1999), which consists of conserved and variable regions. The conserved regions are formed by the cofactor NAD(H) or NADP(H), which is located at the bottom of the substrate-binding site, and the catalytic zinc ion, which is located at the back wall of the substrate-binding site. The left wall is formed by the highly conserved cofactor binding residues. The substrate enters from the front through the substrate access channel. Thus, there are only two variable regions, the ceiling and the right wall of the binding site. Previously, it has been shown that mutating residues located in these two regions changed substrate specificity (Hurley and Bosron 1992; Xie and Hurley 1999; Ziegelmann-Fjeld et al. 2007).
Two classification schemes exist for MDRs. One classification (Nordling et al. 2002; Jörnvall et al. 2003) is based on a consensus evolutionary tree constructed from an alignment of about 100 MDR sequences of six genomes. The MDRs were assigned to eight functional families which belong to two classes: The zinc-containing MDRs include cinnamyl alcohol dehydrogenases (CADs), polyoldehydrogenases (PDHs), dimeric alcohol dehydrogenases (ADHs), and yeast alcohol dehydrogenases/tetrameric alcohol dehydrogenases (YADHs), while the non-zinc-containing MDRs include quinone oxidoreductases (QORs), mitochondrial response proteins (MRF), leukotriene B4 dehydrogenases (LTDs), and acyl-CoA reductases (ACRs) (Nordling et al. 2002; Jörnvall et al. 2003). A second classification (Riveros-Rosas et al. 2003) extended this concept based on a much larger data set of 583 MDRs. They were grouped into three macrofamilies: Macrofamilies I and II include the zinc-containing MDRs, macrofamily III the non-zinc-containing MDRs. Each macrofamily was divided into further subfamilies, which were not consistent with the eight functional families introduced by Nordling et al. (2002).
In the meantime, the number of MDR sequences in public databases has increased more than fivefold, and structures of 42 proteins became available. We took advantage of this wealth of data and established the Medium-Chain Dehydrogenase/Reductase Engineering Database (MDRED), applying the extensible database system DWARF (Fischer et al. 2006). The MDRED provides a predictive classification scheme based on sequence and structure. By a systematic analysis of sequence and structure, conserved motifs and functionally relevant residues were identified.
Results
Database and data content
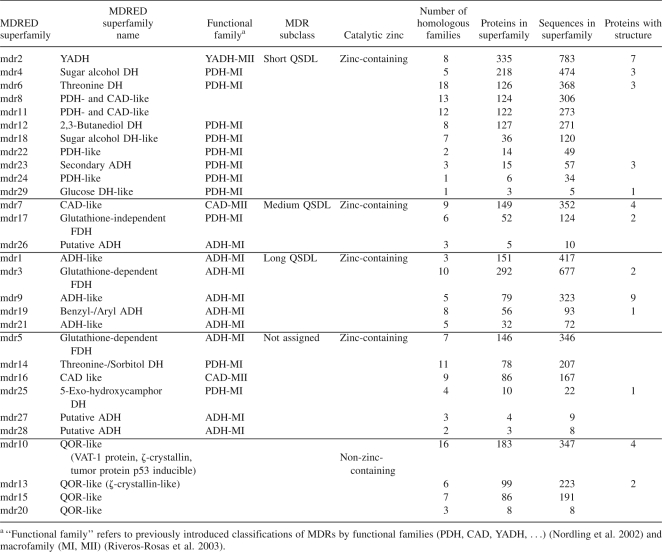
For a systematic analysis of sequence, structure, and function of the huge and diverse protein family of medium-chain dehydrogenases/reductases, the Medium-Chain Dehydrogenase/Reductase Engineering Database (MDRED) has been established. The MDRED contains 6420 sequence entries for 2684 proteins and 257 structure entries for 42 proteins. The MDRs were assigned to 29 superfamilies based on sequence similarity (Table 1). The superfamilies were further divided into 199 homologous families based on multiple sequence alignments of the superfamilies. For 24 homologous families (13 superfamilies), at least one family member with experimentally determined structure is available. A systematic nomenclature mdrx.y was introduced, where x describes the superfamily number and y the homologous family number. For each family, a multiple sequence alignment was performed and a phylogenetic tree was calculated. Functionally relevant residues were annotated. Annotation information was extracted from GenBank and transferred to all family members with a conserved residue at the respective position. All alignments and phylogenetic trees are accessible at http://www.mdred.uni-stuttgart.de. The MDRED can be browsed on the level of family classification, structure, and organisms. Sequence and protein entries can be searched by providing their GI number (general identifier) from NCBI. Protein name, source organism, and links to the respective GenBank entry are provided for each protein. The MDRED supports classification of new sequences by providing a BLAST interface and by pre-calculated HMM profiles for each superfamily and homologous family. The complete data are available via a tar archive.
Table 1.
Superfamilies of the Medium-Chain Dehydrogenase/Reductase Engineering Database and subclassification of zinc-containing MDRs according to the QSDL
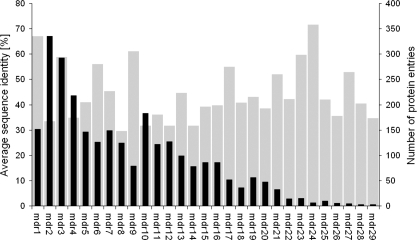
The largest superfamilies are mdr2, mdr3, and mdr4 with 335, 292, and 218 protein entries, respectively, which account for 32% of all protein entries (Fig. 1). Sequence lengths vary from 272 to 437 residues, with an average length of 357. The average sequence identity within each superfamily varies from 30% (superfamily mdr8) to 71% (superfamily mdr24). The superfamilies were grouped into two classes, zinc-containing and non-zinc-containing MDRs, dependent on the presence of a strictly conserved and annotated sequence motif G-H-E of the catalytic zinc binding site. In the non-zinc-containing MDRs, the highly conserved active-site residues asparagine, aspartic acid/glutamic acid, and threonine were annotated. MDRs of superfamily mdr28 are an exception, as they lack both motifs. They were classified as zinc-containing MDRs owing to their global sequence similarity.
Figure 1.
Average sequence identity (gray) and number of protein entries (black) per superfamily.
Non-zinc-containing MDRs
Superfamilies mdr10, mdr13, mdr15, and mdr20 comprise family members of QOR, LTD, MRF, and ACR (Nordling et al. 2002). These superfamilies were assigned to the class of non-zinc-containing MDRs. All non-zinc-containing MDRs are lacking the catalytic zinc binding motif. Most of the non-zinc-containing MDRs are also lacking a sequence motif capable for coordination of a structural zinc atom. However, in some family members of superfamilies mdr15 and mdr20, such a sequence motif was found.
Zinc-containing MDRs
Proteins belonging to the functional family of ADHs (Nordling et al. 2002) and homologs were found in superfamilies mdr1, mdr3, mdr5, mdr9, mdr19, and mdr21, glutathione-dependent formaldehyde dehydrogenases in mdr3 and mdr5, benzyl/aryl ADHs in mdr19, and the majority of YADHs in mdr2. Members of PDHs and CADs were distributed over superfamilies mdr4, mdr6, mdr7, mdr8, mdr11, mdr12, mdr14, mdr17, mdr18, mdr22, mdr23, mdr24, mdr25, and mdr29, with the majority of sugar alcohol dehydrogenases (sorbitol, arabinitol, iditol, idionate, and xylitol dehydrogenases) found in superfamily mdr4. Secondary ADHs were found in superfamily mdr23, glucose dehydrogenases in mdr29, glutathione-independent formaldehyde dehydrogenases in mdr17, 5-exo-hydroxycamphor dehydrogenases in mdr25, and 2,3-butanediol dehydrogenases in mdr12. While threonine dehydrogenases were found in almost all superfamilies of zinc-containing MDRs, most of them as well as sorbitol dehydrogenases were found in superfamilies mdr6 and mdr14.
Subclassification of zinc-containing MDRs
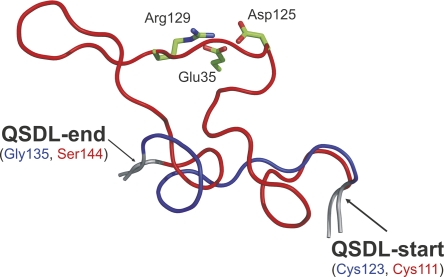
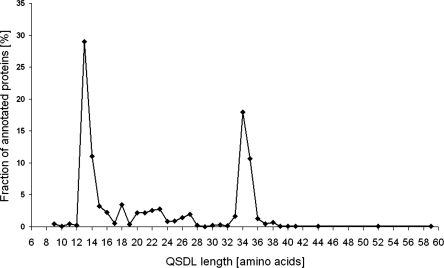
Comparison of 37 superimposed structures of zinc-containing MDRs revealed a highly conserved overall tertiary structure, although the proteins are diverse in sequence. Horse liver ADH (HLADH, PDB entry 1HEU) (Meijers et al. 2001) and the ADH from Aeropyrum pernix (ApADH, PDB entry 1H2B) (Guy et al. 2003) have only a small Cα root mean square deviation of 1.6 Å for 235 out of 343 residues, although these enzymes considerably differ in sequence (only 28% sequence identity). The most variable region is a loop segment located subsequent to the structural zinc binding site. This loop segment varies in sequence, length, and conformation. Because this loop segment was previously postulated to mediate quaternary structure formation of MDRs, we named it quaternary structure determining loop (QSDL). As seen from multiple sequence alignments and superimposition of all available structures of zinc-containing MDRs, the QSDL is flanked by residues that are highly conserved in sequence and structure. These residues were annotated as QSDL-start and QSDL-end (Fig. 2). In HLADH and ApADH, the residue at QSDL-start was a cysteine (position 111 in HLADH and position 123 in ApADH). At QSDL-end, a serine was found in the structure of HLADH at position 144 and a glycine in ApADH at position 135. In all annotated sequences of zinc-containing MDRs, only cysteine and serine were found at position QSDL-start (98% and 2%, respectively). Because it represents the fourth zinc-coordinating residue of the structural zinc binding motif, it could be easily identified on sequence level. QSDL-end is more difficult to find. However, it is embedded in a conserved sequence motif: At position QSDL-end, glycine was found in 75%, and serine and threonine were found in 22% of all sequences. A high frequency of occurrence was found for phenylalanine (52%) and leucine, tyrosine, or glutamine (12% each) at position QSDL-end+2. At position QSDL-end+3, alanine and serine (50% and 30%, respectively) were found; at position QSDL-end+4, glutamic acid and glutamine (62% and 11%, respectively) were found. At position QSDL-end+5, tyrosine was found in 67% of all cases. Using this motif, for 2131 zinc-containing MDRs (92%), QSDL-start and QSDL-end could be annotated. Counting the number of amino acids of each annotated QSDL revealed two peaks in the QSDL length distribution (Fig. 3). Thus, three classes were defined. The class of short QSDL has a QSDL of less than 19 amino acids and comprises 52% of all proteins with annotated QSDL. Only 15% of proteins with annotated QSDLs have a QSDL of medium length of 19 to 31 residues. The class of long QSDL has a QSDL of more than 31 amino acids and comprises 33% of all zinc-containing MDRs with annotated QSDL. An MDRED superfamily was assigned to the class of short QSDL if >90% of its members have a QSDL of less than 19 residues in length. Thus, superfamilies mdr2, mdr4, mdr6, mdr8, mdr11, mdr12, mdr18, mdr22, mdr23, mdr24, and mdr29 were assigned to the class of short QSDL. Likewise, superfamilies mdr7, mdr17, and mdr26 were assigned to the class of medium QSDL, because >90% of the family members have a QSDL length between 19 and 31 residues. Superfamilies mdr1, mdr3, mdr9, mdr19, and mdr21 were assigned to the class of long QSDL, because >90% of proteins possess a QSDL of more than 31 residues. All remaining superfamilies of zinc-containing MDRs that were not assigned to either of these classes were classified as “not assigned,” because their QSDLs could not be annotated for the superfamily members (mdr27, mdr28) or because family members have QSDLs of mixed lengths (mdr5, mdr14, mdr16, and mdr25).
Figure 2.
Structurally conserved loop segments (QSDL) of the class of long QSDL (red) and short QSDL (blue). The representative QSDL for each family is shown. (Red) Horse liver ADH (PDB entry: 1HEU); (blue) Aeropyrum pernix ADH (PDB entry: 1H2B). QSDL-start and QSDL-end are labeled. The conserved salt bridge network in long QSDL MDRs (side chains of Asp125, Arg129, Glu35) are shown as sticks and colored by atom type.
Figure 3.
Fraction of proteins with annotated QSDL for each QSDL length.
A sequence conservation analysis of the QSDL was performed for all proteins of each class of short and long QSDL to identify highly conserved and thus potentially structurally relevant residues located in the QSDL. The QSDL sequences of the class of long QSDL are slightly more conserved than those of the class of short QSDL and contain two highly conserved residues, Asp125 and Arg129 (numbering refers to PDB entry 1HEU). A third highly conserved residue (Glu35) is located in the catalytic domain, pointing toward the QSDL. Owing to their side chain distances, they form a strong, highly conserved salt bridge network, which probably stabilizes the conformation of the QSDL. Such highly conserved and potentially stabilizing residues were not found for the QSDLs of the class of short and medium QSDL.
Shape of the binding site
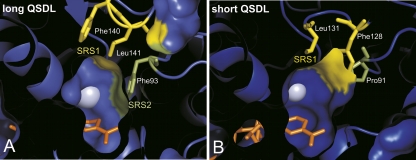
A comparison of 37 structures of zinc-containing MDRs revealed positions that are variable and contribute to the shape of the binding site. The residues at these positions were annotated in all superfamilies where structure information was available. For the class of long QSDL, two neighboring hydrophobic residues in the QSDL are forming the variable ceiling region above the cofactor NAD(P), which was named substrate recognition site 1 (SRS1). In the majority of proteins, a phenylalanine or tyrosine residue followed by leucine, valine, methionine, isoleucine, or phenylalanine was found. In HLADH (PDB entry 1HEU), these residues are Phe140 and Leu141. A third residue (Phe93 in PDB entry 1HEU of HLADH) forms the right wall of the binding site, which was named the substrate recognition site 2 (SRS2) (Fig. 4A). It is located two residues subsequent to a highly conserved proline (Pro91 in PDB entry 1HEU of HLADH) outside the QSDL. Mainly phenylalanine or tyrosine was found at SRS2. In the class of short QSDL, two hydrophobic residues form the binding site at SRS1 (Fig. 4B). These two residues are not adjacent as observed for the class of long QSDL, but separated by two residues. In the structure of ApADH, these residues are Phe128 and Leu131 (PDB entry 1H2B). No conserved residue at SRS2 was found for proteins in the class of short QSDL. However, this region contributes to substrate specificity, which has been demonstrated for yeast ADH, where exchanging tryptophan at position 93 by alanine led to an increased oxidation rate of 2-propanol (Creaser et al. 1990).
Figure 4.
Top view of the MDR binding-site. Representative structures of (A) long QSDL (PDB entry 1HEU) and (B) short QSDL (PDB entry 1H2B). The variable regions SRS1 (yellow) and SRS2 (green) are marked. The (blue) zinc atom is shown as a sphere; the (orange) nicotinamide part of NAD(P) as sticks.
Generally, the more bulky the residues at positions SRS1 and SRS2 are, the smaller the available space for the substrate in the binding site is expected to be. In a few MDRs, a tryptophan residue is found at either SRS1 or SRS2, which restricts the space of the binding site considerably.
Discussion
Classification
The Medium-Chain Dehydrogenase/Reductase Engineering Database (MDRED) has been designed to serve as a navigation and analysis tool of MDRs. The MDRED includes 2684 MDRs in a consistent data structure. It is based on the extensible database system DWARF (Fischer et al. 2006), which integrates information on sequence, structure, and function. The DWARF system has already been applied successfully to build up the Lipase Engineering Database (http://www.led.uni-stuttgart.de) (Pleiss et al. 2000; Fischer and Pleiss 2003) and the Cytochrome P450 Engineering Database (http://www.cyped.uni-stuttgart.de) (Fischer et al. 2007). The MDRED is a platform to analyze sequence–structure–function relationships and to classify new sequences by providing multiple sequence alignments, phylogenetic trees, and family-specific HMM profiles. Multiple sequence alignments with annotated functionally relevant residues are provided for each superfamily and homologous family.
In this study, 2684 MDRs were assigned to 29 superfamilies using a Markov cluster algorithm based on a sequence similarity graph, as implemented in the program TRIBE-MCL (Enright et al. 2002). Owing to the large number of sequences analyzed here, we suggest a subsequent classification of superfamilies into homologous families. The assignment of proteins to homologous families is based on multiple sequence alignments of the superfamilies, whereby a homologous family is defined as a cluster of protein sequences that are more similar to each other than to other protein sequences within the same superfamily. In accordance with previous classifications based on smaller numbers of about 100 MDRs (Nordling et al. 2002; Jörnvall et al. 2003) and of about 500 MDRs (Riveros-Rosas et al. 2003), the MDRs were assigned to two groups of zinc-containing and non-zinc-containing MDRs. The group of non-zinc-containing MDRs corresponds to macrofamily III, as introduced by Riveros-Rosas et al. (2003). The group of zinc-containing MDRs includes functional families of ADHs, PDHs, CADs, and YADHs as defined by Nordling et al. (2002) and corresponds to macrofamilies I and II by Riveros-Rosas et al. (2003). Our classification into zinc-containing and non-zinc-containing MDRs and the assignment of MDRs into superfamilies are consistent with previous classifications. The assignment of MDRs to superfamilies generally corresponds to the classification of Nordling et al. (2002), although more than one superfamily might exist for a single functional family as defined by Nordling et al. (2002) because of the large number of sequences investigated in this study (Table 1). Although the assignment of MDRs into superfamilies globally corresponds to the functional families, some exceptions were found. In two superfamilies (mdr8 and mdr11), proteins from several functional families (PDH and CAD) were found. Therefore, it seems that the specific patterns of PDH and CAD introduced previously to identify functional families (Nordling et al. 2002) are not yet specific enough to distinguish between the two functional families. Interestingly, the functional families PDH and CAD were grouped into different macrofamilies (PDH into macrofamily I and CAD into macrofamily II) by Riveros-Rosas et al. (2003). Thus, the currently defined sequence-specific patterns for PDH and CAD are not consistent with their global sequence similarity based on the large number of MDRs analyzed here.
Functional families according to Nordling et al. (2002) were further subclassified into subfamilies by Riveros-Rosas et al. (2003). This subclassification is generally in accordance with our sequence-based classification into superfamilies. However, owing to the large number of sequences analyzed here, functionally different MDRs were not necessarily found separated into different superfamilies of the MDRED: Threonine dehydrogenases (belonging to the functional family of PDHs) were found in different superfamilies of the MDRED in which also other PDHs were found. Thus, substrate specificity is not always linked to sequence similarity. Our classification by sequence similarity therefore enables the correlation of sequence and function, and helps us to understand the sequence–structure–function relationships of MDRs.
As a new classification criterion, we grouped superfamilies of zinc-containing MDRs into three structural classes based on the QSDL length (short, medium, and long QSDL). This assignment was successful for 86% of all zinc-containing MDRs. Superfamilies of the class of short QSDL comprise the functional families of YADHs, PDHs, and CADs; the class of long QSDL, the functional family of ADHs (Nordling et al. 2002). This assignment of superfamilies to classes based on structural properties further assists the understanding of sequence–structure–function relationships of MDRs.
Application
By systematically comparing sequence and structure of zinc-containing MDRs, a variable loop region was identified that varies in sequence and conformation. This loop (QSDL) mediates quaternary structure and possesses residues relevant for the shape of the substrate-binding site. Depending on the length of QSDL, three classes (class of short, medium, and long QSDL) were introduced. Superfamilies whose members possess a QSDL of a special length are assigned to one of the three classes. A correlation of the quaternary structure of MDRs with the length of their QSDLs was observed. With only one exception, the benzyl alcohol dehydrogenase from Acinetobacter calcoaceticus (PDB entry: 1F8F; http://dx.doi.org/10.2210/pdb1f8f/pdb) (MacKintosh and Fewson 1988), all proteins of the class of long QSDL with known three-dimensional structure are active as dimers, whereas all members of the class of short QSDL are tetramers. This is in accordance with previous observations, where this loop segment was suggested to mediate quaternary structure (Jörnvall 1977; Persson et al. 1994; Norin et al. 1997). The QSDL was successfully annotated in most of the superfamilies of zinc-containing MDRs, enabling the prediction of quaternary structure of more than 2000 MDRs. This prediction is in accordance with experimental data on quaternary structure for secondary ADHs (mdr23), threonine and sorbitol dehydrogenases, glutathione-dependent formaldehyde dehydrogenases (mdr3), hydroxynitrile lyases (mdr3), and the YADH functional families (Riveros-Rosas et al. 2003). However, there are a small number of exceptions: The (R,R)-butanediol dehydrogenase from Saccharomyces cerevisiae (mdr12.1) has been reported to be active as the dimer (Gonzalez et al. 2000), although another homologous (R,R)-butanediol dehydrogenase from Saccharomyces cerevisiae is known to be tetrameric (Heidlas and Tressl 1990). Furthermore, proteins of the family of galactitol 1-phosphate dehydrogenases (mdr18) have been classified in the class of short QSDL according to their QSDL length, although forming dimers (Riveros-Rosas et al. 2003).
MDRs of the class of medium QSDL whose structures are known show both, dimeric and tetrameric quaternary structures, such as the alcohol dehydrogenase from Escherichia coli (PDB entry 1UUF; http://dx.doi.org/10.2210/pdb1uuf/pdb), which has a QSDL length of 21 residues and is active as the dimer, while the formaldehyde dismutase from Pseudomonas putida (PDB entry 2DPH; http://dx.doi.org/10.2210/pdb2dph/pdb) has a QSDL length of 24 residues and is active as the tetramer. Even an active trimeric structure was found within the class of medium QSDL, the mycothiol-dependent formaldehyde dehydrogenase (Norin et al. 1997) with a QSDL of 26 residues in length.
The QSDL is not only involved in quaternary structure formation, but is also part of the substrate-binding site (Shafqat et al. 1999). Residues located within the QSDL contribute to the shape of the binding site at the variable ceiling region above the NAD(P) cofactor (SRS1). Since these residues are located at different positions within the classes of short QSDL and long QSDL, they are generally difficult to identify. By grouping zinc-containing MDRs into three classes, these residues are predictable for most MDRs within the classes of short and long QSDL. For most of the proteins of the class of short and long QSDL, annotation of these relevant residues was possible, which accounts for about 1500 proteins. The majorities of residues found at these positions are hydrophobic and point toward the substrate. Thus, the bulkiness of their side chain is expected to mediate substrate specificity. This was demonstrated for members of the class of short QSDL. Exchanging Trp110 by alanine significantly broadened substrate specificity toward phenyl-substituted alcohols and ketones of the alcohol dehydrogenase of Thermoanaerobacter ethanolicus (Ziegelmann-Fjeld et al. 2007). For members of the class of long QSDL, an increased affinity for the 4-methylpyrazole inhibitor was attributed to the exchange of Met141 by leucine in human σσ alcohol dehydrogenase (Xie and Hurley 1999). Additionally, exchange of hydrophobic residues at the second substrate recognition site (SRS2), which contribute to the shape of the right wall of the binding site, have led to changes in substrate specificity. A significant increase of activity toward bulky secondary alcohols of a double mutant, Phe93Ala/Thr94Ile, of human liver alcohol dehydrogenase as compared to the wild type was explained by removal of steric hindrance mainly caused by Phe93 (Hurley and Bosron 1992). Thus, residues located at positions SRS1 and SRS2 determine the available space within the binding site, and annotation of these residues enables a reliable prediction for a large number of MDRs.
For the first time, a classification of MDRs based on structural differences (length of QSDL) is proposed that is predictive for the quaternary structure of MDRs. In addition, the QSDL contributes amino acids that are relevant for the shape of the binding site (SRS1). These functionally relevant residues are annotated within the database. The MDRED provides a comprehensive resource of information on the MDR family in a consistent format, and thus is a valuable tool for a deeper understanding of biochemical properties of MDRs. By a systematic classification and annotation of MDRs, it facilitates extracting rules for substrate specificity and identifying promising mutation sites in order to engineer proteins with improved properties.
Materials and Methods
Database setup
The MDRED was established by applying the data warehouse system DWARF (Fischer et al. 2006). The DWARF system integrates data on sequence, structure, and functional annotation for protein fold families and provides tools for extracting, transforming, and loading data from public resources to populate a local protein family database. Data and annotation information were extracted from GenBank (Benson et al. 2007) and the Protein Data Bank (Berman et al. 2000). Additional annotation information (zinc-binding site, active-site residues, QSDL-start, QSDL-end, SRS1, and SRS2) was added manually. Proteins were assigned to superfamilies and homologous families based on sequence similarity. The protein entries were classified into superfamilies applying the TribeMCL method (Enright et al. 2002) with an inflation value I = 2. The subsequent classification of each superfamily into homologous families was achieved by multiple sequence alignments and phylogenetic trees, as calculated by CLUSTALW (v1.83) (Thompson et al. 1994) with default parameters.
All sequence entries that share >98% sequence identity and descent from the same organism are considered as a single protein entry in the database. In case of multiple sequence entries for each protein, the longest sequence for one protein entry was assigned as the reference sequence for analysis. All sequences shorter than a minimal length of 272 amino acids and longer than a maximum length of 439 amino acids were discarded. These minimal and maximal values were derived based on the shortest and longest sequence within the database with known 3D structure ± 10% (quinone oxidoreductase from Thermus thermophilus, 302 amino acids; formaldehyde dehydrogenase from Pseudomonas putida, 399 amino acids). For sequence entries where structure information was available, structure data were stored as a structure entry. Secondary structure information was calculated using DSSP (Kabsch and Sander 1983) and displayed within one set of the annotated multiple sequence alignments. Multiple sequence alignments of each family were used for improvement of classification, as well as for enrichment of annotation information.
Web accessibility
Annotated multiple sequence alignments and phylogenetic trees are provided via the online accessible version of the MDRED at http://www.mdred.uni-stuttgart.de. For each alignment, the information for amino acid conservation is given as calculated by PLOTCON (Rice et al. 2000). For each homologous family and superfamily, family-specific HMM profiles, calculated with the HMMER program (http://hmmer.janelia.org/), and phylogenetic trees that are visualized applying the program PHYLODENDRON (http://iubio.bio.indiana.edu/soft/molbio/java/apps) are supplied. All protein entries are linked to the respective GenBank entries, and all annotated multiple sequence alignments, phylogenetic trees, structural monomers, and HMM profiles can be visualized and accessed via the website. Additionally, an archive, comprising sequences, structures, alignments, and phylogenetic trees grouped by families, and a formatted text file listing all protein information can be downloaded.
Data analysis
For structural analysis and visualization, the PyMOL program (DeLano Scientific) and Swiss-PdbViewer (Guex and Peitsch 1997) were used. In case of multiple structure entries for one protein, the structural monomer originating from the wild-type enzyme with the best resolution was used for analysis. All structures were superimposed onto the structure of HLADH with PDB entry 1HEU (Meijers et al. 2001) for analysis. Conservation analysis was performed for each class of short, medium, and long QSDL, using the program Al2Co (Pei and Grishin 2001) based on a multiple sequence alignment including all proteins of each class. Scripts for analysis were written in Perl.
Acknowledgments
Valuable contributions by Ima Avalos Vizcarra and Florian Wagner are acknowledged. This work was financially supported by the Deutsche Bundesstiftung Umwelt.
Footnotes
Reprint requests to: Jürgen Pleiss, Institute of Technical Biochemistry, University of Stuttgart, Allmandring 31, D-70569 Stuttgart, Germany; e-mail: Juergen.Pleiss@itb.uni-stuttgart.de; fax: 49-711-68563196.
Abbreviations: ACR, acyl-CoA reductase; ADH, alcohol dehydrogenase; ApADH, alcohol dehydrogenase from Aeropyrum pernix; CAD, cinnamyl alcohol dehydrogenase; HLADH, horse liver alcohol dehydrogenase; HMM, Hidden Markov model; LTD, leukotriene B4 dehydrogenase; MI, macrofamily I; MII, macrofamily II; MDR, medium-chain dehydrogenase/reductase; MDRED, Medium-Chain Dehydrogenase/Reductase Engineering Database; MRF, mitochondrial response protein; PDH, polyoldehydrogenase; QOR, quinone oxidoreductase; QSDL, quaternary structure determining loop; SRS, substrate recognition site; YADH, yeast alcohol dehydrogenase/tetrameric alcohol dehydrogenase.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035428.108.
References
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Wheeler, D.L. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase T., Jr Alcohol dehydrogenases: Identification and names for gene families. Plant Mol. Biol. Rep. 1999;17:333–350. [Google Scholar]
- Creaser, E.H., Murali, C., Britt, K.A. Protein engineering of alcohol dehydrogenases: Effects of amino acid changes at positions 93 and 48 of yeast ADH1. Protein Eng. 1990;3:523–526. doi: 10.1093/protein/3.6.523. [DOI] [PubMed] [Google Scholar]
- Eklund, H., Nordström, B., Zeppezauer, E., Söderlund, G., Ohlsson, I., Boiwe, T., Söderberg, B.-O., Tapia, O., Brändén, C.-I., Åkeson, Å. Three-dimensional structure of horse liver alcohol dehydrogenase at 2.4 Å resolution. J. Mol. Biol. 1976;102:27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Van Dongen, S., Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, M., Pleiss, J. The Lipase Engineering Database: A navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, M., Thai, Q.K., Grieb, M., Pleiss, J. DWARF—a data warehouse system for analyzing protein families. BMC Bioinformatics. 2006;7:495. doi: 10.1186/1471-2105-7-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, M., Knoll, M., Sirim, D., Wagner, F., Funke, S., Pleiss, J. The Cytochrome P450 Engineering Database: A navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics. 2007;23:2015–2017. doi: 10.1093/bioinformatics/btm268. [DOI] [PubMed] [Google Scholar]
- Gonzalez, E., Fernandez, M.R., Larroy, C., Sola, L., Pericas, M.A., Pares, X., Biosca, J.A. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J. Biol. Chem. 2000;275:35876–35885. doi: 10.1074/jbc.M003035200. [DOI] [PubMed] [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guy, J.E., Isupov, M.N., Littlechild, J.A. The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix . J. Mol. Biol. 2003;331:1041–1051. doi: 10.1016/s0022-2836(03)00857-x. [DOI] [PubMed] [Google Scholar]
- Heidlas, J., Tressl, R. Purification and characterization of a (R)-2,3-butanediol dehydrogenase from Saccharomyces cerevisiae . Arch. Microbiol. 1990;154:267–273. doi: 10.1007/BF00248966. [DOI] [PubMed] [Google Scholar]
- Hurley, T.D., Bosron, W.F. Human alcohol dehydrogenase: Dependence of secondary alcohol oxidation on the amino acids at positions 93 and 94. Biochem. Biophys. Res. Commun. 1992;183:93–99. doi: 10.1016/0006-291x(92)91613-u. [DOI] [PubMed] [Google Scholar]
- Jörnvall, H. Differences between alcohol dehydrogenases—structural-properties and evolutionary aspects. Eur. J. Biochem. 1977;72:443–452. doi: 10.1111/j.1432-1033.1977.tb11268.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall, H., Eklund, H., Branden, C.I. Subunit conformation of yeast alcohol dehydrogenase. J. Biol. Chem. 1978;253:8414–8419. [PubMed] [Google Scholar]
- Jörnvall, H., Hoog, J.O., Persson, B. SDR and MDR: Completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999;445:261–264. doi: 10.1016/s0014-5793(99)00130-1. [DOI] [PubMed] [Google Scholar]
- Jörnvall, H., Nordling, E., Persson, B. Multiplicity of eukaryotic ADH and other MDR forms. Chem. Biol. Interact. 2003;143–144:255–261. doi: 10.1016/s0009-2797(02)00242-9. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- MacKintosh, R.W., Fewson, C.A. Benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II from Acinetobacter calcoaceticus. Purification and preliminary characterization. Biochem. J. 1988;250:743–751. doi: 10.1042/bj2500743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers, R., Morris, R.J., Adolph, H.W., Merli, A., Lamzin, V.S., Cedergren-Zeppezauer, E.S. On the enzymatic activation of NADH. J. Biol. Chem. 2001;276:9316–9321. doi: 10.1074/jbc.M010870200. [DOI] [PubMed] [Google Scholar]
- Nordling, E., Jörnvall, H., Persson, B. Medium-chain dehydrogenases/reductases (MDR). Family characterizations including genome comparisons and active site modeling. Eur. J. Biochem. 2002;269:4267–4276. doi: 10.1046/j.1432-1033.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- Norin, A., Van Ophem, P.W., Piersma, S.R., Persson, B., Duine, J.A., Jörnvall, H. Mycothiol-dependent formaldehyde dehydrogenase, a prokaryotic medium-chain dehydrogenase/reductase, phylogenetically links different eukaroytic alcohol dehydrogenases–primary structure, conformational modelling and functional correlations. Eur. J. Biochem. 1997;248:282–289. doi: 10.1111/j.1432-1033.1997.00282.x. [DOI] [PubMed] [Google Scholar]
- Pei, J., Grishin, N.V. AL2CO: Calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- Persson, B., Zigler J.S., Jr, Jörnvall, H. A super-family of medium-chain dehydrogenases/reductases (MDR). Sub-lines including zeta-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductase enoyl reductases, VAT-1 and other proteins. Eur. J. Biochem. 1994;226:15–22. doi: 10.1111/j.1432-1033.1994.tb20021.x. [DOI] [PubMed] [Google Scholar]
- Pleiss, J., Fischer, M., Peiker, M., Thiele, C., Schmid, R.D. Lipase engineering database—understanding and exploiting sequence–structure–function relationships. J. Mol. Catal., B Enzym. 2000;10:491–508. [Google Scholar]
- Rice, P., Longden, I., Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Riveros-Rosas, H., Julian-Sanchez, A., Villalobos-Molina, R., Pardo, J.P., Pina, E. Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur. J. Biochem. 2003;270:3309–3334. doi: 10.1046/j.1432-1033.2003.03704.x. [DOI] [PubMed] [Google Scholar]
- Shafqat, J., Hoog, J.O., Hjelmqvist, L., Oppermann, U.C., Ibanez, C., Jörnvall, H. An ethanol-inducible MDR ethanol dehydrogenase/acetaldehyde reductase in Escherichia coli: Structural and enzymatic relationships to the eukaryotic protein forms. Eur. J. Biochem. 1999;263:305–311. doi: 10.1046/j.1432-1327.1999.00323.x. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, P.T., Hurley, T.D. Methionine-141 directly influences the binding of 4-methylpyrazole in human σσ alcohol dehydrogenase. Protein Sci. 1999;8:2639–2644. doi: 10.1110/ps.8.12.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelmann-Fjeld, K.I., Musa, M.M., Phillips, R.S., Zeikus, J.G., Vieille, C. A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and stereoselective on phenylacetone and benzylacetone. Protein Eng. Des. Sel. 2007;20:47–55. doi: 10.1093/protein/gzl052. [DOI] [PubMed] [Google Scholar]