Abstract
In the type III secretion system (T3SS) of Aeromonas hydrophila, the putative needle complex subunit AscF requires both putative chaperones AscE and AscG for formation of a ternary complex to avoid premature assembly. Here we report the crystal structure of AscE at 2.7 Å resolution and the mapping of buried regions of AscE, AscG, and AscF in the AscEG and AscEFG complexes using limited protease digestion. The dimeric AscE is comprised of two helix–turn–helix monomers packed in an antiparallel fashion. The N-terminal 13 residues of AscE are buried only upon binding with AscG, but this region is found to be nonessential for the interaction. AscE functions as a monomer and can be coexpressed with AscG or with both AscG and AscF to form soluble complexes. The AscE binding region of AscG in the AscEG complex is identified to be within the N-terminal 61 residues of AscG. The exposed C-terminal substrate-binding region of AscG in the AscEG complex is induced to be buried only upon binding to AscF. However, the N-terminal 52 residues of AscF remain exposed even in the ternary AscEFG complex. On the other hand, the 35-residue C-terminal region of AscF in the complex is resistant to protease digestion in the AscEFG complex. Site-directed mutagenesis showed that two C-terminal hydrophobic residues, Ile83 and Leu84, of AscF are essential for chaperone binding.
Keywords: type III secretion system, chaperones AscE and AscG, limited protease digestion, induced folding, needle subunit AscF
Aeromonas hydrophila is a ubiquitous Gram-negative bacterium that can cause motile aeromonad septicemia in both fish and human (Thune et al. 1993; Austin and Adams 1996). The main clinical symptoms associated with Aeromonas infection are gastroenteritis, wound infections, and systemic illness (Janda 2001). For survival, many Gram-negative bacteria use type III secretion systems (T3SSs) to deliver effectors into host cells to exploit host cellular functions (Finlay and Cossart 1997). AexT, an ADP-ribosyltransferase toxin (Braun et al. 2002), and AopP, a protein that inhibits the NF-κB signaling pathway (Fehr et al. 2006), are two examples of T3SS effectors delivered by the T3SS of Aeromonas salmonicida. A T3SS gene cluster has also been located in A. hydrophila AH-1 by using homology-based analysis and shown to be necessary for its pathogenesis (Yu et al. 2004). At least three T3SS-secreted proteins (or effector proteins) are found in the extracellular proteome of a T3SS-negative regulator mutant but not in a T3SS-deficient mutant (Yu et al. 2007). One of these effector proteins showed homology with the AexT/AexU effector, which has been reported recently in A. hydrophila strains AH-3 (Vilches et al. 2007) and SSU (Sha et al. 2007; Sierra et al. 2007).
The T3SS is composed of a base that spans both membranes, a needle complex that protrudes outward from the bacterial surface (Cornelis and Van Gijsegem 2000), and various chaperones, effectors, and translocators that are necessary for the assembly and function of the T3SS (Ghosh 2004). The T3SS needle complex is composed essentially of one low-molecular-weight protein, for example, PrgI in Salmonella typhimurium (Kimbrough and Miller 2000), MxiH in Shigella flexneri (Blocker et al. 2001), YscF in Yersinia enterocolitica (Hoiczyk and Blobel 2001), and PscF in Pseudomonas aeruginosa (Pastor et al. 2005), which polymerizes to form a structure of 60–100 nm in length. In A. hydrophila AH-1, AscF has been identified as the putative subunit of the T3SS needle complex based on its sequence homology with these low-molecular-weight proteins. Polymerized T3SS needles form a conduit for the transport of specific effector proteins through a pore within the eukaryotic plasma membrane into the eukaryotic cytoplasm, where they interfere with the host cell-signaling pathway (Hoiczyk and Blobel 2001). The NMR solution structures of the T3SS needle subunits, Burkholderia pseudomallei BsaL (Zhang et al. 2006) and S. typhimurium PrgI (Wang et al. 2007), as well as the crystal structure of S. flexneri MxiH (Deane et al. 2006), have been determined. BsaLCΔ5 (lacking its five C-terminal residues), MxiHCΔ5, and PrgICΔ5 all form a similar antiparallel two-helix bundle with the two α-helices joined by a conserved four-residue [P-(S/D)-(D/N)-P] turn.
To avoid premature oligomerization of the needle complex subunit inside the bacterial cell, chaperones are needed to keep the subunit in a soluble and monomeric form. In P. aeruginosa, PscE and PscG interact to form a heteromolecular chaperone, PscEG, which is required to trap PscF in a monomeric state by forming a 1:1:1 ternary PscE-PscF-PscG complex. Neither PscE nor PscG could rescue polymerized PscF when the proteins were expressed and purified separately (Quinaud et al. 2005). The crystal structure of YscE from Yersinia pestis has been reported as a dimeric helical protein (Phan et al. 2005). Recently, the crystal structure of a YscEFG complex has been determined, showing that YscE interacts with the N-terminal tetratricopeptide repeat motif of YscG. YscG binds tightly to the C-terminal half of YscF, which adopts an α-helical hairpin conformation (Sun et al. 2008). The analogous crystal structure of the PscE-PscF55-85-PscG complex has also been determined, showing that the PscE-PscG heterodimeric chaperone folded in the form of a cupped hand with the C terminus of PscF engulfed within the hydrophobic groove of PscG (Quinaud et al. 2007). Here we report the crystal structure of A. hydrophila AH-1 AscE (PDB ID: 2Q1K) refined at 2.7 Å resolution. Additionally, using partial protease digestion, mass spectrometry, and N-terminal sequencing, we have determined exposed regions in AscE and AscG that are induced to be buried only upon interacting with their respective binding partners. Our findings suggested that the conformation of the buried part of AscG in the AscEG complex may not be the same as that of the same region in the AscEFG complex.
Results
Structure of A. hydrophila AH-1 AscE
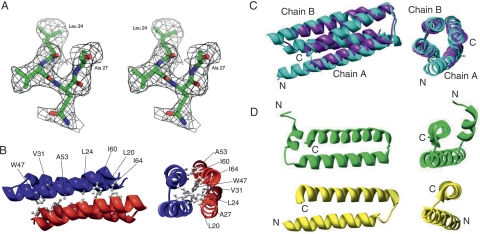
The structure of recombinant AscE from A. hydrophila AH-1 was solved by the multi-wavelength anomalous dispersion (MAD) method from the synchrotron data. The model has been refined with good stereochemical parameters (Table 1). There are four molecules of AscE in each of the asymmetric units. The first 13 N-terminal residues, Met1 to Asp13, did not show interpretable electron density and thus were not modeled. Each monomer comprises residues from Pro14 to Lys65. The oligomerization state of AscE in solution was verified with Dynamic Light Scattering (DLS) and gel filtration experiments and found to be consistent with the observation of a dimeric crystal structure (Fig. 1B). Each monomer of AscE consists of two antiparallel α-helices, an N-terminal α-helix (residues Pro14 to Leu36), and a C-terminal α-helix (residues Gln41 to Lys65). These two α-helices are connected by a loop (residues Arg37 to Thr40) forming a helix–turn–helix conformation (Fig. 1B). Two monomers of the dimeric AscE molecules packed in an antiparallel head-to-tail manner with ∼990 Å2 (or 23.13% of total surface area of each molecule) surface area on each molecule involved in the dimeric interface, as calculated by the program GRASP (Nicholls et al. 1991). Close contacts between monomers of the dimeric AscE are maintained by extensive hydrophobic interactions from the side chains of residues Leu20, Leu24, Ala27, Val31, Trp47, Ala53, Ile60, and Ile64 on both monomers of the dimer and one hydrogen-bonding contact (between Glu50-OE1 and Gly57-O, 3.18 Å). The hydrophobic interactions are primarily responsible for stabilizing the dimeric architecture of AscE (Fig. 1B).
Table 1.
Crystallographic data and refinement statistics
Figure 1.
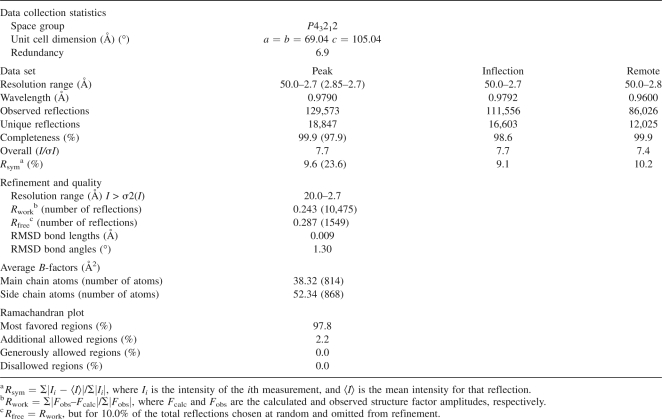
Crystal structure and the simulated-annealing F o –F c omit map in the conserved region of AscE. (A) The map is contoured at a level of 2.0 σ. Residues Leu24 to Ala27 and all atoms within 3.0 Å of Leu24 to Ala27 were omitted prior to refinement. (B) Ribbon representation of the crystal structure of the dimeric AscE from residue Pro14 to Glu65 at two different angles. The hydrophobic residues (Leu20, Leu24, Ala27, Val31, Trp47, Ala53, Ile60, and Ile64) that form an interlocking network at the dimeric interface of the protein are shown in a ball-and-stick model. The figure was generated with the program Chimera (Pettersen et al. 2004). (C) Overlay of the crystal structures of AscE (purple) and YscE (chain A and B) (cyan) viewed at two different angles. The dimers of AscE and YscE overlay with an RMSD of 2.2 Å for 101 Cα atoms using DaliLite pairwise comparison of protein structure. (D) Ribbon representation of the structures of PscE (green) and YscE (yellow) as in the crystal structures of the complexes PscE-PscF55–85-PscG and YscEFG viewed at two different angles (Quinaud et al. 2007; Sun et al. 2008). Figure 1A was prepared using the program PyMOL (DeLano Scientific). Figure 1B–D was prepared using the program Chimera (Pettersen et al. 2004).
Sequence and structural homology of AscE
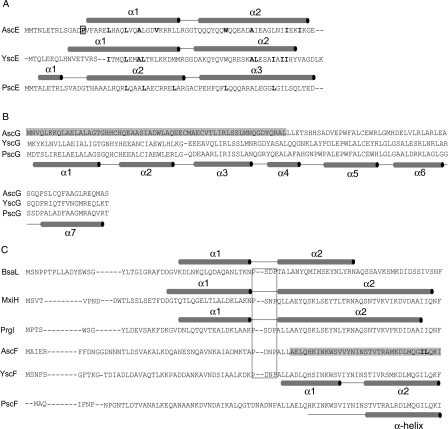
Sequence alignment of A. hydrophila AH-1 AscE with the homologous YscE from Y. pestis (38.9% identity and 63.0% similarity, from residues Pro14 to Glu67) and PscE from P. aeruginosa (33.3% identity and 57.4% similarity, from residues Pro14 to Glu67) shows relatively high sequence homology. Identical residues, however, are not confined to a particular region of the protein and are not conserved completely in YscE and PscE (Fig. 2). AscE is structurally similar to YscE (PDB ID: 1ZW0) (Phan et al. 2005). The monomers (chain A of AscE and YscE) superimposed with a root mean square deviation (RMSD) of 1.7 Å for 49 Cα atoms, while the dimers (chain A and B of AscE and YscE) superimposed with an RMSD of 2.2 Å for 101 Cα atoms using the DaliLite pairwise comparison of protein structures (http://www.ebi.ac.uk/DaliLite/index.html) (Fig. 1C). However, the N-terminal helix (α1) of AscE is much shorter compared to that of YscE (Fig. 2A). This may be due to the presence of residue Pro14, which disrupts the α-helix, hence making the first 13 residues disordered and resulting in no observable electron density in this region (Fig. 1B). Furthermore, the two α-helices in AscE are connected by a short turn of four residues (RRGT), while the flexible loop of eight residues (MRGGDAKQ) in YscE is much longer. Recently, the crystal structure of YscE in complex with YscG and YscF was also reported (PDB ID: 2P58) (Sun et al. 2008). The YscE subunit of the YscEFG complex is composed of two α-helices, residues 10–33 and residues 37–60, connected by a three-residue loop (Fig. 1D). Comparison of YscE in the YscEFG complex with that of the YscE dimer showed that the predominate differences lie in the relative position of the two helices and the conformation of the loop between them. Interestingly, the first nine residues of YscE are disordered in the complex (Sun et al. 2008). In this aspect, YscE in the YscEFG complex is more similar to the crystal structure of AscE. An analogous crystal structure of the PscE–PscF55–85–PscG complex is also available (PDB ID: 2UWJ) (Quinaud et al. 2007). The functional form of PscE when in complex with PscG and PscF consisted of three α-helices (residues 6–13, 17–41, and 44–69) separated by two loops of five and three residues, respectively (Figs. 1D and 2A). The location of PscE corresponding to residue Pro14 of AscE is at the loop region that connected helix 1 to helix 2 at a 45° angle (Fig. 1D). The residue Pro14 is absent in PscE and YscE but present in AscE of other Aeromonas sp., for example, A. hydrophila AH-3 (AAS91830) and A. salmonicida subsp. salmonicida (CAE83119).
Figure 2.
Protein sequence alignment of A. hydrophila AH-1. Protein sequences of (A) AscE, (B) AscG, and (C) AscF were aligned with related proteins from Pseudomonas aeruginosa (PscE, PscG, and PscF), Yersinia pestis (YscE, YscG, and YscF), Shigella flexneri (MxiH), Salmonella typhimurium (PrgI), and Burkholderia pseudomallei (BsaL) using CLUSTLAW (Thompson et al. 1994). (A) The secondary structures as determined in the crystal structures of AscE, YscEFG, and PscE-PscF55-85-PscG are shown above their respective protein sequences. The hydrophobic residues in the dimeric interface are bold-faced and residue Pro14, which is conserved among AscE in Aeromonas species is bold-faced and boxed. (B) The proposed AscE binding region of AscG is shaded in gray. The secondary structure of PscG as determined in the complex formed between PscE, PscF55–85, and PscG is shown below the protein sequence of PscG. (C) Secondary structures as determined in the NMR structures of BsaL and PrgI and the crystal structure of MxiH are shown above their protein sequences. The conserved P-(S/D)-(D/N)-P motifs that form the interhelical turn are boxed. The proposed chaperone region of AscF is shaded in gray and the two hydrophobic residues (Ile83 and Leu84) of the C-terminal “ILQKI” sequence that are essential for chaperone binding are shown in bold-face type. The secondary structure of YscF as determined in the YscEFG complex is shown below the protein sequence of YscF. The secondary structure of PscF as determined in the complex formed between PscE, PscF55-85, and PscG is shown below the protein sequence of PscF.
AscE is a dimer with exposed N-terminal region
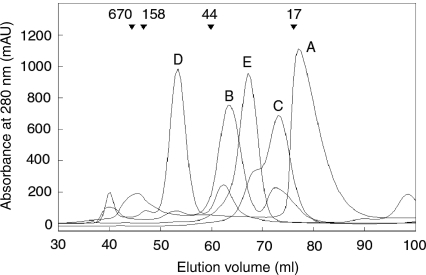
The thermal denaturation of AscE as followed by far-UV circular dichroism shows that the AscE dimer unfolds in a two-state reaction with a Tm of 52.7°C, which is very close to the Tm of 53°C as determined for PscE (Quinaud et al. 2005). Using gel filtration chromatography, we have verified that AscE exists as a dimer in solution, which agrees with the dimeric architecture as suggested by crystal structure. AscE (7.5 kDa monomeric molecular weight (MW), 15 kDa dimeric MW) eluted at a volume slightly less than that corresponded to a protein standard of 17 kDa (Fig. 3). We have also performed cross-linking experiments on AscE and found that the only species after cross-linking corresponds to the molecular weight of a dimeric AscE (data not shown).
Figure 3.
Gel filtration elution profiles. (A) AscE dimer, 15.12 kDa; (B) His-tagged AscEG complex, 22.12 kDa; (C) AscEG complex after partial protease digestion, 15.94 kDa; (D) His-tagged AscEFG complex, 31.74 kDa; and (E) His-tagged AscEFG complex after partial protease digestion, 26.53 kDa. The down arrows above the elution profiles label the elution volumes of known protein standards with their corresponding molecular weights. Note that His-tagged AscEG (B) and His-tagged AscEFG (D) appear to have a larger elution volume than their corresponding molecular weight, which is likely because of the presence of exposed region in the protein complexes.
Although AscE is a dimer in solution, the homologous PscE or YscE functions as a monomer by forming a 1:1:1 ternary complex with PscG and PscF or YscG and YscF, respectively (Quinaud et al. 2005, 2007; Sun et al. 2008). The crystal structure of AscE reveals the hydrophobic nature of the dimeric interface, and exposure of any of the hydrophobic residues will render the monomeric AscE unstable in solution. Formation of the dimer will keep these residues buried and increase the solubility of AscE. We speculate that these hydrophobic regions of the dimeric interface are involved in protein–protein interaction within the AscEG and AscEFG complexes. Indeed, the hydrophobic surfaces of helices Hb and Hc of the PscE monomer are recognized by a hydrophobic platform formed by helices H1 and H2 of PscG (Quinaud et al. 2007). The two N-terminal helices of YscG, α1 and α2, are packed against the two helices of YscE through hydrophobic interactions (Sun et al. 2008).
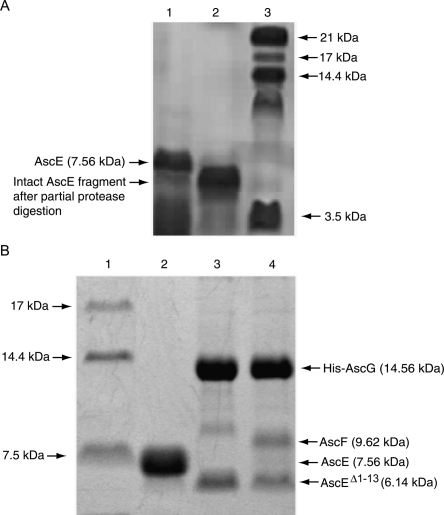
As the first 13 N-terminal residues, Met1 to Asp13, of AscE did not show any interpretable electron density map in the crystal structure, we speculate that this region of the protein was disordered and highly exposed. Using partial protease digestion, N-terminal sequencing, and mass spectrometry, we, indeed, observed that this region can be easily digested away, leaving an intact fragment starting from residue Pro14 (Fig. 4A).
Figure 4.
SDS Tricine PAGE showing partial protease digestion of AscE and coexpression of AscEΔ1–13 with AscG or AscG and AscF. (A) Purification and partial protease digestion of the AscE dimer. (Lane 1) Purified AscE dimer; the molecular weight of the monomer is ∼7.56 kDa; (lane 2) AscE after partial protease digestion. The intact AscE fragment has a smaller molecular weight, with the N-terminal 13 residues digested as confirmed by N-terminal sequencing; and (lane 3) protein molecular weight marker. (B) Coexpression of AscEΔ1–13 with AscG or AscG and AscF. (Lane 1) Protein molecular weight marker; (lane 2) purified AscE dimer; (lane 3) coexpression of AscEΔ1–13 with AscG; (lane 4) coexpression of AscEΔ1–13 with AscG and AscF. The AscEΔ1–13 N-terminal truncation mutant (6.14 kDa) can be coexpressed and copurified with His-AscG or both His-AscG and AscF to form a soluble complex.
The C-terminal substrate-binding region of AscG is exposed
AscG is predicted to be an all α-helical protein with six putative α-helices. Sequence alignment shows that it has an identity of 50.9% and 49.1% with the homologous PscG and YscG, respectively (Fig. 2). YscG is a putative Syc-like chaperone that directly binds to YscE, which is a cytoplasmic component of the T3SS (Day et al. 2000). YscG functions as a monomer as no YscG–YscG interaction was detected using the yeast two-hybrid system (Day et al. 2000). It is also shown that PscE, instead of acting as a substrate for the chaperone PscG, interacts with PscG to form a heteromolecular chaperone that is required to bind and stabilize PscF (Quinaud et al. 2005, 2007).
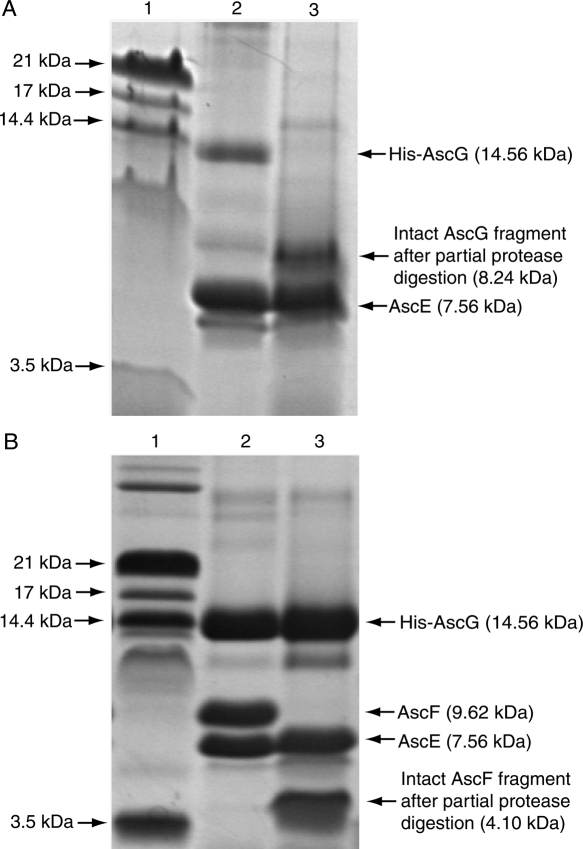
AscE and AscG can be coexpressed together and copurified as a soluble complex. The gel-filtration profile of the AscEG complex, however, showed that the complex eluted out at a volume corresponding to a larger molecular weight. This is likely due to the presence of a highly exposed region in the protein that increased its hydrodynamic radius. Partial protease digestion can be used to easily remove this exposed region and the digested AscEG complex eluted out at the expected volume (Fig. 3). By performing mass spectrometry and N-terminal sequencing, we have confirmed that this exposed region is located on AscG in the AscEG complex (Fig. 5A). AscE, on the other hand, is stable and resistant to partial protease digestion after forming a complex with AscG (Fig. 5A). Based on N-terminal sequencing (GSSHHH) of the intact region of AscG that remains bound to AscE after digestion, the AscE binding region of AscG is mapped to be at the N-terminal 61 residues of AscG (MW of 8.24 kDa with the His-tag) (Figs. 2 and 5A). This intact fragment can be observed consistently when different kinds of proteases were used for digestion (Supplemental Fig. S1), strongly suggesting that the C-terminal region of AscG is highly exposed in the AscEG complex. It should be noted that overexpression of AscG alone as a His-tagged protein resulted in insoluble inclusion bodies.
Figure 5.
SDS Tricine PAGE showing purification and partial protease digestion of the AscEG and AscEFG complexes. (A) Partial protease digestion of the AscEG complex. (Lane 1) Protein molecular weight marker; (lane 2) His-tagged AscG coexpressed and copurified with AscE to form a soluble AscEG complex. Note that the band for AscG represents AscG (12.93 kDa) with an intact His-tag (1.63 kDa); (lane 3) AscEG after partial protease digestion. The intact AscG fragment contains the N-terminal His-tag and the N-terminal 61 residues of AscG (theoretical MW of 8.24 kDa) with a molecular weight of 8.27 kDa (based on MALDI-TOF MS) as confirmed by N-terminal sequencing. (B) Partial protease digestion of the AscEFG complex. (Lane 1) Protein molecular weight marker; (lane 2) His-tagged AscG coexpressed and copurified with AscE and AscF to form a soluble ternary AscEFG complex; (lane 3) AscEFG after partial protease digestion. The intact AscF fragment has a molecular weight of 4.10 kDa (based on MALDI-TOF MS) and contains the C-terminal 35 residues of AscF (theoretical MW of 4.07 kDa) as confirmed by N-terminal sequencing.
Also noticeable is that the N-terminal 13 residues of AscE became buried and protected from protease digestion after formation of the AscEG complex (Fig. 5A). This indicates that these N-terminal residues were induced to be buried upon binding to AscG. The situation could be similar to PscE in the PscEFG complex in which the N-terminal residues of PscE formed an additional helix, Ha (Quinaud et al. 2007). In contrast, the first nine N-terminal residues of YscE are disordered in the YscEFG complex (Sun et al. 2008). These residues that formed the Ha of PscE, however, are shown not to be essential for formation of the PscE–PscF55–85–PscG complex (Quinaud et al. 2007). Interestingly, we also found that AscE lacking the first 13 residues (AscEΔ1–13) can still be coexpressed with AscG or AscG and AscF forming soluble complexes (Fig. 4B). This further suggested that the N-terminal residues of AscE are not essential for formation of the AscEG or AscEFG complex, although they were induced to be buried by interaction with AscG.
The C-terminal region of AscG is induced to be buried upon binding AscF
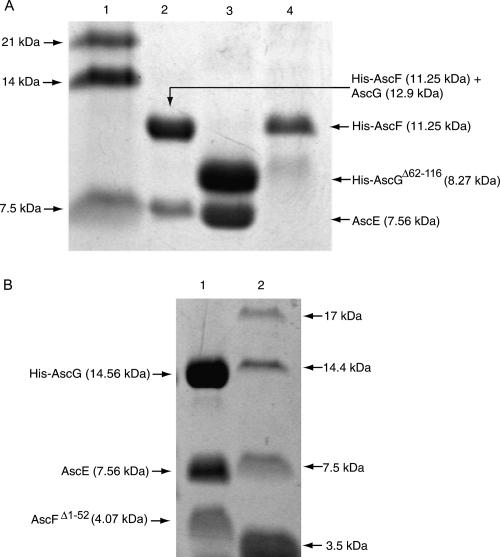
The C-terminal region of AscG consisted of 55 residues that were exposed in the AscEG complex, but they became buried and resistant to partial protease digestion only upon formation of the ternary complex with AscF (Fig. 5B). To confirm that the 55-residue C-terminal region of AscG is essential for interaction with AscF in the ternary complex, we have prepared a C-terminal truncation mutant of AscG by deleting residues Leu62 to Ser116 (AscGΔ62–116) and attempted to coexpress it with AscE and AscF. The molecular weight of His-AscGΔ62–116 (8.27 kDa), however, is very close to AscF (9.62 kDa), thus making it harder to detect the formation of the ternary complex. In addition, AscE will be copurified with His-AscG, full-length or C-terminal truncated, irrespective of the binding between AscG and AscF. So, we decided to place the His-tag at the N terminus of AscF and coexpress it with AscE and AscG. If there is a formation of the ternary complex upon coexpression, AscE will be copurified with His-AscF through its interaction with AscG. Figure 6A shows that the full-length AscG can interact with His-AscF and that AscE can be copurified because of such an interaction. The band of full-length AscG (12.9 kDa) overlapped with that of His-AscF (11.25 kDa), but mass spectrometry confirmed the presence of both species. In contrast, coexpression of AscGΔ62–116 with His-AscF and AscE did not lead to formation of a ternary complex, clearly showing that the 55-residue C-terminal region of AscG is essential for substrate binding (Fig. 6A). The AscGΔ62–116 C-terminal truncation mutant, however, can coexpress with AscE to form a stable and soluble complex that can be copurified (Fig. 6A).
Figure 6.
Coexpression of truncated fragments of AscG or AscF in the ternary complex. (A) Coexpression of AscGΔ62–116 with AscE or AscE and AscF. (Lane 1) Protein molecular weight marker; (lane 2) coexpression of full-length AscG with AscE and His-AscF. Note that the His-tag (1.63 kDa) is located at the N terminus of AscF (9.62 kDa) resulting in a fusion protein band (11.25 kDa) that overlapped with AscG (12.9 kDa). The formation of the ternary complex is indicated by copurification of AscE (7.56 kDa). (Lane 3) The His-AscGΔ62–116 C-terminal truncation mutant (8.27 kDa) can be coexpressed and copurified with AscE as a stable soluble complex; (lane 4) coexpression of the AscGΔ62–116 C-terminal truncation mutant with AscE and His-AscF. No ternary complex can be formed, as indicated by absence of the band corresponding to AscE. (B) Coexpression of AscFΔ1–52 with AscE and AscG. (Lane 1) The AscFΔ1–52 N-terminal truncation mutant (4.07 kDa) can be coexpressed and copurified with both AscE and His-AscG to form a soluble complex; (lane 2) protein molecular weight marker.
The chaperone-binding region of AscF
AscF alone is not stable, and attempts to overexpress His-tagged AscF protein resulted in an aggregate that is prone to degradation. Expression of GST-AscF yielded soluble fusion protein, but AscF precipitated upon the removal of the GST-tag by thrombin cleavage. This agrees with the observation that both homologs of AscF, MxiHCΔ5 and PrgICΔ5, are not very stable. The Tms of PrgICΔ5 and MxiHCΔ5 are 37°C and 42°C, respectively. The ΔG of both proteins is <2.0 kcal/mol, and their relative low m-values obtained for urea denaturation suggest that there is only a small change in solvent-accessible surface area (Darboe et al. 2006). It is suggested that assembly of the needle subunits into their native helically packed complexes (Blocker et al. 2003) may result in a much more stable structure than the monomeric subunit.
AscF can be coexpressed with both AscE and AscG and copurified as a soluble ternary complex. As in the case for the AscEG complex, a gel-filtration elution profile showed that AscEFG complex eluted at a volume corresponding to a larger molecular weight. This suggested that certain regions of the complex are exposed, resulting in an increased overall hydrodynamic radius of the protein. When this exposed region is removed by partial protease digestion, the digested AscEFG complex eluted at the expected volume (Fig. 3). Tricine-SDS gel showed that this exposed region is located on AscF in the AscEFG complex. Note that both AscE and AscG are stable in the ternary AscEFG complex and resistant to partial protease digestion (Fig. 5B). Using mass spectrometry and N-terminal sequencing (AELQHK), the region of AscF that remains intact and is bound to the chaperone is mapped to the 35-residue C-terminal region of the protein (MW of 4.10 kDa). At the same time, the N-terminal part of the AscF protein, from residues Met1 to Leu52, in the ternary complex remains exposed and can be digested away easily (Fig. 5B). The intact chaperone-binding region of AscF can be observed to remain consistent when different kinds of proteases were used for digestion (Supplemental Fig. S2). Formation of the ternary complex allows the more hydrophobic C-terminal residues (based on Kyte-Doolittle hydropathy plot) to be buried to prevent aggregation and premature association of AscF. The 35-residue C-terminal region of AscF can also be coexpressed with AscE and AscG to form a stable and soluble ternary complex (Fig. 6B). Our findings agree with the crystal structure of PscE–PscF55–85–PscG in which the N-terminal 54 residues were required to be removed by digestion with papain before the complex could be crystallized. The 30 C-terminal residues of PscG are involved in the interaction with residues 55–67 of PscF, which are in an extended conformation, as well as residues 68–85, which fold into an α-helix (Quinaud et al. 2007). In the YscEFG complex, the 49 N-terminal residues of YscF are disordered but not degraded, while the C-terminal residues 50–87 adopt an α-helical hairpin structure with the two helices connected by a five-residue loop (residues 64–68) (Sun et al. 2008).
It is worth mentioning that AscF cannot be coexpressed with just AscG to form a soluble complex. When the two proteins were coexpressed using the pETDuet-1 vector, AscG will be expressed as an inclusion body. We have also attempted to coexpress AscE and AscF; however, no complex between the two proteins can be formed. This suggested that both AscE and AscG are required to form a soluble ternary complex with AscF.
The C-terminal hydrophobic residues of AscF are essential for chaperone binding
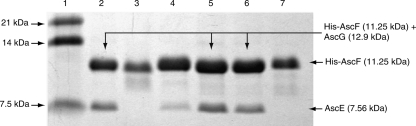
Previous studies have shown that short deletions of five residues in the C terminus of Shigella T3SS needle protein MxiH can render it unable to polymerize (Kenjale et al. 2005). Such C-terminal deletions can also yield soluble and monomeric forms of MxiH and PrgI, needle components of the T3SS from Shigella and Salmonella (Darboe et al. 2006). To determine whether the C-terminal residues are also essential for chaperone binding by the T3SS needle subunit protein, we have coexpressed AscF with the last seven C-terminal residues deleted (AscFΔ81–87) together with AscE and AscG. Results show that this short stretch of C-terminal residues is essential for chaperone binding by AscF (Fig. 7). This short stretch contains mostly of hydrophobic residues in the sequence “ILQKI” that is also conserved in PscF. To determine whether the interaction between AscF and the chaperone is conducted mainly through these hydrophobic residues, we have constructed single-site mutants (I83A, L84A, and I87A), double-site mutants (I83A_L84A, I83A_I87A, L84A_I87A), and a triple-site mutant (I83A_L84A_I87A) of AscF, and coexpressed them with both AscE and AscG. None of the single mutants affected formation of the ternary AscEFG complex (data not shown). However, mutation of both residues Ile83 and Leu84 can significantly reduce chaperone binding by AscF and inhibit formation of the ternary complex. Residue Ile87 is also involved in chaperone binding, but not to the same extent as either residue Ile83 or Leu84. Mutation of all the three hydrophobic residues leads to complete abrogation of chaperone binding (Fig. 7).
Figure 7.
The C-terminal hydrophobic residues (Ile83 and Leu84) of AscF are essential for chaperone binding. (Lane 1) Protein molecular weight marker; (lane 2) coexpression of full-length His-AscF with AscE and AscG. The formation of the ternary complex is indicated by copurification of AscE (7.56 kDa). (Lane 3) Coexpression of the His-AscFΔ81–87 C-terminal truncation mutant with AscE and AscG. No ternary complex can be formed, as indicated by the absence of the band corresponding to AscE. (Lanes 4–7) Coexpression of His-AscF I83A_L84A, His-AscF I83A_I87A, His-AscF L84A_I87A, and His-AscF I83A_L84A_I87A, respectively, with AscE and AscG. Mutation of both residues Ile83 and Leu84 are necessary to reduce the chaperone binding of AscF significantly. Residue Ile87 is also involved in the interaction but not to the same extent as residues Ile83 and Leu84.
Discussion
It is reported that in the T3SS of P. aeruginosa, PscE and PscG prevent PscF from polymerizing prematurely and keep it in a secretion-prone conformation. The T3SS needle subunit, PscF, requires both cytoplasmic partners, PscE and PscG, which trap PscF in a ternary 1:1:1 complex, thus blocking it in a monomeric state (Quinaud et al. 2005). This is also likely to be the case in the ternary AscEFG complex in A. hydrophila. Recently, the crystal structures of the PscE–PscF55–85–PscG complex (Quinaud et al. 2007) and the YscEFG complex have both been determined (Sun et al. 2008). However, what still remains unknown is the structure of the intermediate EG complex, which is unlikely to be the same as that in the EFG complex (C. Chatterjee, Y.W. Tan, and Y.K. Mok, unpubl.). Using a T3SS of A. hydrophila, we studied the locations of regions that are highly exposed in the homologous AscE and AscEG complex, and they were buried upon binding to their respective partners. As significant structural changes may occur upon interaction, the location of those regions can only be identified by examining the individual protein or complex before the interaction occurs.
Our crystal structure of AscE shows that it is a dimer comprised of a pair of antiparallel amphipathic α-helices in each monomer with a strong hydrophobic dimeric interface. To form a complex with AscG as a monomer, the hydrophobic dimeric interface from AscE will need to be bound and buried by AscG. The 13-residue N-terminal region of AscE is disordered and exposed but becomes stable and resistant to protease digestion after forming a complex with AscG or with both AscG and AscF (Fig. 5A,B). Our result shows that this region can be involved in interaction with AscG and changed from an exposed conformation to become buried upon formation of the AscEG complex. In contrast, the same region in YscE assumed a helical conformation and formed a single long helix with the N-terminal helix in the absence of YscG (Phan et al. 2005) but became disordered in the YscEFG complex (Sun et al. 2008). In the PscE–PscF55–85–PscG complex, this region of PscE formed an additional short helix of only six residues with residue Met2 interacting with PscF (Quinaud et al. 2007). The existence of such an exposed region could not be determined by just examining the structure of the final ternary complex. However, we found that this exposed region of AscE is not essential for its interaction with AscG or formation of the AscEFG complex. AscE lacking the first 13 residues (AscEΔ1–13) could be coexpressed with AscG to form a soluble complex of AscEΔ1–13–AscG (Fig. 4B). Our finding agrees with the observation that the N-terminal 11 residues of PscE can be truncated without affecting formation of the PscE12–67–PscF–PscG complex. It is also shown that a P. aeruginosa ΔpscE mutant carrying PscE12–67 complemented plasmid is fully cytotoxic (Quinaud et al. 2007).
Using partial protease digestion, mass spectrometry, and N-terminal sequencing, we have established that the N-terminal 61 residues of AscG is the “AscE binding region” that is necessary and sufficient to form a 1:1 complex with AscE (C. Chatterjee, Y.W. Tan, and Y.K. Mok, unpubl.). The N-terminal 61 residues of AscG are aligned to helices H1–H3 and up to the middle of helix H4 of PscG as in the crystal structure of the PscE–PscF55–85–PscG complex. Helices H1 and H2 are mainly involved in interaction with PscE, while H1 and H3 are involved in forming a concave hydrophobic surface with H5 and H7 for interaction with PscF. Helix H4 is located at the outer surface of the structure, sandwiched between helices H3 and H5, and is not involved in interaction with PscF or PscE. In the AscEG complex in the absence of AscF, it is not likely that helices H3 and H4 of AscG will assume a conformation similar to those in the PscE–PscF55–85–PscG complex.
We have also mapped the 55-residue C-terminal region of AscG to be the “substrate-binding region” and shown it to be essential for interaction with AscF. This region is exposed when AscG forms a complex with AscE and can be easily digested away by protease in the absence of AscF. Upon binding with AscF, this region becomes resistant to protease digestion and likely induced to be buried during the process. The existence of this induced folding process at the C-terminal region of AscG could not be detected by examining the crystal structure of the PscEFG or YscEFG complex (Quinaud et al. 2007; Sun et al. 2008). AscG is necessary for the interaction with AscF. However, there seems to be no strong interaction between AscF and AscE. When the C-terminal substrate-binding region of AscG is deleted, AscF cannot form a ternary complex with the AscEGΔ62–116 complex. This indicates that the interaction between AscE and AscF, if it does exist in the ternary complex, should be much weaker and nonessential compared to that between AscE and AscG. This is confirmed in the YscEFG complex, in which there is very little direct interaction between YscF and YscE in the complex (Sun et al. 2008). AscG is either unstable or improperly folded by itself, and it cannot be expressed as a soluble protein. The binding of AscE seems to stabilize AscG and enables it to interact with AscF using its C-terminal region. Thus, AscE can be a potential regulator of the chaperone activity of AscG and enable it to bind to AscF only when necessary; further experiments are required to establish this role.
For AscF, the chaperone-binding region has been mapped to the C-terminal 35 residues that remained intact after partial protease digestion of the ternary AscEFG complex. In the crystal structure of the PscE–PscF55–84–PscG complex, residues 55–67 assumed an extended conformation, while residues 68–85 formed a helix that used hydrophobic residues Val70, Leu74, Ile77, Met78, Ile81, and Leu82 for interaction with PscG (Quinaud et al. 2007). In contrast, the C-terminal residues 50–87 of YscF adopted an α-helical hairpin conformation in the YscEFG complex, interacting with the hydrophobic concave surface of YscG (residues Ile66, Ile71, Met75, Met78, Ile82, Leu83, and Phe86) (Sun et al. 2008). The C-terminal five residues of MxiH and PrgI are reported to be essential in oligomerization and are hydrophobic in nature. When these five residues are removed, they can be expressed as soluble proteins but will not be able to assemble (Darboe et al. 2006). As these C-terminal residues are highly hydrophobic, they are likely to be protected and bound by the chaperone to prevent premature oligomerization inside the cell. Indeed, using site-directed mutagenesis, we have confirmed that the hydrophobic residues Ile83 and Leu84 of the C-terminal sequence “ILQKI” of AscF, which aligned with Ile81 and Leu82 of PscF as well as Ile82 and Leu83 of YscF, are not just important for oligomerization but also essential for chaperone binding. Ile83 of AscF is also highly conserved among all the known T3SS needle complex subunits.
While the C-terminal chaperone-binding region of AscF is buried and resistant to protease digestion in the ternary AscEFG complex, the N-terminal 52 residues of AscF remain exposed and susceptible to protease digestion in the ternary complex. The N-terminal part of the needle complex subunit is more varied and may be species-specific. For instance, complementation studies of the PscF deletion mutant with YscF showed that the first 11 N-terminal residues of PscF are species-specific and essential for the assembly and function of the needle components (Pastor et al. 2005). Substitution of the N-terminal 12 amino acid residues of YscF impaired the secretion of YopB and YopD only and led to a calcium independency phenotype (Allaoui et al. 1995). The reason that this N-terminal region of AscF is kept in an exposed and disordered state could be to allow AscF to travel through the conduit of the growing T3SS needle more effectively for assembly. A similar example can be observed in the assembly of bacterial flagellum. The flagellar filament is a helical assembly of a single protein, flagellin, with roughly 11 subunits per 2 turns of the 1-start helix (Samatey et al. 2001; Yonekura et al. 2003). Flagellin may transit partially unfolded (Samatey et al. 2001) through the channel to its tip, where it refolds and inserts into the growing filament (Yonekura et al. 2000). Another probable explanation of an exposed N-terminal region of AscF is that it carries an export signal that can be recognized by the export apparatus. The C-terminal half of the bacteria flagellum anti-sigma factor FlgM is flexible and becomes structured when bound to its target σ28. However, the N-terminal part of FlgM remains unfolded, and this unfolded region contains an export signal that is likely recognized by proteins near the entrance of the export apparatus to facilitate export through the membrane (Daughdrill et al. 1997).
Although the N-terminal part of AscF is exposed when in complex with the AscEG chaperone, it is likely that it will assume a well-structured form after being released from the chaperone. The NMR structure of BsaLΔ5 shows that BsaL has a helix–turn–helix core domain with two well-defined α-helices (residues 30–46 and residues 51–68) joined by an ordered four-residue linker (Zhang et al. 2006). This interhelical turn, P-(S/D)-(D/N)-P motif, is also conserved in AscF and believed to constrain the orientation of the two helices (Fig. 2). The two-helix bundle is stabilized by interhelix hydrophobic contacts. The flanking regions to these two helices are not completely disordered, but adopt a partial helical conformation (Zhang et al. 2006). It is proposed that the N-terminal 10–25 residues and the C-terminal 5 residues are involved in association of the subunit (Zhang et al. 2006). The subunits are believed to be associated with a helical packing as MxiH, the monomer of the S. flexneri T3SS needle, assembles into a helical structure with parameters similar to flagella (Cordes et al. 2005). Indeed, docking of the structure of MxiH into the electron microscopy density of the Shigella T3SS needle showed that both the N and C terminus of MxiH are important for packing and stability of the needle (Deane et al. 2006).
In conclusion, we have determined the structure of AscE and mapped the AscE binding region of AscG and found that the C-terminal region of AscG is exposed and will only attain a buried conformation when in complex with AscF. The boundary of this exposed region suggested that AscG in the AscEG complex may not attain a similar conformation as PscG in the PscE–PscF55–85–PscG complex or YscG in the YscEFG complex. AscF uses the C-terminal hydrophobic residues for chaperone binding, but its N-terminal region remains exposed even in the ternary AscEFG complex. AscE may function to stabilize AscG by interacting with the chaperone using its dimeric interface. This may enable AscG to bind the C terminus of AscF and prevent its premature assembly. Understanding of the various interactions within the ternary complex may provide potential targets for development of diagnostics and therapeutics against T3SS-dependent bacterial infection.
Materials and Methods
Cloning of AscE, AscE L9M_L58M, AscEG, and AscEFG
The gene for AscE from Aeromonas hydrophila AH-1 was subcloned between the BamHI and EcoRI sites of a modified pET-32a vector (Novagen; with thioredoxin and S-tag removed). The AscE L9M_L58M double mutant was generated by using the PCR-based overlap extension method (Ho et al. 1989) with two separate rounds of PCR for each mutation. The L9M mutant was generated first, and then its plasmid DNA was used as a template to generate the second mutation, L58M. The DNA insert carrying the double mutation was subcloned into the same site of the same vector as the wild type AscE for protein expression. For coexpression of the AscE-AscG (AscEG) complex, full-length AscE was subcloned at the second multiple-cloning site (MCS2) of the coexpression vector, pETDuet-1 (Novagen), using restriction enzymes NdeI and XhoI. The pETDuet-1 vector encodes for ampicillin resistance. Full-length AscG was subsequently subcloned at the first multiple-cloning site (MCS1), which contains the 6× His-tag, of the same vector using BamHI and EcoRI and allowing the coexpression of both proteins from the same plasmid. The ternary complex of AscE–AscF–AscG (AscEFG) was constructed by subcloning AscF in replacement of AscE at the MCS2 of the same pETDuet-1 vector with AscG already subcloned at the MCS1. AscE was cloned at the MSC2 of pACYCDuet-1 vector (Novagen) using restriction enzymes NdeI and XhoI. The pACYCDuet-1 vector carries the genes for chloramphenicol resistance. The genes for the six-His-tag were encoded upstream of AscG. The pETDuet-1 vector containing AscG (MCS1) and AscF (MCS2) and the pACYCDuet-1 vector containing AscE (MCS2) were both transformed into the same BL-21 (DE3) cells for coexpression of three proteins with ampicillin and chloramphenicol as antibiotics for selection. For expression of AscEFG with His-tag on AscF rather than on AscG, the position of the two genes in pETDuet-1 is swapped.
Expression and purification of AscE, AscE L9M_L58M, AscEG, and AscEFG
A single ampicillin-resistant colony of Escherichia coli BL21 (DE3) cells transformed with the suitable plasmid was used to inoculate 50 mL of Luria Broth supplemented with 100 μg/mL ampicillin. For expression of Se-Met-labeled AscE L9M_L58M, the plasmid was transformed into B834 (DE3) pLysS E. coli cells, and the cells were cultured in Le Master medium supplemented with the appropriate amount of amino acids and 25 mg/mL seleno-L-methionine (Sigma-Aldrich). Thirty-four micrograms per milliliter chloramphenicol was added to the bacterial culture carrying the plasmid pACYCDuet-1. The culture was grown overnight at 37°C by shaking at 200 rpm. Ten milliliters of the overnight culture was used for inoculating 1 L of fresh Luria Broth or Le Master medium, and cells were grown to early log phase (OD600 = 0.6). Isopropyl-β-D-thiogalactoside was added to a final concentration of 0.3 mM to induce expression, and the cells were grown for an additional 6 h at 25°C. Cells were harvested by centrifugation at 6000 rpm for 15 min. The cell pellet obtained from 1 L of culture was resuspended in 30 mL of Ni-binding buffer (20 mM Tris-HCL, pH 7.5, 200 mM NaCl, 5% glycerol, 10 mM β-mercaptoethanol, and 5 mM imidazole) along with one tablet of Complete EDTA-free Cocktail Protease Inhibitor (Roche). Cells were lyzed using a sonicator, and cell debris was removed by centrifugation at 18,000 rpm for 30 min. The supernatant was purified using Ni-NTA beads (QIAGEN). Ni-binding buffer with 0.5 M imidazole was used to elute proteins from Ni-NTA beads. For the wild-type and mutant AscE, the His-tags were removed by thrombin digestion. Proteins were further purified on a pre-packed HiLoad 16/60 Superdex 75 prep grade (GE Healthcare) gel filtration column on the AKTA Fast Protein Liquid Chromatography system (GE Healthcare) using gel filtration buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5% glycerol, and 10 mM β-mercaptoethanol). Eluted proteins were stored at −80°C for subsequent analysis.
Limited protease digestion
All the proteases used for limited digestion were from Sigma. Small-scale digestions using different amounts of proteases for different periods of time were performed before scaling up of the reaction. For small-scale digestions, 1 mg/mL protein was digested with chymotrypsin, papain, thermolysin, or elastase in a protein-to-protease ratio of 100:1 or 200:1 for 5 h at room temperature. Ten microliters of the digestion mixture was taken at hourly intervals and analyzed on a 15% Tricine-SDS gel to determine the degree of digestion. Limited protease digestion was performed on the AscE, AscEG, and AscEFG complexes in the same buffer as used for gel filtration chromatography. Purified AscE was digested with papain at a molar ratio of 100:1 for 1 h at 25°C. For large-scale digestion, purified AscEG complex was concentrated to 9 mg/mL and subsequently incubated with chymotrypsin at a molar ratio of 200:1 for 5 min at 25°C before loading onto the HiLoad 16/60 Superdex 75 prep grade (GE Healthcare) gel filtration column to remove the protease and to obtain the digested complex. Purified AscEFG complex was concentrated to 10 mg/mL and incubated with papain at a molar ratio of 100:1 for 8 min at 25°C. The reaction was terminated by passing the digested proteins through a gel filtration column as described for AscEG.
Mass spectrometry and N-terminal sequencing
Protease-digested AscEG and AscEFG complexes that contain intact domains still bound to AscE or AscEG, respectively, were collected after gel filtration chromatography. These samples were blotted onto a MALDI target plate, with sinapinic acid as a matrix, and the molecular weight of each component in the complex was determined using Voyager-DE STR Mass Spectrometer (Applied Biosystems) with a voltage of 2100 V and laser intensity of 2500. The incorporation of Se-Met in AscE L9M_L58M was ascertained by comparing the molecular weight of the labeled protein with that of the unlabeled AscE L9M_L58M. As for the N-terminal sequence determination, protease-digested samples were separated on a 12% Tricine-SDS gel, and the protein fragments were blotted onto a PVDF membrane using a Mini Trans-blot Electrophoretic Transfer Cell (Bio-Rad). Trans-blotting was performed at 4°C using a voltage of 100 V for 1 h. Protein bands were visualized with Coomassie staining. Membrane-bound bands were then excised and analyzed on an Applied Biosystems 494A Procise protein sequencer.
Crystallization, data collection, structure solution, and refinement
Both untagged native as well as Se-Met labeled AscE were dialyzed in buffer consisting of 20 mM Tris-HCl pH 7.5, 200 mM NaCl, 3% glycerol, and 5 mM DTT, and concentrated to 10 mg/mL. Dynamic Light Scattering (DLS) and native gels were used to check the homogeneity of concentrated proteins. Initial crystallization condition was identified by using Hampton Research screens and was further optimized to a final condition of 10% PEG 4000, 1.4 M NaCl, and 13 mM Tris (2-carboxyethyl) phosphine hydrochloride. A brief phasing was attempted using AscE with the naturally occurring Met1 and Met2 substituted with Se-Met. However, it failed to provide adequate anomalous signals, especially for the region preceding Pro14. Based on the structural analysis of YscE (Phan et al. 2005), residues Leu9 and Leu58 were mutated to Met, and subsequently the crystals of Se-Met-labeled mutant provided adequate phase information to solve the structure of AscE.
Double mutant (L9M and L58M) Se-Met crystals of AscE were crystallized essentially in the same condition as the wild-type AscE using the hanging-drop vapor diffusion method at room temperature. Diffraction-quality crystals were formed in 3 d, with the smallest dimension measuring ∼0.3 mm. These crystals belonged to the space group P43212 with four molecules in the asymmetric unit. The cell parameters were a = b = 69.09, c = 105.04 Å. The Matthews coefficient (Vm) was 2.11 Å3/Da, and the solvent content, 42% (Matthews 1968). The crystals were cryo-protected by stepwise (5%–10%) increase in the glycerol concentration with the reservoir condition. The crystals were taken from the cryo-condition, and flash cooled in a N2 cold stream at 100 K. These double mutant Se-Met AscE crystals diffracted up to 2.7 Å in the Synchrotron beamlines X29, NSLS, Brookhaven National Laboratory, and a complete MAD data set was collected (Table 1) using Quantum 4-CCD detector (Area Detector Systems Corp.). Data were processed and scaled using the program HKL2000 (Otwinowski and Minor 1997).
Of the expected eight selenium sites in the asymmetric unit, four were located by the program BnP (Weeks et al. 2002). The N-terminal Se-Met of each molecule was disordered. The initial phases were further improved by RESOLVE (Terwilliger 1999), which improved the overall figure of merit (FOM) to 0.75. The RESOLVE built ∼65% of the molecule. The remaining parts of the model were built manually using the program O (Jones et al. 1991). Further cycles of model building alternating with refinement using the program CNS (Brünger et al. 1998) resulted in the final model, with an R-factor of 0.243 (R free = 0.287) to 2.7 Å resolution with reflections I > σI were used in the refinement. Throughout the refinement, non-crystallographic symmetry (NCS) restraints were used. The final model comprises 208 residues in the asymmetric unit (Pro14–Glu65 of each monomer), and owing to limited resolution, no water molecules were added. The N-terminal His-tag and the first 13 residues were not visible in the electron density map. PROCHECK (Laskowski et al. 1993) analysis shows >97.8% of them in the most favored region with no residues in the disallowed regions of the Ramachandran plot. A simulated annealing F o−F c omit map of the conserved region of AscE is shown in Figure 1A.
Protein Data Bank deposition
The coordinates of AscE have been deposited in the Protein Data Bank (http://www.pdb.org) (Berman et al. 2000) under accession code 2Q1K.
Electronic supplemental material
SDS gel electrophoresis showing limited protease digestions of AscEG and AscEFG complexes using different proteases at two different concentrations are available, and mass spectrometry spectra of limited protease-digested AscEG and AscEFG.
Acknowledgments
The authors thank X12C beamline and Anand Saxena of the National Synchrotron Light Source, Brookhaven National Laboratory, for data collection. We also extend our gratitude to Michelle Mok for useful discussions and Shashikant Joshi for proofreading the manuscript. This work was supported by the Academic Research Fund (ARF), National University of Singapore (NUS) to Y.K.M. (R-154-000-246-112) and to J.S. (R-154-000-254-112), and by the BMRC grant from the Agency for Science Technology and Research (A*STAR), Singapore to K.Y.L. (04/1/21/19/346). Y.K.M., J.S., and K.Y.L. are members of the SBPR research group of the OLS of NUS.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Yu-Keung Mok, Department of Biological Sciences, Faculty of Science, National University of Singapore, Science Drive 4, Singapore 117543; e-mail: dbsmokh@nus.edu.sg; fax: (65) 6779-2486; or J. Sivaraman, Department of Biological Sciences, Faculty of Science, National University of Singapore, Science Drive 4, Singapore 117543; e-mail: dbsjayar@nus.edu.sg; fax: (65) 6779-2486.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036798.108.
References
- Allaoui, A., Schulte, R., Cornelis, G.R. Mutational analysis of the Yersinia enterocolitica virC operon: Characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol. Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- Austin, B., Adams, C. Fish pathogen. In: Austin B., et al., editors. The genus Aeromonas. John Wiley and Sons; New York: 1996. pp. 197–229. [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker, A., Jouihri, N., Larquet, E., Gounon, P., Ebel, F., Parsot, C., Sansonetti, P., Allaoui, A. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- Blocker, A., Komoriya, K., Aizawa, S.-I. Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proc. Natl. Acad. Sci. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, M., Stuber, K., Schlatter, Y., Wahli, T., Kuhnert, P., Frey, J. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida . J. Bacteriol. 2002;184:1851–1858. doi: 10.1128/JB.184.7.1851-1858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cordes, F.S., Daniell, S., Kenjale, R., Saurya, S., Picking, W.L., Picking, W.D., Booy, F., Lea, S.M., Blocker, A. Helical packing of needles from functionally altered Shigella type III secretion systems. J. Mol. Biol. 2005;354:206–211. doi: 10.1016/j.jmb.2005.09.062. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R., Van Gijsegem, F. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- Darboe, N., Kenjale, R., Picking, W.L., Picking, W.D., Middaugh, C.R. Physical characterization of MxiH and PrgI, the needle component of the type III secretion apparatus from Shigella and Salmonella . Protein Sci. 2006;15:543–552. doi: 10.1110/ps.051733506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughdrill, G.W., Chadsey, M.S., Karlinsey, J.E., Hughes, K.T., Dahlquist, F.W. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, σ28 . Nat. Struct. Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Day, J.B., Guller, I., Plano, G.V. Yersinia pestis YscG protein is a Syc-like chaperone that directly binds YscE. Infect. Immun. 2000;68:6466–6471. doi: 10.1128/iai.68.11.6466-6471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane, J.E., Roversi, P., Cordes, F.S., Johnson, S., Kenjale, R., Daniell, S., Booy, F., Picking, W.D., Picking, W.L., Blocker, A.J., et al. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc. Natl. Acad. Sci. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, D., Casanova, C., Liverman, A., Blazkova, H., Orth, K., Dobbelaere, D., Frey, J., Burr, S.E. AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-κB signalling pathway. Microbiology. 2006;152:2809–2818. doi: 10.1099/mic.0.28889-0. [DOI] [PubMed] [Google Scholar]
- Finlay, B.B., Cossart, P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- Ghosh, P. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, J.K., Pullen, J.K., Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hoiczyk, E., Blobel, G. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. 2001;98:4669–4674. doi: 10.1073/pnas.071065798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J.M. Aeromonas and Plesiomonas . In: Sussman M., editor. Molecular medical microbiology. Academic Press; San Diego, CA: 2001. pp. 1237–1270. [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kenjale, R., Wilson, J., Zenk, S.F., Saurya, S., Picking, W.L., Picking, W.D., Blocker, A. The needle component of the type III secretion of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- Kimbrough, T.G., Miller, S.I. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., Honig, B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter J., et al., editors. Methods in enzymology. Academic Press; San Diego, CA: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pastor, A., Chabert, J., Louwagie, M., Garin, J., Attree, I. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 2005;253:95–101. doi: 10.1016/j.femsle.2005.09.028. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phan, J., Austin, B.P., Waugh, D.S. Crystal structure of the Yersinia type III secretion protein YscE. Protein Sci. 2005;14:2759–2763. doi: 10.1110/ps.051706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinaud, M., Chabert, J., Faudry, E., Neumann, E., Lemaire, D., Pastor, A., Elsen, S., Dessen, A., Attree, I. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa . J. Biol. Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- Quinaud, M., Plé, S., Job, V., Contreras-Martel, C., Simorre, J.-P., Attree, I., Dessen, A. Structure of the heterotrimetric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatey, F.A., Imada, K., Nagashima, S., Vonderviszt, F., Kumasaka, T., Yamamoto, M., Namba, K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- Sha, J., Wang, S.F., Suarez, G., Sierra, J.C., Fadl, A.A., Erova, T.E., Foltz, S.M., Khajanchi, B.K., Silver, A.C., Graf, J., et al. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—Part I. Microb. Pathog. 2007;43:127–146. doi: 10.1016/j.micpath.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Sierra, J.C., Suarez, G., Sha, J., Foltz, S.M., Popov, V.L., Galindo, C.L., Garner, H.R., Chopra, A.K. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila—Part II. Microb. Pathog. 2007;43:147–160. doi: 10.1016/j.micpath.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Sun, P., Tropea, J.E., Austin, B.P., Cherry, S., Waugh, D.S. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 2008;377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. Reciprocal-space solvent flattening. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thune, R.L., Stanley, L.A., Cooper, K. Pathogenesis of Gram-negative bacterial infections in warmwater fish. Annu. Rev. Fish Dis. 1993;3:37–68. [Google Scholar]
- Vilches, S., Wilhelms, M., Yu, H.B., Leung, K.Y., Tomás, J.M., Merino, S. Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microb. Pathog. 2007;44:1–12. doi: 10.1016/j.micpath.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Ouellette, A.N., Egan, C.W., Rathinavelan, T., Im, W., De Guzman, R.N. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, C.M., Blessing, R.H., Miller, R., Mungee, R., Potter, S.A., Rappleye, J., Smith, G.D., Xu, H., Furey, W. Towards automated protein structure determination: BnP, the SnB-PHASES interface. Z. Kristallogr. 2002;217:686–693. [Google Scholar]
- Yonekura, K., Maki, S., Morgan, D.G., DeRosier, D.J., Vonderviszt, F., Imada, K., Namba, K. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- Yonekura, K., Maki-Yonekura, S., Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Yu, H.B., Srinivasa Rao, P.S., Lee, H.C., Vilches, S., Merino, S., Tomas, J.M., Leung, K.Y. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 2004;72:1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.B., Kaur, R., Lim, S., Wang, X.H., Leung, K.Y. Characterization of extracellular proteins produced by Aeromonas hydrophila AH-1. Proteomics. 2007;7:436–449. doi: 10.1002/pmic.200600396. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Wang, Y., Picking, W.L., Picking, W.D., DeGuzman, R.N. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei . J. Mol. Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]