Abstract
The genetic code is established by the aminoacylation reactions of aminoacyl tRNA synthetases, where amino acids are matched with triplet anticodons imbedded in the cognate tRNAs. The code established in this way is so robust that it gave birth to the entire tree of life. The tRNA synthetases are organized into two classes, based on their active site architectures. The details of this organization, and other considerations, suggest how the synthetases evolved by gene duplications, and how early proteins may have been statistical in nature, that is, products of a primitive code where one of several similar amino acids was used at a specific position in a polypeptide. The emergence of polypeptides with unique, defined sequences—true chemical entities—required extraordinary specificity of the aminoacylation reaction. This high specificity was achieved by editing activities that clear errors of aminoacylation and thereby prevent mistranslation. Defects in editing activities can be lethal and lead to pathologies in mammalian cells in culture. Even a mild defect in editing is casually associated with neurological disease in the mouse. Defects in editing are also mutagenic in an aging organism and suggest how mistranslation can lead to mutations that are fixed in the genome. Thus, clearance of mischarged tRNAs by the editing activities of tRNA synthetases was essential for development of the tree of life and has a role in the etiology of diseases that is just now being understood.
Keywords: tRNA synthetases, editing, mistranslation, aminoacylation, misaminoacylation, error correction
The genetic code is a universal algorithm for relating nucleotide triplets in genes and mRNAs to the 20 canonical amino acids. Although different algorithms, or codes, were doubtless tested during a long period of chemical evolution, the modern code proved so robust that, once established, it gave rise to the entire tree of life with the three great kingdoms—archae, bacteria, and eukarya. All competing coding schemes were driven out, because they could not compete with the power, elegance, and simplicity of what we now know as the universal genetic code. This code was present at the root, or base, of the tree, before the last common ancestor gave rise, over billions of years, to the wonders of a planet teaming with life (Fig. 1).
Figure 1.
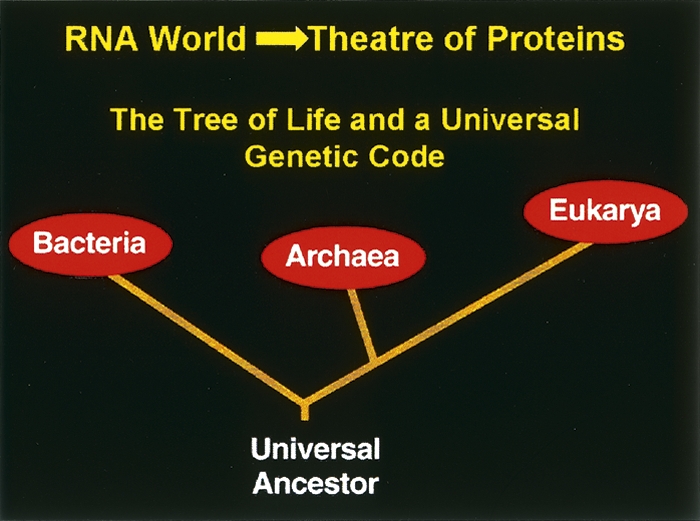
The tree of life. The tree was made possible by a robust genetic code that was needed for the transition from the RNA world to the theatre of proteins.
Aminoacylation reactions of tRNA synthetases establish the genetic code, by matching amino acids with tRNAs that have the triplet anticodons of the code. For each amino acid, there is one synthetase that, in most instances, catalyzes a two step reaction where the amino acid AA is condensed with ATP to form a tightly bound aminoacyl adenylate E(AA-AMP), with the simultaneous release of pyrophosphate PPi. The activated amino acid is then transferred from AMP to the 3′-end of its cognate tRNA to give AA-tRNA and AMP.
These reactions (Equations 1 and 2) establish the rules of the genetic code, and for that reason, tRNA synthetases are at the center of investigations into the origin and development of the code and living systems.
Some of the tRNA synthetases have an editing activity that is encoded by a separate active site. This site clears amino acids when they are attached to the wrong tRNA. When that editing activity is disturbed, mistranslation occurs because the mischarged amino acid is not cleared. Mistranslation coming from disruption of the editing activity not only is toxic to bacteria but leads to serious pathologies in mammalian cells. The degree of mistranslation that causes a pathological effect can be small. For example, a mutation that produces only a twofold decrease in the activity for editing leads to a heritable ataxia and neurodegeneration in the mouse. Significantly, and in a different vein, mistranslation is mutagenic in aging bacteria. This mutagenicity comes from the gradual accumulation of errors (from mistranslation) in the components of the replication apparatus. Summarized here is the historical context that led to such studies and a recapitulation of the most recent results connecting mistranslation to disease and mutagenesis. This summary is taken from, and updated from, a portion of the Stein and Moore Award Lecture delivered in July 2007.
Design and development of tRNA synthetases
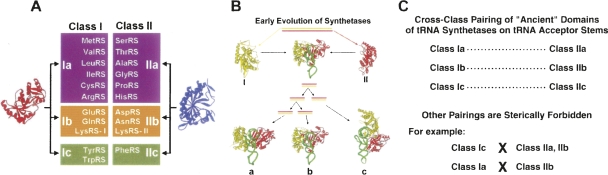
As ancient proteins, synthetases are believed to have been present in the last common ancestor of the tree of life. The activity for aminoacylation is thought to have emerged in the form of a protein early in the transition from the putative RNA world to the theatre of proteins. (Prior to proteins as catalysts of aminoacylation, the reactions are thought to have been catalyzed by ribozymes. Given that the aminoacyl linkage is higher in energy than the amide bond, side-by-side docking of two aminoacyl RNAs can give rise spontaneously to a peptide bond.) During this evolutionary process, the synthetases were split into two distinct groups—named class I and class II—of 10 enzymes each. This classification is based on the architectures of the two distinct active sites (Fig. 2A) (Ludmerer and Schimmel 1987; Cusack et al. 1990; Eriani et al. 1990). Thus, the catalytic domains of the class I enzymes are made up of a Rossmann nucleotide binding fold that promotes the activation reaction of Equation 1 and, in addition, provides determinants needed for binding the 3′-end of the tRNA in a position to receive the activated aminoacyl group. In contradistinction, the class II enzymes have a core made up of a seven-stranded β-structure with flanking α-helices. Although LysRS is occasionally found in class I (it is typically a class II enzyme), the class to which an enzyme is assigned is fixed through evolution. In spite of many efforts, no structural relationship, or evolutionary connection, between the classes has been detected. For these reasons, the two classes are believed to have been present at the time of the last common ancestor.
Figure 2.
Classes of aminoacyl tRNA synthetases and their implications for development of the genetic code. (A) The two major classes can be organized into subclasses that hold enzymes that are most closely related to each other in their sequences. Significantly, the subclasses also group tRNA synthetases according to their amino acid chemical types. Data adapted from Ribas de Pouplana and Schimmel (2004a). (B) Evolution of the two classes of synthetases from an ancestral gene where complementary strands of the gene code for, respectively, class I (red) and II (green) proteins. Subsequent gene duplications gave rise to the subclasses (depicted in A). Data adapted from Ribas de Pouplana and Schimmel (2004b). (C) The ancient catalytic domains of the synthetases can dock to opposite sides of the tRNA acceptor stems (as shown in B), in a way that is highly specific and that recapitulates the organization of subclasses in A.
The distinct structures and sequences of the members of the two classes have left open the question of the origin of the two classes. In spite of this uncertainty, what is clear is that, starting with an ancestral gene, each class expanded by gene duplications to give 10 enzymes. The most provocative hypothesis, which builds off the idea of gene duplications, comes from Rodin and Ohno (1995). They showed that, in the conserved motifs of the active site regions, the coding sequences of class I enzymes have statistically significant complementarity to those for class II (Fig. 2B). Later, Carter and Duax (2002) reported that complementary genes in the same DNA region in Achlya klebsiana code for proteins (not synthetases) that have the same folds as class I and II synthetases. This analysis has been extended to demonstrate that a specific region of the coding sequence of a “minimalist” class I TrpRS is complementary to its “counterpart” in the coding sequence of class II HisRS (Pham et al. 2007). These results are consistent with the idea that early genomes (such as RNA genomes) used both strands of the duplex to code for proteins, resulting in the synthesis of “complementary” proteins, like the class I and class II synthetases. Over the eons, these genes retained complementarity, as they expanded through gene duplications. Separately, a scheme for early tRNAs being encoded by opposite strands of an RNA genome is also plausible, based on powerful statistical analysis of tRNA sequences (Rodin et al. 1993, 1996).
Subclasses and connection to pairing of synthetases on tRNA acceptor stems
The synthetases can be arranged into three subclasses within each class. The subclasses represent enzymes that are more closely related to each other than to other enzymes in the same class. This natural sorting of the enzymes means that each subclass had an ancestor that, in turn, came from the “master gene” for the class.
The most striking feature of the subclasses is that members of each subclass “pair” across from each other in a specific way (Schimmel and Ribas de Pouplana 2001). Remarkably, subclass c has synthetases for aromatic amino acids, where subclass Ic has TyrRS and TrpRS (which are closely related to each other) is across from subclass IIc that has PheRS. Subclasses Ib, IIb capture the carboxyl side-chain amino acids and the amidated (NH2) derivatives. (These subclasses also have LysRS that, while mostly in class II, is in rare instances found in class I.) Finally, subclass I, IIa has many of the hydrophobic amino acids (Fig. 2A).
All of these observations about the classes and subclasses of tRNA synthetases can be pulled together with a single hypothesis. The hypothesis is that the synthetases were created in pairs—one from each subclass—and that two members of the same pair bound to opposite sides of the acceptor helix domains of the early tRNAs. A test of this hypothesis is to see if the historical catalytic domain of, say, a class Ia enzyme can be fit onto the minor groove side of the acceptor helix, without having clash with a class IIa enzyme bound on the major groove side. This hypothesis was tested by taking advantage of synthetase–tRNA crystal structures available for complexes involving members of all six subclasses. When this analysis is carried out, the cross-class simultaneous binding of two synthetases to one tRNA acceptor helix domain is highly specific—that is, a class Ia enzyme can pair with a IIa member, Ib with IIb, and Ic with IIc (Fig. 2C). Other combinations are forbidden by steric clashes, such as Ic with IIa or IIb, and Ia with IIb. Thus, the “ancient” or historical domains of the synthetases can be paired in a precise way on tRNA acceptor helix domains, possibly to be chaperone-like and cover and protect the RNA in an early environment where temperatures were extreme and RNA was susceptible to degradation.
Thus, the specific cross-class pairing of synthetases on tRNA acceptor stems goes hand-in-hand with the Rodin and Ohno idea that each class (I or II) of synthetases had an ancestor that expanded through gene duplications to give 10 enzymes.
Possible relationship of synthetase subclasses to early genetic code
That synthetases can be clustered into subclasses based not only on sequence relatedness but also on groupings according to specific chemical types of amino acids, suggests an early, primitive genetic code that used the same groupings. For example, the six subclasses of synthetases could correspond to six early groupings of interchangeable amino acids from which the highly differentiated modern code emerged. Thus, the early code could produce proteins as statistical polypeptides, where any of several similar amino acids could be used at a specific codon. Statistical polypeptides provide the opportunity to explore all sorts of specific protein sequences in a natural environment. They also enable cells to adapt to complex, sometimes challenging habitats. (Thus, a protein microspecies might be able to exploit a rare chemical as a building block or metabolite, which, in turn, could assure the survival of the organism. Specific experiments have shown that, under certain conditions, the capacity to make statistical proteins can confer a growth advantage [Bacher et al. 2007].) In that connection, the conversion from a primitive to a precise genetic code would create selective pressure on synthetases to develop more specificity of amino acid recognition.
Role of editing reactions for appearance of a precise genetic code
Editing reactions were needed to convert from a statistical to a precise code. The clearance (or removal) of mischarged tRNAIle by isoleucyl–tRNA synthetase was the first reaction to be discovered (Eldred and Schimmel 1972). Mischarging occurs because valine is mistakenly activated (Equation 1) to form Val-AMP bound to isoleucyl-tRNA synthetase. The misactivated valyl moiety is then transferred to tRNAIle. After that, IleRS catalyzes the deacylation of Val-tRNAIle.
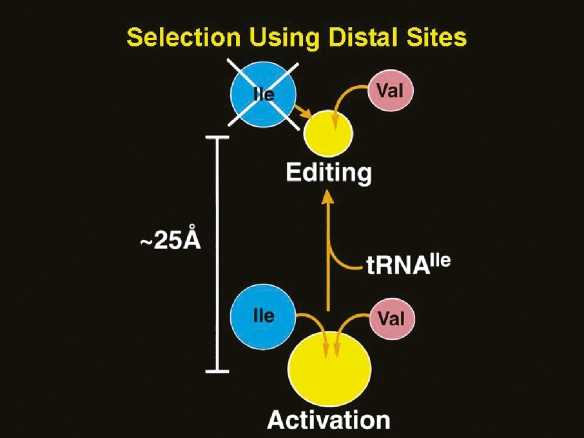
IleRS misactivates valine (which has an isopropyl side-chain that lacks one methylene group compared to the isobutyl side-chain of isoleucine) with a frequency of ∼1:200. (Other work, done at the time of the discovery of IleRS-catalyzed deacylation of Val-tRNAIle, suggested the editing reaction was more general [Schreier and Schimmel 1972; Yarus 1972].) The existence of a distinct site for editing was established by genetics (Schmidt and Schimmel 1994). Thus, mutational isolation of a distinct site for editing was accomplished by experiments showing that mutations that attenuated editing had no effect on aminoacylation and that, conversely, mutations that weakened aminoacylation did not impair deacylation of Val-tRNAIle by IleRS. (In addition, without the advantages of genetic technology, biochemical experiments inferred two distinct sites for editing [Schmidt and Schimmel 1995].) Later, the site for editing was cloned and expressed as a separate domain (Lin et al. 1996). X-ray crystallography subsequently showed that the site for editing was ∼30 Å from the site for amino acid activation (Fig. 3) (Nureki et al. 1998).
Figure 3.
Illustration of distinct active sites for amino acid activation and editing. The two sites, in the example of IleRS, are separated by ∼25 Å. A misactivated amino acid, such as valine, is translocated from the site for activation to the site for editing, where it is cleared. This translocation requires tRNA. The two distinct sites were first identified by chemical and genetic methods. Adapted from Schimmel (2008) (reprinted with permission from the American Society for Biochemistry and Molecular Biology ©2008).
Editing results from action of an RNP complex, where tRNAIle triggers the translocation of misactivated valine as Val-AMP (pretransfer editing) or Val-tRNAIle (post-transfer editing) from the active site for aminoacylation to the site for editing. (“Pre” and “post” refer to “before” and “after” transfer of the valyl moiety from the adenylate to the tRNA.) Discrete nucleotide determinants within tRNAIle were shown to be needed for the translocation reaction. That translocation was the rate determining step in editing was shown in further studies, using kinetic analysis and fluorescence energy transfer methods (Nomanbhoy et al. 1999; Hendrickson et al. 2002; Hendrickson and Schimmel 2003).
Rationale for how mistranslation from editing defect can connect to disease
The aforementioned studies raised the possibility for diseases related to mistranslation from defective editing by a tRNA synthetase. Because mutations in the site for editing do not disrupt the essential aminoacylation function, mild defects in editing can in principle be vertically transmitted in the population. Diseases arising from misfolded protein aggregates and autoimmunity seemed like logical candidates. The pioneering work of Weigle (1962, 1965) was of particular interest. In that early work, anthranilic acid was coupled to thyroglobulin and injected into rabbits. The injected rabbits raised antibodies against the anthranilic acid hapten conjugated to the protein. This result was expected but not anticipated was the finding that, over time, the rabbits developed thyroiditis. This pathology developed because the immune system gradually elicited antibodies directed against the native protein (thyroglobin) itself. In the same or similar vein, small amounts of mistranslation might yield immunogenic protein variants. As the immune system reacts to these variants by producing antibodies, the system can be imagined, over time, to develop antibodies against the native protein itself, giving rise to an autoimmune condition.
Demonstration of connection to disease: Trans-dominant effects of an editing defect in mammalian cells in culture
Investigations were done in a mammalian cell culture system with editing-defective ValRS, to see whether broad pathological effects could be seen from a severe editing defect (Nangle et al. 2006). At the active site for aminoacylation, ValRS misactivates the sterically similar (to valine) threonine and catalyzes production of Thr-tRNAVal. Editing occurs at a distinct second active site located in an insertion known as connective polypeptide 1 (CP1). This insertion is conserved throughout the three kingdoms of the tree of life. Specific mutations in CP1 of Escherichia coli ValRS disrupt the efficient hydrolysis of mischarged tRNAs, such as Thr-tRNAVal. Because the misacylated tRNA is not cleared, it is picked up and used directly for decoding mRNA. Significantly, editing-defective mutants are fully competent for aminoacylation (Fig. 4A). However, when the gene for ValRS is deleted from the chromosome, cell growth can be sustained, with a gene encoding an editing-defective, aminoacylation-proficient ValRS. With a ΔvalS null background in E. coli, when cells were grown in the presence of elevated amounts of a noncognate amino acid that is misactivated by ValRS (i.e., Thr and L-α-aminobutyric acid [α-Abu]), editing-defective mutants of ValRS attenuated cell viability. A direct correlation was seen between the magnitude of the editing defect measured in vitro and the degree of sensitivity to noncognate amino acid addition that was observed in vivo (Doring et al. 2001; Nangle et al. 2002).
Figure 4.
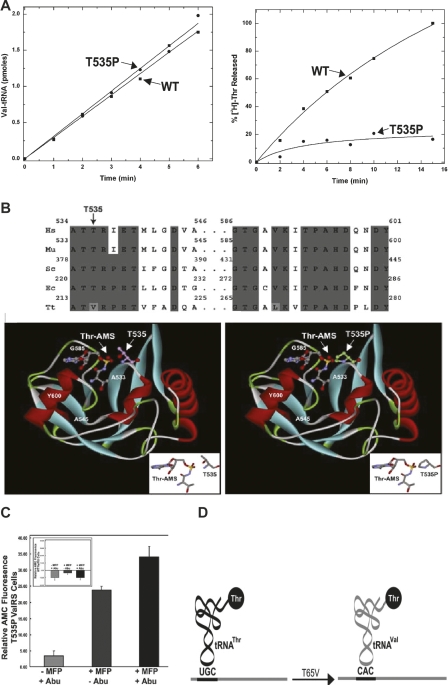
Analysis of editing-defective murine T535P ValRS. (A) The mutation does not affect aminoacylation (left) but severely attenuates the clearance of Thr-tRNAVal (right). (B) Highly conserved sequences of the region around T535P of murine ValRS and the placement of the mutation in the structure of the cocrystal of ValRS with a threonyl-AMP analog. (C) Visualization of the activation of caspase-3, using a fluorescence assay, in mouse 3T3 fibroblasts harboring the editing-defective T535P ValRS. (D) A simple biosensor to detect by fluorescence the substitution of Thr for Val at codon 65 in the enhanced form of green fluorescent protein (EGFP). Fluorescence is greatly enhanced by the insertion of Thr (from Thr-tRNAVal) instead of Val. Adapted from Nangle et al. (2006) (reprinted with permission from Elsevier ©2006).
Of the mutant enzymes, T222P E. coli ValRS was the most editing-defective (Doring et al. 2001; Nangle et al. 2002). T222 of E. coli ValRS is mostly conserved through evolution—of the known sequences, Thermus thermophilus and some species of Deinococcus, among a few other organisms, have replaced this conserved T with the isosteric V (Fig. 4B). Interestingly, the T. thermophilus ValRS editing domain complexed with a Thr-AMP analog (N-[L-threonyl]sulfamoyl)adenosine was crystallized and the structure was solved (Fukunaga and Yokoyama 2005). With the structure, the severe phenotype of the T222P substitution in E. coli ValRS could be rationalized. When a P replaces T222, assuming no structural rearrangement, the misactivated substrate is partly occluded. (Of course, P would undoubtedly produce a significant distortion of the binding pocket.) The corresponding substitution in murine ValRS is T535P Mus musculus ValRS. This analysis gave a clear structural rationale for the deficiency of the previously studied T222P substitution of E. coli ValRS. It also suggested that a similar substitution in M. musculus ValRS would be deleterious to editing. As expected, both wild-type and T535 mutant mouse ValRS were functional for aminoacylation. In contrast, T535P ValRS was editing-defective.
For an in vivo study, an inducible (mifepristone [MFP] transgene of both wild-type and T535P ValRS was constructed. Prolonged induction of the transgenes incorporated into stably transfected cells showed a dramatic, dominant-negative, change in cell morphology associated with the cells harboring editing-defective (but not wild-type) enzyme. For example, many cells lost their attachment to the culture flask and, alternatively, floated in the media. In addition to not forming a confluent monolayer, many cells (seen microscopically) had altered morphologies, including cell contraction and membrane blebbing. Cell shape was little affected, however, in cells induced to express the transgene encoding wild-type mouse ValRS. When the cells were grown in the presence of α-Abu (Nangle et al. 2006), a natural noncoding amino acid that is activated by ValRS, the aforementioned effects were more exacerbated.
Mammalian cell pathology linked to polyubiquitinylation and apoptosis
When the endoplasmic reticulum accumulates unfolded or misfolded proteins, cell death can occur through a pathway for apoptosis. One of the hallmarks of apoptosis is an increase in polyubiquitinylation of unfolded protein substrates. These polyubiquitinated substrates are then targeted for degradation by the proteasome. Indeed, in comparison to uninduced cells and cells expressing wild-type ValRS, a marked increase in polyubiquitinylation was seen in cells expressing T535P ValRS.
The activation of caspase-3 (a marker for cells under going apoptosis) can be studied as a way to monitor apoptosis. Caspase-3 is produced as an inactive zymogen. During apoptosis, it is converted by proteolytic activation to a functional cysteine protease (Nicholson et al. 1995). Cleavage by activated caspase-3 can be followed with a fluorescence assay, where a linked fluorophore was liberated upon cleavage of a caspase-3-specific peptide (Fig. 4C). These results linked genetic code ambiguity, from an editing-defect, to an apoptotic response involving caspase-3 activation.
Misincorporation in vivo detected with a biosensor
The enhanced form of green fluorescent protein (EGFP) from the Pacific Northwest jelly fish Aequorea Victoria has a Thr at position 65 of its T-Y-G consensus sequence. This tripeptide sequence is needed for the enhanced fluorescence (Prasher et al. 1992; Cody et al. 1993; Cormack et al. 1996). When substitutions are introduced at this consensus sequence, the fluorescence from GFP is sharply reduced. Thus, with T65V EGFP, the degree of misincorporation at a particular codon in EGFP can be followed (Fig. 4D).
Using fluorescence spectrophotometric analysis of cell lysates, together with fluorescence microscopy, a gain in EGFP fluorescence in cells expressing T535P but not wild-type ValRS could be demonstrated. This analysis showed an ∼16.7% gain in fluorescence in cells coexpressing T535P ValRS and T65V EGFP. No gain of significance was seen in cells expressing wild-type ValRS. The degree of misincorporation seen in these experiments was estimated to be on the order of 10%. This degree of misincorporation is similar to that observed with bacteria that harbor an editing-defective tRNA synthetase. Compared to bacteria, mammalian cells are far more sensitive to mistranslation. Toxic effects in bacteria are only observed when the chromosome-encoded wild-type allele is knocked out. In that situation, only the editing-defective enzyme is expressed and cell growth is studied in the presence of a noncognate amino acid that can no longer be cleared by editing (Doring et al. 2001; Nangle et al. 2002). In the mammalian system, deleterious effects were seen even without exogenous addition of noncognate amino acid to the growth medium, so that the editing defect was trans-dominant over the wild-type alleles encoded by the chromosome (Nangle et al. 2006).
Given the potency of the editing defect in mammalian cells in culture, the possibilities of diseases in whole organisms arising from even mild defects in an editing activity seemed realistic and gave motivation to investigate an editing defect in the mouse.
Demonstration of connection to disease: Neurodegeneration caused by an editing-defective alanyl-tRNA synthetase
The possibility of linking an editing defect to disease was realized in the mouse. Severe neurodegeneration and ataxia was seen in mice with a heritable mutation in the editing domain of murine AlaRS. The collaborative work with Ackerman group at Jackson Laboratory showed that the mutation weakened the ability of the enzyme to distinguish Ser or Gly from Ala. As a consequence, mischarged Ser-tRNAAla and Gly-tRNAAla were not cleared efficiently (Lee et al. 2006). The consequence of the mutation is mistranslation followed by up-regulation of cytoplasmic protein chaperones and induction of the unfolded protein response. The observed ataxia correlated with intracellular accumulation of misfolded proteins in neurons. Purkinje cells of the cerebellum were especially sensitive. (The cerebellum undergoes extensive deterioration as the editing-deficient mouse ages.) Importantly, the aminoacylation activity is unaffected, and the editing defect itself is small (for example, only a twofold change in activity for clearance of Ser-tRNAAla). Presumably, more pronounced defects in the editing activity of AlaRS, such as the severely defective editing mutant of ValRS studied in cells in culture (see above), would be lethal.
Mistranslation is normally prevented by reiterative use of the same G:U pair in tRNAAla
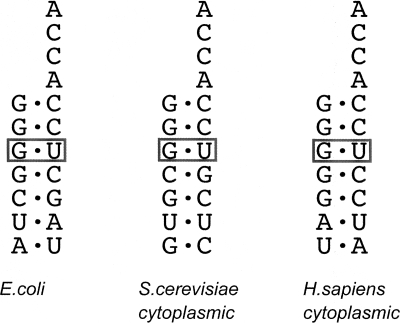
For AlaRS, a single specific G:U base pair (G3:U70) marks a tRNA for aminoacylation (Hou and Schimmel 1988, 1989; McClain and Foss 1988). Thus, mistranslation resulting from glycine or serine being joined to the G3:U70-containing tRNAs is prevented by the editing activity that clears the mischarged amino acid. The assumption has been that the specificity for recognition of tRNAAla for editing was provided by the same structural determinants as used for aminoacylation. In contrast to this assumption, the distinct editing and aminoacylation domains each, and separately, recognize the same G:U base pair, to distinguish tRNAAla from all other tRNAs (Fig. 5;) (Beebe et al. 2008). Thus, an artificial recombinant fragment, encoding just the editing domain, targets mischarged tRNAAla, using a structural motif unrelated to that for aminoacylation. Recognition by the recombinant fragment is sensitive to G:U. The structural motifs for certain editing domains, including that of AlaRS, are also found naturally in genome-encoded protein fragments that are widely distributed in evolution (Ahel et al. 2003; An and Musier-Forsyth 2004, 2005). An example is AlaXp, a genome-encoded fragment of the editing domain of AlaRS (Beebe et al. 2008). AlaXps also recognize mischarged tRNAAla in a G:U-sensitive way. Thus, mistranslation is suppressed by three different complexes with the same tRNA. These complexes guard against mistaking glycine or serine for alanine.
Figure 5.
The G3:U70 base pair marks a tRNA for acceptance of alanine. G:U recognition occurs with determinants in the catalytic domain of AlaRS. Remarkably, the editing domain is also sensitive to the same G:U base pair. Thus, two distinct domains in AlaRSs recognize the same base pair.
A rationale for mutagenesis from mistranslation
Aging is associated with the accumulation of mutations that arise from direct environmental insults, including oxidative events that are not accurately repaired. In addition, as an organism ages, mistranslation could play a role in producing mutations. These mutations come from occasional errors of translation made in the polymerases or the repair enzymes. These sporadically produced variants may then make errors of replication or repair that become fixed in the genome. These considerations led to experiments in bacteria to see whether more mutations were accumulated in aging bacteria that harbored an editing-defective tRNA synthetase.
Mutagenesis in aging bacteria from an editing-defective isoleucyl-tRNA synthetase
For these experiments, mutagenesis in aging colonies (MAC) of E. coli was studied. In this work, bacteria were plated on solid media for either one or seven days. Then, the frequency of spontaneous rifampicin-resistant (RifR) mutants in the population after each time period was determined. After one day, the difference, if any, in the appearance of RifR colonies was negligible in the editing-defective strain. While the frequency of spontaneous mutations increased with time for normal and editing-defective strains, the frequency was substantially higher by 7 d for the editing-defective strain.
In other work, some of the genes associated with the SOS response were shown to be involved in MAC (Taddei et al. 1995, 1997; Bjedov et al. 2003). Activation of the SOS response comes from the presence of single-stranded DNA (Sutton et al. 2000; Goodman 2002). This response can be experimentally triggered by exposure of cells to ciprofloxacin (Ci) and related quinolone antibiotics, which target DNA gyrase and topoisomerase IV (Drlica and Zhao 1997)—two enzymes needed to relax supercoiling during DNA replication. By binding to these proteins, Ci prevents rejoining of DNA ends. The free DNA ends are then bound by RecA protein, which, in turn, induces the SOS response through LexA. Accumulation of CiR mutants after exposure to ciprofloxacin is diagnostic for the SOS response (Piddock and Wise 1987; Drlica and Zhao 1997; Riesenfeld et al. 1997; Cirz et al. 2005; Cirz and Romesberg 2006).
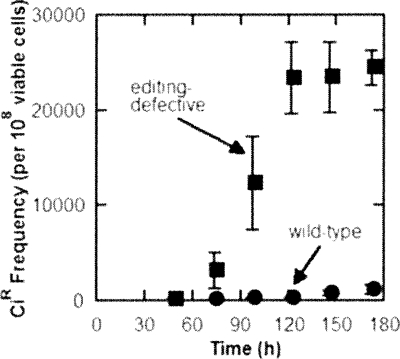
Editing-defective and wild-type strains show a sharp difference in the accumulation of CiR mutants over time (Fig. 6). For example, 2 d of growth on minimal media with Ci was sufficient for the editing-defective strain to acquire a much greater mutation rate. This observation is consistent with editing-defective strains accumulating CiR mutations at an increased rate, as a result of exposure to the antibiotic and to the associated induction of the SOS response. Indeed, with a lexA-deficient, editing-defective strain, the mutation rate was reduced greatly. Thus, the inability to induce the SOS response resulted in a dramatically diminished accumulation of CiR mutants (Bacher and Schimmel 2007).
Figure 6.
In aging bacteria that are grown in the presence of ciprofloxin, mutations accumulate in enzymes needed to relax supercoiling of DNA. These mutations are diagnostic for induction of the SOS response and the error-prone DNA repair systems. Adapted from Bacher and Schimmel (2007) (reprinted with permission from The National Academy of Sciences ©2007).
In summary, an editing-defect that causes mistranslation can, over time, lead to a higher mutation rate that is due to the induction of error-prone DNA polymerases.
Conclusions
Editing plays a far greater role in sustaining cell homeostasis than was appreciated when the editing activities were first observed from direct biochemical experiments. It is now clear from experiments in bacteria and mammals that mistranslation is a serious problem for cell growth and survival, and that the establishment and maintenance of activities for editing were essential for development of the tree of life. That a spontaneous heritable mutation in the editing domain of a synthetase has been annotated in the mouse raises the possibility that such mutations will be found in the human population. But, from what was learned from studies in the mouse, these mutations will doubtless be mild, because more severe mutations would be lethal. And yet, even mild mutations may have profound consequences in the development of specific diseases, such as the neurological disorder seen in the editing-deficient sti mouse.
Not understood is why some amino acids pose more difficulties than others. Of all of the amino acids, the most challenging appears to be alanine. Confusion of glycine or serine for alanine has resulted in three checkpoints that guard against this confusion (see above). For example, the separate and widely distributed genome-encoded, editing-proficient AlaXp homologs of the editing domain of AlaRS are not seen for the counterpart CP1 editing domains of IleRS, ValRS, or LeuRS. One speculation is that protein misfolding is more severe, on average, when Ser or Gly is substituted for Ala than when Val, for example, is substituted for Ile (when IleRS fails to clear misactivated valine). But regardless of the explanation of the need for genome-encoded fragments of the editing domains for some, but not other, synthetases, the dependence of the development of the tree of life on the editing activities of tRNA synthetases, and the connection to disease, is now well established.
Acknowledgments
This work was supported by grants from the National Institutes of Health, The Skaggs Institute for Chemical Biology and by a fellowship from the National Foundation for Cancer Research. The presentation above is based on, and updated from, a portion of the Stein and Moore Award Lecture of July 2007.
Footnotes
Editor's Note: Dr. Schimmel is the recipient of the 2007 Stein and Moore Award, the highest recognition given by the Protein Society. This review develops themes from his Award Lecture.
Reprint requests to: Paul Schimmel, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA; e-mail: schimmel@scripps.edu; fax: (858) 784-8990.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037242.108.
References
- Ahel, I., Korencic, D., Ibba, M., Söll, D. Trans-editing of mischarged tRNAs. Proc. Natl. Acad. Sci. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, S., Musier-Forsyth, K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J. Biol. Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- An, S., Musier-Forsyth, K. Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase.YbaK.tRNA ternary complex. J. Biol. Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- Bacher, J.M., Schimmel, P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc. Natl. Acad. Sci. 2007;104:1907–1912. doi: 10.1073/pnas.0610835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher, J.M., Waas, W.F., Metzgar, D., de Crecy-Lagard, V., Schimmel, P. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi . J. Bacteriol. 2007;189:6494–6496. doi: 10.1128/JB.00622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe, K., Mock, M., Merriman, E., Schimmel, P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- Bjedov, I., Tenaillon, O., Gerard, B., Souza, V., Denamur, E., Radman, M., Taddei, F., Matic, I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- Carter, C.W., Duax, W.L. Did tRNA synthetase classes arise on opposite strands of the same gene? Mol. Cell. 2002;10:705–708. doi: 10.1016/s1097-2765(02)00688-3. [DOI] [PubMed] [Google Scholar]
- Cirz, R.T., Romesberg, F.E. Induction and inhibition of ciprofloxacin resistance-conferring mutations in hypermutator bacteria. Antimicrob. Agents Chemother. 2006;50:220–225. doi: 10.1128/AAC.50.1.220-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz, R.T., Chin, J.K., Andes, D.R., de Crecy-Lagard, V., Craig, W.A., Romesberg, F.E. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody, C.W., Prasher, D.C., Westler, W.M., Prendergast, F.G., Ward, W.W. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- Cormack, B.P., Valdivia, R.H., Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Berthet-Colominas, C., Hartlein, M., Nassar, N., Leberman, R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Doring, V., Mootz, H.D., Nangle, L.A., Hendrickson, T.L., de Crecy-Lagard, V., Schimmel, P., Marliere, P. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- Drlica, K., Zhao, X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldred, E.W., Schimmel, P.R. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 1972;247:2961–2964. [PubMed] [Google Scholar]
- Eriani, G., Delarue, M., Poch, O., Gangloff, J., Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fukunaga, R., Yokoyama, S. Structural basis for non-cognate amino acid discrimination by the valyl-tRNA synthetase editing domain. J. Biol. Chem. 2005;280:29937–29945. doi: 10.1074/jbc.M502668200. [DOI] [PubMed] [Google Scholar]
- Goodman, M.F. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Hendrickson, T.L., Schimmel, P. Transfer RNA-dependent amino acid discrimination by aminoacyl-tRNA synthetases. In: La Pointe J., Brakier-Gingras L., editors. Translational mechanisms. Kluwer Academic/Plenum Publishers; New York: 2003. pp. 34–64. [Google Scholar]
- Hendrickson, T.L., Nomanbhoy, T.K., de Crecy-Lagard, V., Fukai, S., Nureki, O., Yokoyama, S., Schimmel, P. Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol. Cell. 2002;9:353–362. doi: 10.1016/s1097-2765(02)00449-5. [DOI] [PubMed] [Google Scholar]
- Hou, Y.M., Schimmel, P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Hou, Y.M., Schimmel, P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989;28:6800–6804. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- Lee, J.W., Beebe, K., Nangle, L.A., Jang, J., Longo-Guess, C.M., Cook, S.A., Davisson, M.T., Sundberg, J.P., Schimmel, P., Ackerman, S.L. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lin, L., Hale, S.P., Schimmel, P. Aminoacylation error correction. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- Ludmerer, S.W., Schimmel, P. Gene for yeast glutamine tRNA synthetase encodes a large amino-terminal extension and provides a strong confirmation of the signature sequence for a group of the aminoacyl-tRNA synthetases. J. Biol. Chem. 1987;262:10801–10806. [PubMed] [Google Scholar]
- McClain, W.H., Foss, K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket.”. Science. 1988;241:1804–1807. doi: 10.1126/science.2459773. [DOI] [PubMed] [Google Scholar]
- Nangle, L.A., De Crecy Lagard, V., Doring, V., Schimmel, P. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem. 2002;277:45729–45733. doi: 10.1074/jbc.M208093200. [DOI] [PubMed] [Google Scholar]
- Nangle, L.A., Motta, C.M., Schimmel, P. Global effects of mistranslation from an editing defect in mammalian cells. Chem. Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Nicholson, D.W., Ali, A., Thornberry, N.A., Vaillancourt, J.P., Ding, C.K., Gallant, M., Gareau, Y., Griffin, P.R., Labelle, M., Lazebnik, Y.A., et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy, T.K., Hendrickson, T.L., Schimmel, P. Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol. Cell. 1999;4:519–528. doi: 10.1016/s1097-2765(00)80203-8. [DOI] [PubMed] [Google Scholar]
- Nureki, O., Vassylyev, D.G., Tateno, M., Shimada, A., Nakama, T., Fukai, S., Konno, M., Hendrickson, T.L., Schimmel, P., Yokoyama, S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- Pham, Y., Li, L., Kim, A., Erdogan, O., Weinreb, V., Butterfoss, G.L., Kuhlman, B., Carter C.W., Jr A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol. Cell. 2007;25:851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Piddock, L.J., Wise, R. Induction of the SOS response in Escherichia coli by 4-quinolone antimicrobial agents. FEMS Microbiol. Lett. 1987;41:289–294. [Google Scholar]
- Prasher, D.C., Eckenrode, V.K., Ward, W.W., Prendergast, F.G., Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana, L., Schimmel, P. Aminoacyl tRNA synthetases as potential markers for the development of the genetic code. In: Keinan E., et al., editors. Life sciences for the 21st century. Wiley-VCH; Weinheim: 2004a. pp. 81–92. [Google Scholar]
- Ribas de Pouplana, L., Schimmel, P. Aminoacylations of tRNAs: recordkeepers for the genetic code. In: Nierhaus K.H., Wilson D.N., editors. Protein synthesis and ribosome structure. Wiley-VCH; Weinheim: 2004b. pp. 169–184. [Google Scholar]
- Riesenfeld, C., Everett, M., Piddock, L.J., Hall, B.G. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 1997;41:2059–2060. doi: 10.1128/aac.41.9.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin, S.N., Ohno, S. Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 1995;25:565–589. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- Rodin, S., Ohno, S., Rodin, A. Transfer RNAs with complementary anticodons: could they reflect early evolution of discriminative genetic code adaptors? Proc. Natl. Acad. Sci. 1993;90:4723–4727. doi: 10.1073/pnas.90.10.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin, S., Rodin, A., Ohno, S. The presence of codon-anticodon pairs in the acceptor stem of tRNAs. Proc. Natl. Acad. Sci. 1996;93:4537–4542. doi: 10.1073/pnas.93.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel, P. An editing activity that prevents mistranslation and connection to disease. J. Biol. Chem. 2008 doi: 10.1074/jbc.X800007200. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel, P., Ribas de Pouplana, L. Formation of two classes of tRNA synthetases in relation to editing functions and genetic code. Cold Spring Harb. Symp. Quant. Biol. 2001;66:161–166. doi: 10.1101/sqb.2001.66.161. [DOI] [PubMed] [Google Scholar]
- Schmidt, E., Schimmel, P. Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science. 1994;264:265–267. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- Schmidt, E., Schimmel, P. Residues in a class I tRNA synthetase which determine selectivity of amino acid recognition in the context of tRNA. Biochemistry. 1995;34:11204–11210. doi: 10.1021/bi00035a028. [DOI] [PubMed] [Google Scholar]
- Schreier, A.A., Schimmel, P.R. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- Sutton, M.D., Smith, B.T., Godoy, V.G., Walker, G.C. The SOS response: Recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- Taddei, F., Matic, I., Radman, M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, F., Halliday, J.A., Matic, I., Radman, M. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Mol. Gen. Genet. 1997;256:277–281. doi: 10.1007/s004380050570. [DOI] [PubMed] [Google Scholar]
- Weigle, W.O. Termination of acquired immunological tolerance to protein antigens following immunization with altered protein antigens. J. Exp. Med. 1962;116:913–928. doi: 10.1084/jem.116.6.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle, W.O. The induction of autoimmunity in rabbits following injection of heterologous or altered homologous thyroglobulin. J. Exp. Med. 1965;121:289–308. doi: 10.1084/jem.121.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus, M. Phenylalanyl-tRNA synthetase and isoleucyl-tRNA Phe: A possible verification mechanism for aminoacyl-tRNA. Proc. Natl. Acad. Sci. 1972;69:1915–1919. doi: 10.1073/pnas.69.7.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]