Abstract
The identification of sequences involved in binding to erythrocytes is an important step for understanding the molecular basis of merozoite–erythrocyte interactions that take place during invasion of the Plasmodium falciparum malaria parasite into host cells. Several molecules located in the apical organelles (micronemes, rhoptry, dense granules) of the invasive-stage parasite are essential for erythrocyte recognition, invasion, and establishment of the nascent parasitophorous vacuole. Particularly, it has been demonstrated that rhoptry proteins play an important role in binding to erythrocyte surface receptors, among which is the PfRhopH3 protein, which triggers important immune responses in patients from endemic regions. It has also been reported that anti-RhopH3 antibodies inhibit in vitro invasion of erythrocytes, further supporting its direct involvement in erythrocyte invasion processes. In this study, PfRhopH3 consecutive peptides were synthesized and tested in erythrocyte binding assays for identifying those regions mediating binding to erythrocytes. Fourteen PfRhopH3 peptides presenting high specific binding activity were found, whose bindings were saturable and presented nanomolar dissociation constants. These high-activity binding peptides (HABPs) were characterized by having α-helical structural elements, as determined by circular dichroism, and having receptors of a possible sialic acid-dependent and/or glycoprotein-dependent nature, as evidenced in enzyme-treated erythrocyte binding assays and further corroborated by cross-linking assay results. Furthermore, these HABPs inhibited merozoite in vitro invasion of normal erythrocytes at 200 μM by up to 60% and 90%, suggesting that some RhopH3 protein regions are involved in the P. falciparum erythrocyte invasion.
Keywords: malaria, Plasmodium falciparum, RhopH3 protein, high-activity binding peptides
Malaria is a tropical disease caused by parasites of the genus Plasmodium, being P. falciparum, P. vivax, P. ovale, and P. malariae the only species capable of infecting humans (Thwing et al. 2007). Of these four species, P. falciparum is responsible for the majority of cases, reaching an alarming number of 300–500 million cases per year and having the highest mortality rate worldwide. In Africa alone, more than a million children under the age of five die from malaria every year (Breman et al. 2004; Summer et al. 2005; WHO 2005).
During the last decades, developing new strategies for the control of this terrible disease has become an urgent need due to the alarming development of the parasite's resistance to antimalarial drugs and the mosquito's resistance to insecticides (Greenwood and Mutabingwa 2002; Lanteri et al. 2007). Such new strategies demand a profound understanding of the parasite's invasion pathways, the molecular characterization of proteins involved in the parasite's infection process, as well as their interaction with host cells (Greenwood and Mutabingwa 2002; Lanteri et al. 2007).
The malaria life cycle begins when the sporozoite stages of the P. falciparum parasite developing inside the salivary glands of an infected Anopheles female mosquito are transmitted to humans during a blood meal. Once inside the bloodstream, sporozoites travel to the liver, where they multiply and transform into infectious stages named merozoites. Each merozoite released from the liver invades an erythrocyte, inside which it grows and multiplies. Once the intraerythrocytic cycle is completed, the infected erythrocyte membrane lyses, releasing newly formed merozoites that go on to invade more erythrocytes by means of specialized invasion processes (Fujioka and Aikawa 2002; Bannister and Mitchell 2003).
Erythrocyte invasion is therefore one of the most important steps during the parasite's life cycle and is directly mediated by three important secretory organelle systems: the rhoptries, micronemes, and dense granules (Preiser et al. 2000). The Plasmodium parasite rhoptries are located at the merozoite's apical end; their content is discharged onto the erythrocyte membrane once the merozoite binds to the erythrocyte surface. They disappear shortly after the merozoite internalization, and then reappear in every newly formed merozoite with each intraerythrocytic cycle (Preiser et al. 2000; Topolska et al. 2004; Kats et al. 2006).
Different protein complexes have been found in these discharges, including the Rhop complex, which is in turn made up of two large protein groups. The first one is a low molecular weight complex (RhopL) constituted by rhoptry-associated proteins such as RAP1-83 kDa, RAP2-40 kDa, and RAP3-37 kDa, while the second one has high molecular weight (RhopH) and is composed of RhopH1/Clag (cytoadherence-linked asexual gene)-140 kDa, RhopH2-130 kDa, and RhopH3-110 kDa proteins, which are believed to participate in erythrocyte invasion processes (Cooper et al. 1988; Sam-Yellowe and Ndengele 1993; Kaneko 2007). In general, the RhopH proteins are characterized by having a signal sequence, constituted approximately by the first 15–24 residues, as well as conserved cysteine (Cys) domains (Kaneko 2007). Additionally, they associate in a noncovalent and stable manner with a glycosylphosphatidylinositol (GPI)-tail membrane-anchored protein named the rhoptry-associated membrane antigen (RAMA) (Sam-Yellowe and Perkins 1991; Sam-Yellowe 1993; Topolska et al. 2004).
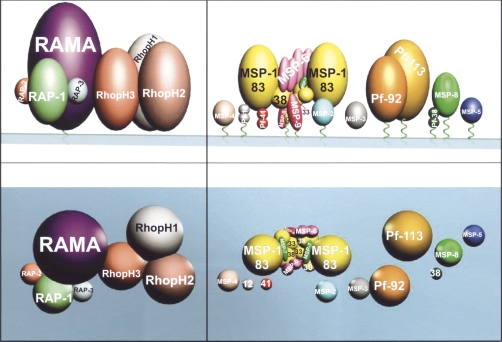
The RhopH3 protein has been found to be located in detergent resistant membrane (DRM) lipid rafts (obtained by isolation with Triton-X100), which are suggested to serve as signaling platforms for receptor molecules and their accessory proteins, and thus have an essential role in cell adhesion and motility (Wang et al. 2003; Sanders et al. 2005). A model of the localization of RhopH3 can be seen in Figure 1, showing its direct interaction with the RAMA protein (Topolska et al. 2004), and the interaction of the complex formed by both proteins with other rhoptry proteins such as the RAP-1, -2, and -3 proteins.
Figure 1.
Membrane and detergent-resistant lipid raft-like membrane-associated proteins (DRM). Molecule sizes are drawn at their approximate molecular weight. (Left panel) RAMA anchored to the membrane via a GPI tail and proteins noncovalently associated with RAMA and involved in merozoite invasion of erythrocytes (recognized in DRM proteomes). All high (RhopH) and low (RAP-1, -2, -3) molecular weight rhoptry protein members are also shown. (Top panel) Lateral view of the hypothetical organization of these proteins, (bottom panel) view from the top. (Right panel) DRM rafts formed by MSP-1 83 kDa, 30 kDa, and 38 kDa fragments (yellow) and noncovalently associated molecules such as MSP-6 (fuchsia) and MSP-7 (green). MSP-1 33 kDa (yellow) and the only 19 kDa (yellow) fragment anchored to the merozoite membrane via a GPI tail are also displayed. Other GPI tail (green twists traversing the pale green membrane) anchored membrane surface proteins such as MSP-2 (clear blue), MSP-3 (dark gray), MSP-4 (clear brown), MSP-5 (dark blue), MSP-8 (green), and Pf113 (dark gold), Pf92 (brown), Pf41 (red), and Pf12 (gray), recently identified in DRM proteome analysis, are also shown. Figure adapted from Pinzón et al. (2008) and reprinted with permission from Elsevier © 2008.
Both the RhopH and RhopL complexes are precipitated by antisera directed against their individual components, and studies have indicated that antibodies raised against proteins from both complexes are associated with resistance to in vivo infection (Ridley et al. 1990; Yang et al. 1996). Additionally, the RhopH complex confers partial protection in Aotus monkeys (Siddiqui et al. 1987), and mAbs inhibit parasite growth and/or merozoite in vitro invasion of erythrocytes (Cooper et al. 1988; Sam-Yellowe and Perkins 1990).
RhopH3 is a 110-kDa protein mainly encoded by a seven-exon gene. The exons have lengths of 63, 111, 961, 63, 57, 771, and 668 bp in P. falciparum, while the six intron sequences span a total of 1355 bp (Brown and Coppel 1991; Mongui et al. 2007). It has been demonstrated that RhopH3 appears 30 h after infection in the schizont stage and that anti-RhopH3 antibodies react primarily to epitopes located in the C-terminal region (Brown and Coppel 1991; Yang et al. 1996). Serological studies using human sera of malaria-infected individuals from different geographic regions show that the C terminus of RhopH3 is structurally conserved among different geographical and laboratory P. falciparum isolates (Yang et al. 1996; Wang et al. 2006).
Recently, in our Institute, a homolog of the P. falciparum RhopH3 protein was identified and characterized in P. vivax, the second malaria-causing parasite species, which revealed the codification of both proteins by homologous chromosome regions. Furthermore, the PfRhopH3 and PvRhopH3 amino acid alignment showed 68.6% similarity and 52.7% identity between both homolog proteins, which are considerably higher values than those of other parasite proteins involved in erythrocyte invasion (Mongui et al. 2007).
Due to its role in erythrocyte binding and its presence in the erythrocyte membrane following invasion, RhopH3 has been considered to be an ideal candidate for vaccine studies (Sam-Yellowe et al. 1988; Sam-Yellowe and Perkins 1990, 1991). This study defines the specific regions of PfRhopH3 mediating binding to erythrocytes, characterizes the nature of their possible receptors, and assesses their ability for inhibiting parasite invasion. The results demonstrate that some regions of this protein are suitable for being included in the design of an antimalarial, subunit-based, chemically synthesized vaccine.
Results
Determining RhopH3 HABPs
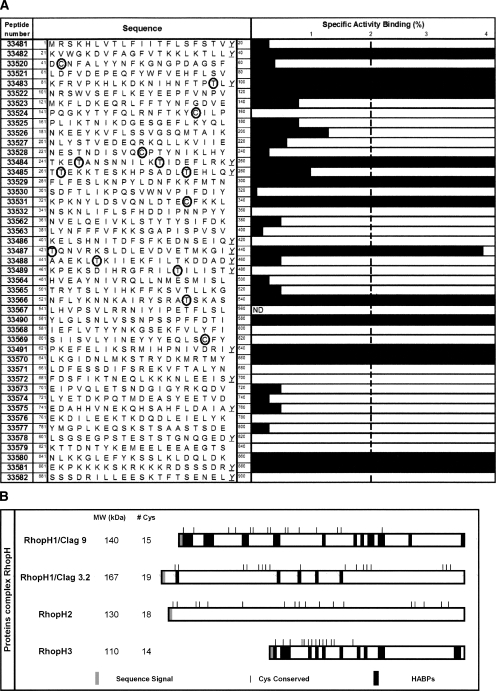
Binding assays were used for determining which of the 45 RhopH3 synthetic peptides had specific erythrocyte binding activity (GenBank number accession gi: 124506661 of strain 3D7). Those peptides having ≥2% specific binding activity were considered to be high-activity binding peptides (HABPs) (Fig. 2A).
Figure 2.
(A) Erythrocyte binding assays using RhopH3 peptides. The length of the black bar indicates the slope of the specific binding graph of each peptide. Peptides are numbered according to investigator's numbering system. (ND) Not determined. (B) Schematic representation of conserved Cys residues in high molecular weight rhoptry complex. RhopH1/Clag 9 (C.G. Pinzon, H. Curtidor, and M.E. Patarroyo, unpubl.), RhopH1/Clag 3.2 (Ocampo et al. 2005), RhopH2 (protein in study), and RhopH3.
The results show that the RhopH3 protein has 14 HABPs distributed throughout the sequence of the protein. These HABPs were 33482, 33521, 33483, 33522, 33484, 33529, 33531, 33487, 33566, 33490, 33491, 33570, 33580, and 33581. The code number is assigned to each peptide according to our Institute's numbering system and does not correspond to its localization within the protein sequence.
Binding assay with scramble HABPs
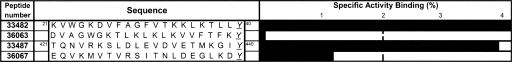
Two HABPs showing different degrees of high erythrocyte binding activity were selected according to the slopes obtained for their binding profiles (data not shown). HABP 33482, localizing at the N-terminal region, had a considerably high specific binding activity, reporting a slope >70%, while HABP 33487 was located in the RhopH3 central region and had a slope slightly >4%. Figure 3 shows the sequences of the scramble peptides together with the sequence of their corresponding original HABP (Stothard 2000).
Figure 3.
Erythrocyte binding profile for two HABPs compared with that of their corresponding scramble peptide. Peptides are numbered according to the investigator's numbering system. Scramble peptides' binding activities were much lower than those of their native HABPs, or were even undetectable.
Saturation assay
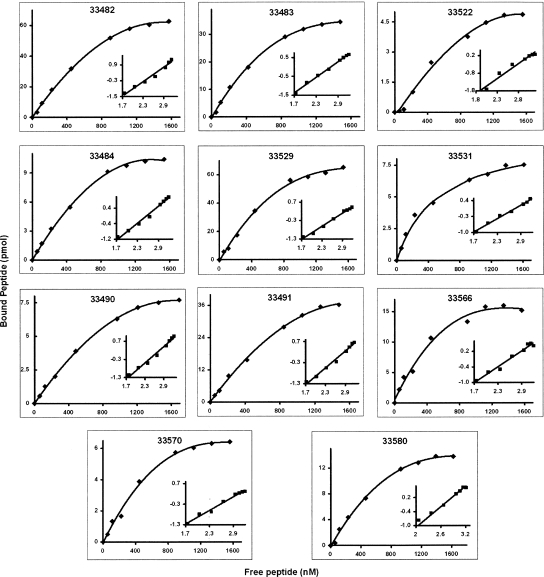
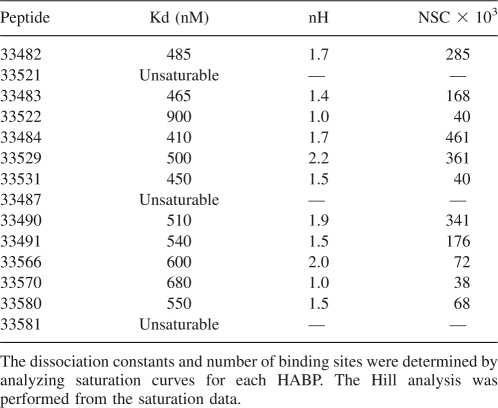
Saturation assays allowed the determination of the kinetics constant, Hill coefficients (Hulme 1993), and number of binding sites per cell for all HABPs (Fig. 4). Values for HABPs' dissociation constants were between ∼410 and 900 nM, suggesting a low dissociation for the receptor–peptide complex. Hill coefficient values were >1 (1.4 and 2.2), indicating a positive cooperativity, while nearly 40,000 and 461,000 binding sites were found per cell for RhopH3 HABPs (Table 1). Most HABP interactions with erythrocytes were saturable under the assay conditions (Fig. 4). The three HABPs (33487, 33521, and 33481) whose erythrocyte binding was not saturable are not shown in this figure.
Figure 4.
Saturation curves for HABPs 33482, 33483, 33484, 33490, 33491, 33522, 33529, 33531, 33566, 33570, and 33580. Increasing amounts of radiolabeled peptide were added in the presence or absence of unlabeled peptide. The curve represents specific binding of labeled peptide to human erythrocytes. The abscissa of the Hill plot (inset graph) is log F and the ordinate is log (B/Bmax − B). (F) Free peptide, (B) amount of bound peptide, (Bmax) maximum amount of bound peptide.
Table 1.
Dissociation constants (Kd), Hill coefficients (nH), and number of binding sites per cell (NSC) for RhopH3 HABPs
Merozoite invasion inhibition assay
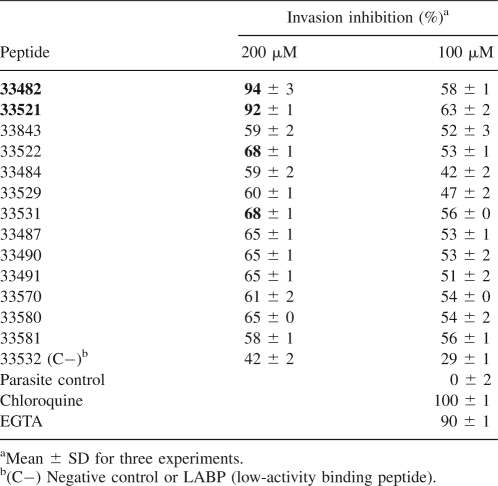
To determine whether RhopH3 HABPs had any effect on merozoite invasion of erythrocytes, each HABP was independently assessed in in vitro cultures of P. falciparum, using chloroquine and EGTA as inhibition positive controls while the low-activity binding peptide (LABP) 33532 served as a negative control. The merozoite invasion inhibition percentage of each HABP is shown in Table 2 together with their respective standard deviations. As shown in this table, merozoite invasion was mainly inhibited at a 200 μM concentration, and these inhibition percentages ranged between 60% and 94%. The highest inhibition was achieved by HABPs 33482 and 33521, while the remaining HABPs showed an intermediate inhibition.
Table 2.
RhopH3 peptide inhibition of parasite's erythrocyte invasion
CD measurement
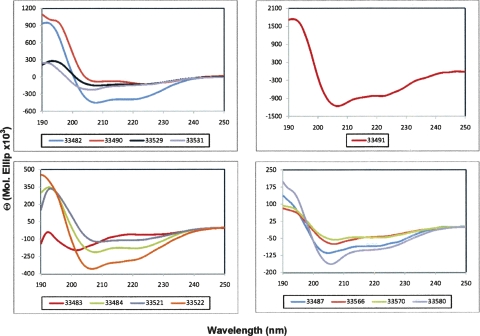
The circular dichroism (CD) spectra of all PfRhopH3 protein HABPs were studied for determining their secondary structure. The spectra obtained were measured in water and 30% TFE/water (Fig. 5). All peptides' CD profiles showed a clear shift toward an ordered structure when measured in the more hydrophobic medium, which was mostly an α-helical structure characterized by two minima at 209 and 222 nm (Fig. 5).
Figure 5.
CD spectra of RhopH3 HABPs (30% TFE/water). All HABPs clearly showed a helical conformation.
Effect of enzymatic treatment on RhopH3 HABP binding
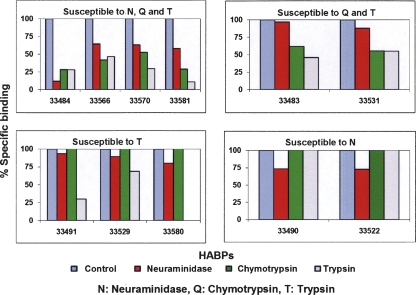
In order to determine whether the HABPs of the PfRhopH3 protein interacted directly with a particular erythrocyte membrane's receptor site, human erythrocytes were treated with two different proteases (chymotrypsin and trypsin) and a neuraminidase prior to performing erythrocyte-binding assays. The percentage of specific binding to enzyme-treated erythrocytes was determined for each HABP using untreated erythrocytes as a total binding control (100% specific binding). Five different enzyme-associated binding patterns were seen (Fig. 6). Erythrocyte binding of four HABPs (33484, 33566, 33570, and 33581) was susceptible to neuraminidase, chymotrypsin, and trypsin treatment, whereas it was susceptible only to chymotrypsin and trypsin for two HABPs (33483 and 33531). The binding of HABPs 33491, 33529, and 33580 was susceptible to trypsin, whereas the binding of HABPs 33490 and 33522 was susceptible to chymotrypsin (Fig. 6). Binding was not affected by any enzymatic treatment for three HABPs (33482, 33487, and 33521).
Figure 6.
Effect of enzymatic treatment on HABPs' erythrocyte binding. HABPs are organized according to their erythrocyte-binding pattern to each of the assessed enzymes. (Top right panel) Those HABPs whose erythrocyte binding was susceptible to neuraminidase, chymotrypsin, and trypsin. (Top left panel) Those HABPs whose receptors were affected by chymotrypsin and trypsin. (Bottom panels) Trypsin- (left) and chymotrypsin (right)-susceptible HABPs. Those HABPs whose binding behavior was not affected by any enzymatic treatment are not shown.
Cross-linking assays
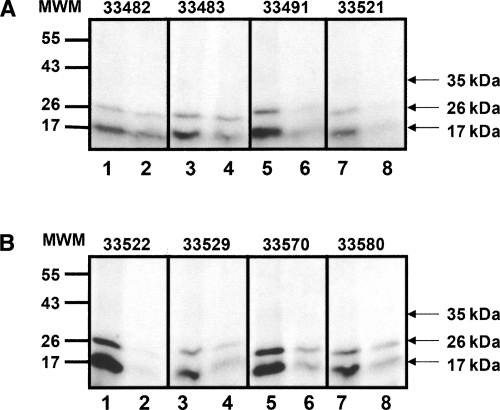
As shown in Figure 7, HABPs 33482, 33483, 33491, 33521, 33522, 33529, 33570, and 33581 bound strongly and specifically to protein erythrocyte membrane receptors. Such proteins have molecular masses of ∼35, 26, and 17 kDa. Additionally, HABPs binding to these bands disappeared when the binding assay was performed in the presence of the corresponding nonlabeled peptide (Fig. 7A,B).
Figure 7.
Cross-linking assays. Ligand–receptor complexes were obtained by using erythrocyte membrane proteins cross-linked with radiolabeled versions of each HABP. (Lanes 1,3,5,7 in A,B) Total binding for peptides 33482, 33483, 33491, 33521, 33522, 33529, 33570, and 33580, respectively. (Lanes 2,4,6,8 in A,B) Inhibited binding for peptides following the same order as above. The images were acquired by using BioRad Quantity One (Quantitation Software) and were not further manipulated.
Discussion
The RhopH protein complex has been reported as one of the most abundant rhoptry proteins that participate in invasion of erythrocytes by Plasmodium merozoites. Proteomic analysis has identified that all the RhopH protein complex belonging to the detergent resistant membrane (DRM) proteins associates with lipid rafts, along with other rhoptry proteins such as the RAMA protein and low molecular weight proteins such as RAP-1, -2, and -3 and other surface proteins such as MSP-1,-2 -4, -5, and -6 (Sanders et al. 2005). Also, proteins such as Pf92, Pf 41, and Pf 12 have been identified recently as part of the DRM (Sanders et al. 2005; Fig. 1).
The gene encoding RhopH3 has been characterized in several Plasmodium species (P. falciparum, P. yoelii, P. chabaudi, P. berghei, and P. vivax) (Brown and Coppel 1991; Anthony et al. 2000; Sam-Yellowe et al. 2000; Shirano et al. 2001; Mongui et al. 2007), and different studies suggest that this protein could be included in an antimalarial vaccine (Siddiqui et al. 1987; Yang et al. 1996). Strong immune response against RhopH3 has been observed in malaria-infected people, and anti-RhopH3 antibodies inhibit merozoite in vitro invasion of erythrocytes (Campbell et al. 1984; Cooper et al. 1988). In addition, it has been shown that the PfRhopH3 protein binds to mouse erythrocytes (Sam-Yellowe and Perkins 1990). Immunization studies using recombinant RhopH3 proteins from murine malarial parasites such as P. yoelii and P. berghei have shown that RhopH3 not only is immunogenic but also is capable of protecting mice against an otherwise lethal challenge with these parasite species (Wang et al. 2006).
Fourteen HABPs were found in RhopH3 distributed along the entire amino acid sequence (Fig. 2A). Altogether, these HABPs represented ∼31% of the entire RhopH3 protein, the majority of which presented a high specific erythrocyte binding activity notably >4%. This protein's C terminus is a serine- and lysine-region conserved among different P. falciparum strains that have been shown to be highly antigenic (Yang et al. 1996). It was precisely within this region (389K–L895) where seven HABPs of the RhopH3 protein were located (33487, 33566, 33490, 33491, 33570, 33580, 33581). Regarding the C-terminal region, multiple epitopes have been identified within it using mouse mAb (Doury et al. 1997). Interestingly, HABPs 33580 (841NLKKGLEFYKSSLKLDQLDK 860) and 33581 (861 EKPKKKKSKRKKKRDSSSDRY880) contained a portion of an epitope identified by Doury et al. (1997) (the epitope's portion is shown in bold type and underlined), which makes these two HABPs attractive and good candidates to be included in later studies for developing an antimalarial vaccine.
When the sequences of HABPs 33482, 33487, and 33570 were modified regarding the order of their amino acids, but retaining their net charge and final amino acid composition (scramble peptides), the specific binding to erythrocytes decreased to such an extent that the peptides could no longer be considered as having a high binding activity. These results show that the HABPs' specific binding depends specifically on their amino acid sequence and not on their composition, since variations in such sequences altered their erythrocyte binding activity, which in turn demonstrates the specificity of our methodology for identifying HABPs. (Fig. 3).
Saturation assays show that most of the peptides have dissociation constant ranging between 410 and 900 nM, and therefore their bindings are saturable, which suggests high-affinity interactions (Fig. 4). There were 40,000–461,000 binding sites (Table 1), indicating that erythrocyte receptors were available in considerable amounts. HABPs 33487, 33521, and 33581 showed a clear nonsaturation behavior (Table 1), possibly indicating that these HABPs have a tendency for binding to several different receptors on the erythrocyte membrane, since no single receptor site is completely occupied.
According to the CD characterization, all HABPs present preferentially α-helical structures with two minima around 209 and 222 nm (Fig. 5). However, HABPs 33483, 33529, 33531, 33487, 33566, and 33580 possess minor random-coil structural features with a 70%–85% α-helix percentage (Fig. 5). These results are in full agreement with those obtained by deconvolution programs such as Selcon3, Continll, and Cdsstr (Sreerama et al. 1999, 2000).
According to Sam-Yellowe and Perkins, erythrocyte binding of rhoptry proteins and subsequent invasion is blocked when they are treated with trypsin and chymotrypsin but not with neuraminidase, indicating that their proteic receptors are exposed and accessible on the erythrocyte surface (Sam-Yellowe and Perkins 1990). Our results strongly support the aforementioned, since >70% of PfRhopH3 HABPs showed a preference for non-glycosylated proteic receptor sites. However, HABP 33484 binding was affected when erythrocytes were treated with neuraminidase (Fig. 6), suggesting its interaction with oligosaccharide-type sialic acid-dependent receptors. HABPs 33566, 33570, and 33581 showed a similar behavior but to a lesser extent, since their binding was reduced only 25% (Fig. 6). The binding of HABPs 33484, 33566, 33570, and 33581 was affected by the three enzymes assessed. In particular, neuraminidase treatment decreased HABP 33484 binding ∼88% but only 36%–42% for the remaining three HABPs. Incubation with chymotrypsin decreased HABPs' specific binding ∼47%–72%, while trypsin treatment diminished binding to ∼53%–89%. These results suggest the interaction of these HABPs both with glycosidic and non-glycosidic receptors such as glycophorins, band 3 protein, and other membrane proteins such as putative receptors “E,” “Y,” and “Z” (Baum et al. 2005).
HABPs 33483 and 33531 binding was noticeably susceptible to incubation with chymotrypsin- and trypsin-treated erythrocytes, since their specific binding decreased 38%–44% and 45%–54%, respectively, with each enzyme (Fig. 6). Such behavior suggests a proteic receptor on erythrocyte membrane for these HABPs.
HABPs 33491, 33529, and 33580 showed an increased tendency for binding to glycoproteic receptors such as glycophorin A and C, since these proteins are strongly affected by the enzymatic activity of trypsin. HABPs 33490 and 33522 binding was slightly affected when erythrocytes were treated with neuraminidase and had a preference for oligosaccharide receptors containing sialic acid residues (Baum et al. 2005).
Additionally, it was found that HABPs 33482, 33487, and 33521 binding to their receptors was not affected by any of the enzymatic treatments performed (data not shown). The results suggest that these HABPs bound with affinity to receptor sites located on the surface of erythrocytes and to cryptic receptors that were exposed after being enzymatically cleaved. Such receptors could include erythrocyte surface proteins such as band 4.9, 4.1, ankyrin, and other internal membrane proteins (Hogh et al. 1994).
The cross-linking assay was performed for eight of the 14 RhopH3 HABPs (33482, 33483, 33491, 33521, 33522, 33529, 33570, and 33580) (Fig. 7A,B). These HABPs were shown to bind specifically to two erythrocyte membrane proteins having apparent molecular weights of 26 and 17 kDa. HABP 33570 recognized an additional 55-kDa band (slightly noticeable), whereas HABP 33522 recognized this same band as well as an extra 43-kDa band (slightly noticeable) (Fig. 7B). The autoradiography revealed that binding in the presence of nonradiolabeled HABPs (odd numbers, Fig. 7A,B) decreased for all tested HABPs. Nevertheless, such a reduction in their binding inhibition was evident only for five of these eight HABPs (33482, 33491, 33521, 33522, and 33529), whereas it was not clearly inhibited in HABPs 33483, 33570, and 33580 (Fig. 7A,B). Additionally, HABPs 33521, 33522, and 33570 recognized a 35-kDa band.
Enzymatic treatment assays revealed the presence of different receptor sites for each HABP, whereas the cross-linking assays evidenced the recognition of similar receptors for all the assessed HABPs. This result could be explained by considering that the 17-, 26-, and 35- kDa receptors were sensitive to treatment with proteases and therefore exposed new receptors sites when they were cleaved. Both assays suggested receptors of glycoproteic nature.
A possible functional role for PfRhopH3 HABPs during erythrocyte invasion could also be suggested since all HABPs inhibited merozoite invasion in vitro at 100–200 μM (Table 2) up to 60%. Interestingly, HABPs 33482 and 33521 inhibit invasion by 91%, indicating that these peptides are possibly competing with native parasite proteins for receptor sites on the erythrocyte surface and therefore blocking merozoite invasion more effectively. The peptide concentration needed for inhibiting merozoite invasion is relatively high (200 μM), possibly due to a direct competition between the peptide and the native parasite proteins, since the latter bound with more affinity than the individual peptides. In all cases, HABPs inhibited merozoite invasion in a concentration-dependent manner. Seventy percent of PfRhopH3 HABPs showed a moderate inhibition percentage (60%), which indicates that affinity for erythrocyte receptor sites of these HABPs is lower, compared with that of native parasite proteins, and that the latter compete more strongly possibly due to their larger size. HABPs 33522 and 33531 inhibited in vitro invasion up to 70%, suggesting a significant competition with the parasite protein.
In conclusion, the RhopH3 protein involved in merozoite invasion of erythrocytes contains 14 peptides that present a high and specific binding activity to erythrocytes. The majority of HABPs' binding was saturable and sensitive to different enzymatic treatments. Two of these HABPs (33482 and 33521) inhibited the parasite's erythrocyte invasion in vitro by 92%, which is possibly associated with a relevant role during merozoite invasion of erythrocytes.
The results of the erythrocyte binding assays done with HABP homologous peptides having a scramble sequence showed that HABP binding ability is dependent on the peptide's specific structural conformation and not on its peptide composition. However, previous studies (Espejo et al. 2001; Rodriguez et al. 2003) have shown that modifying a HABP's structure in any of its residues could render it more sensitive to the enzyme treatment and more capable of inhibiting in vitro invasion and eliciting an immune response against P. falciparum. Therefore, such modified HABPs would be good candidates for the chemically synthesized, subunit-based, multi-epitopic, multistage antimalarial vaccine that is currently being developed in our Institute.
Materials and Methods
Synthesizing RhopH3 peptides
20-mer nonoverlapping peptides were synthesized using the solid-phase multiple peptide system (Merrifield 1963; Houghten 1985); t-Boc protected amino-acids (Bachem), MBHA resin (0.5 meq/g), and the Low-High HF cleavage technique were used (Tam et al. 1983). They were subsequently purified by RP-HPLC, lyophilized, and then characterized by MALDI-TOF mass spectrometry. All synthesized peptides were at least 90% pure. A tyrosine (Tyr) residue was added to the C terminus of those peptides that did not contain this amino acid in their sequences to enable 125I-labeling. Figure 2A shows synthesized sequences in one-letter code. The cysteine (Cys) residues were substituted for threonine (Thr) residues to avoid difficulties in its synthesis, such as dimerization and/or cyclation.
Radiolabeling
According to a previously described methodology, the purified peptides were radiolabeled and tested (Yamamura et al. 1978; Urquiza et al. 1996; Rodriguez et al. 2000; Curtidor et al. 2001). Briefly, 5 μL of purified peptide, at a 1 mg/mL concentration in HBS (HEPES buffered saline), was treated with 0.3 μmol chloramine T (2.75 μg/μL) and 5 μL of Na125I (100 mCi/mL, MB Biomedicals) for ∼15 min. This reaction was stopped by adding 0.18 μmol sodium metabisulphite as the reducing agent (Slater 1990). Radiolabeled peptides were purified by size exclusion chromatography on a Sephadex G10 column (10 cm × 5 mm) (Pharmacia) and then quantified on a gamma counter (Auto Gamma Counter Cobra II Packard).
Erythrocyte binding assay
Human erythrocytes (2 × 107 cells), obtained from healthy donors, were washed in HBS buffer and then incubated for 90 min at room temperature with increasing concentrations of radiolabeled peptide (0–560 nM) in the absence (total binding) or presence (nonspecific binding) of 4 nmol unlabeled peptide (Rodriguez et al. 2003; Ocampo et al. 2004). HBS was added to reach a 200-μL final volume. The cells were then washed twice with HBS to remove unbound peptide. Cell-bound radiolabeled peptide was quantified in an automatic gamma counter (Auto Gamma Counter Cobra II Packard). The assay was done in triplicate in identical conditions.
Peptide binding activity was defined as being the amount of peptide (pmol) that specifically bound to erythrocyte per added peptide (pmol). High-activity binding peptides (HABPs) were defined as those peptides showing an activity ≥2%, according to a previously established criterion (Urquiza et al. 1996; Rodriguez et al. 2000, 2003; Curtidor et al. 2001; Ocampo et al. 2004).
Binding assay with scramble HABPs
Two scramble HABP analogs (i.e., peptides having the same amino acid composition as RhopH3 HABPs but with random sequences) were synthesized (Stothard 2000) to determine whether HABP binding to red blood cells was due to their specific amino acid sequences or just their amino acid composition. These peptides were tested in binding assays done in triplicate (described in the previous section).
Saturation assays
A modified erythrocyte binding assay was used to determine binding constants for erythrocyte interaction with each HABP. These saturation assays were done by incubating 1.5 × 107 cells with radiolabeled peptide concentrations ranging from 0 to 1600 nM and 24 μM unlabeled peptide at a 255-μL HBS final volume. Cells were then washed twice with HBS, and a gamma counter was used for measuring cell-bound radiolabeled peptide (Urquiza et al. 1996; Ocampo et al. 2004).
CD spectroscopy
CD assays were performed at room temperature on nitrogen-flushed cells using a Jasco J-810 spectro-polarimeter for determining HABPs secondary structure. Spectra were recorded at a wavelength interval of 190–260 nm. Peptides were analyzed in 30% v/v TFE aqueous solution on a quartz 1-cm path length rectangular cell thermostated at 20°C (Compton and Johnson Jr. 1986; Sreerama et al. 1999).
Cross-linking assays
Following a conventional binding assay that had been carried out with 4.2 × 106 erythrocytes in HBS buffer pH 7.4, HABPs were cross-linked with 50 μL (1 mg/mL) of bis sulphosuccinymidyl suberate BS3 (Sigma-Aldrich) for 1 h at 4°C and the reaction was then stopped by adding Tris-HCl buffer (pH 7.4). Samples were centrifuged at 1000g for 5 min and the supernatant was discarded. Subsequently, lysis buffer (5 mM Tris HCl, 7 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 0.1 mM phenylmethylsulfonul fluoride [PMSF]) and Laemmli buffer were added, and then centrifuged at 15,000g for 15 min. Cross-linked proteins were separated by SDS-PAGE on 12% gels and then were exposed on BioRad Imaging Screen K (BioRad Molecular Imager FX; BioRad Quantity One, Quantitation Software) for 5–8 d (Puentes et al. 2000; Lopez et al. 2006). The apparent molecular weights of those proteins that cross-linked to radiolabeled peptides were determined using molecular weight markers as reference patterns (Fermentas Life Sciences).
Erythrocyte enzymatic treatment
6.0 × 107 erythrocytes suspended in HBS buffer (pH 7.4) were treated independently with ∼150 μU/mL neuraminidase (ICN 9001-67-6), and trypsin (Sigma T-1005) or chymotrypsin (Sigma C-4129) at a final 1 mg/mL concentration, for 1 h at 37°C. Excess enzyme was removed by washing twice with HBS buffer and centrifuging at 1000g for 3 min. After enzyme treatment, these erythrocytes were tested in traditional binding assays with each HABP as described above and then analyzed by a gamma counter, using untreated 2.0 × 107 erythrocytes in HBS as a positive control (Pinzón et al. 2008).
Merozoite invasion inhibition assay
Sorbitol-synchronized P. falciparum (FCB-2 strain) cultures were incubated until late schizont stage at final 0.5% parasitaemia and 5% haematocrit in RPMI 1640 + 10% O + plasma (Lambros and Vanderberg 1979). The cultures were seeded in 96-well cell culture plates (Nunc) in the presence of 100 and 200 μM HABPs, testing each peptide in triplicate. After incubation for 18–20 h at 37°C in a 5% O2/5% CO2/90% N2 atmosphere, the supernatant was removed and the cells were stained with 15 μg/mL hydroethydine, incubated for 30 min at 37°C, and washed thrice with PBS. The suspensions were analyzed by flow cytometry using a FACsort in Log FL2 data mode and CellQuest software (Becton Dickinson immunocytometry system) (Wyatt et al. 1991). Infected and uninfected erythrocytes treated with 5 mM ethylene glycol tetraacetic acid (EGTA) and 2.9 mM chloroquine were used as positive invasion inhibition controls. Untreated infected erythrocytes were used as a negative control.
Acknowledgments
This research project was supported by COLCIENCIAS contract RC-2008. Nora Martinez's collaboration in translating and revising this manuscript is greatly appreciated.
Footnotes
Reprint requests to: Manuel Elkin Patarroyo, Carrera 50, No. 26-00, Bogotá 020304, Colombia; e-mail: mepatarr@mail.com; fax: 57-1-3244672/73 Ext. 108.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035923.108.
References
- Anthony, R.N., Yang, J., Krall, J.A., Sam-Yellowe, T.Y. Sequence analysis of the Rhop-3 gene of Plasmodium yoelii . J. Eukaryot. Microbiol. 2000;47:319–322. doi: 10.1111/j.1550-7408.2000.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Bannister, L., Mitchell, G. The ins, outs and roundabouts of malaria. Trends Parasitol. 2003;19:209–213. doi: 10.1016/s1471-4922(03)00086-2. [DOI] [PubMed] [Google Scholar]
- Baum, J., Maier, A.G., Good, R.T., Simpson, K.M., Cowman, A.F. Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLoS Pathog. 2005;1:e37. doi: 10.1371/journal.ppat.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breman, J.G., Alilio, M.S., Mills, A. Conquering the intolerable burden of malaria: What's new, what's needed: A summary. Am. J. Trop. Med. Hyg. 2004;71:1–15. [PubMed] [Google Scholar]
- Brown, H.J., Coppel, R.L. Primary structure of a Plasmodium falciparum rhoptry antigen. Mol. Biochem. Parasitol. 1991;49:99–110. doi: 10.1016/0166-6851(91)90133-q. [DOI] [PubMed] [Google Scholar]
- Campbell, G.H., Miller, L.H., Hudson, D., Franco, E.L., Andrysiak, P.M. Monoclonal antibody characterization of Plasmodium falciparum antigens. Am. J. Trop. Med. Hyg. 1984;33:1051–1054. doi: 10.4269/ajtmh.1984.33.1051. [DOI] [PubMed] [Google Scholar]
- Compton, L.A., Johnson W.C., Jr Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A., Ingram, L.T., Bushell, G.R., Fardoulys, C.A., Stenzel, D., Schofield, L., Saul, A.J. The 140/130/105 kilodalton protein complex in the rhoptries of Plasmodium falciparum consists of discrete polypeptides. Mol. Biochem. Parasitol. 1988;29:251–260. doi: 10.1016/0166-6851(88)90080-1. [DOI] [PubMed] [Google Scholar]
- Curtidor, H., Urquiza, M., Suarez, J.E., Rodriguez, L.E., Ocampo, M., Puentes, A., Garcia, J.E., Vera, R., Lopez, R., Ramirez, L.E., et al. Plasmodium falciparum acid basic repeat antigen (ABRA) peptides: Erythrocyte binding and biological activity. Vaccine. 2001;19:4496–4504. doi: 10.1016/s0264-410x(01)00202-x. [DOI] [PubMed] [Google Scholar]
- Doury, J.C., Goasdoue, J.L., Tolou, H., Martelloni, M., Bonnefoy, S., Mercereau-Puijalon, O. Characterisation of the binding sites of monoclonal antibodies reacting with the Plasmodium falciparum rhoptry protein RhopH3. Mol. Biochem. Parasitol. 1997;85:149–159. doi: 10.1016/s0166-6851(96)02819-8. [DOI] [PubMed] [Google Scholar]
- Espejo, F., Cubillos, M., Salazar, L.M., Guzman, F., Urquiza, M., Ocampo, M., Silva, Y., Rodriguez, R., Lioy, E., Patarroyo, M.E. Structure, immunogenicity, and protectivity relationship for the 1585 malarial peptide and its substitution analogues. Angew. Chem. Int. Ed. 2001;40:4654–4657. doi: 10.1002/1521-3773(20011217)40:24<4654::aid-anie4654>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Fujioka, H., Aikawa, M. Structure and life cycle. Chem. Immunol. 2002;80:1–26. doi: 10.1159/000058837. [DOI] [PubMed] [Google Scholar]
- Greenwood, B., Mutabingwa, T. Malaria in 2002. Nature. 2002;415:670–672. doi: 10.1038/415670a. [DOI] [PubMed] [Google Scholar]
- Hogh, B., Petersen, E., Crandall, I., Gottschau, A., Sherman, I.W. Immune responses to band 3 neoantigens on Plasmodium falciparum-infected erythrocytes in subjects living in an area of intense malaria transmission are associated with low parasite density and high hematocrit value. Infect. Immun. 1994;62:4362–4366. doi: 10.1128/iai.62.10.4362-4366.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghten, R.A. General method for the rapid solid-phase synthesis of large numbers of peptides: Specificity of antigen–antibody interaction at the level of individual amino acids. Proc. Natl. Acad. Sci. 1985;82:5131–5135. doi: 10.1073/pnas.82.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme, E.C. Receptor–ligand interactions: A practical approach. IRL Press; Oxford, UK: 1993. [Google Scholar]
- Kaneko, O. Erythrocyte invasion: Vocabulary and grammar of the Plasmodium rhoptry. Parasitol. Int. 2007;56:255–262. doi: 10.1016/j.parint.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Kats, L.M., Black, C.G., Proellocks, N.I., Coppel, R.L. Plasmodium rhoptries: How things went pear-shaped. Trends Parasitol. 2006;22:269–276. doi: 10.1016/j.pt.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Lambros, C., Vanderberg, J.P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Lanteri, C.A., Johnson, J.D., Waters, N.C. Recent advances in malaria drug discovery. Recent Patents Anti-Infect. Drug Disc. 2007;2:95–114. doi: 10.2174/157489107780832640. [DOI] [PubMed] [Google Scholar]
- Lopez, R., Valbuena, J., Rodriguez, L.E., Ocampo, M., Vera, R., Curtidor, H., Puentes, A., Garcia, J., Ramirez, L.E., Patarroyo, M.E. Plasmodium falciparum merozoite surface protein 6 (MSP-6) derived peptides bind erythrocytes and partially inhibit parasite invasion. Peptides. 2006;27:1685–1692. doi: 10.1016/j.peptides.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Merrifield, R.B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
- Mongui, A., Perez-Leal, O., Rojas-Caraballo, J., Angel, D.I., Cortes, J., Patarroyo, M.A. Identifying and characterising the Plasmodium falciparum RhopH3 Plasmodium vivax homologue. Biochem. Biophys. Res. Commun. 2007;358:861–866. doi: 10.1016/j.bbrc.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Ocampo, M., Curtidor, H., Vera, R., Valbuena, J.J., Rodriguez, L.E., Puentes, A., Lopez, R., Garcia, J.E., Tovar, D., Pacheco, P., et al. MAEBL Plasmodium falciparum protein peptides bind specifically to erythrocytes and inhibit in vitro merozoite invasion. Biochem. Biophys. Res. Commun. 2004;315:319–329. doi: 10.1016/j.bbrc.2004.01.050. [DOI] [PubMed] [Google Scholar]
- Ocampo, M., Rodriguez, L.E., Curtidor, H., Puentes, A., Vera, R., Valbuena, J.J., Lopez, R., Garcia, J.E., Ramirez, L.E., Torres, E., et al. Identifying Plasmodium falciparum cytoadherence-linked asexual protein 3 (CLAG 3) sequences that specifically bind to C32 cells and erythrocytes. Protein Sci. 2005;14:504–513. doi: 10.1110/ps.04883905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzón, C.G., Curtidor, H., Bermudez, A., Forero, M., Vanegas, M., Rodriguez, J., Patarroyo, M.E. Studies of Plasmodium falciparum rhoptry-associated membrane antigen (RAMA) protein peptides specifically binding to human RBC. Vaccine. 2008;26:853–862. doi: 10.1016/j.vaccine.2007.11.086. [DOI] [PubMed] [Google Scholar]
- Preiser, P., Kaviratne, M., Khan, S., Bannister, L., Jarra, W. The apical organelles of malaria merozoites: Host cell selection, invasion, host immunity and immune evasion. Microbes Infect. 2000;2:1461–1477. doi: 10.1016/s1286-4579(00)01301-0. [DOI] [PubMed] [Google Scholar]
- Puentes, A., Garcia, J., Vera, R., Lopez, Q.R., Urquiza, M., Vanegas, M., Salazar, L.M., Patarroyo, M.E. Serine repeat antigen peptides which bind specifically to red blood cells. Parasitol. Int. 2000;49:105–117. doi: 10.1016/s1383-5769(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Ridley, R.G., Takacs, B., Etlinger, H., Scaife, J.G. A rhoptry antigen of Plasmodium falciparum is protective in Saimiri monkeys. Parasitology. 1990;101:187–192. doi: 10.1017/s0031182000063228. [DOI] [PubMed] [Google Scholar]
- Rodriguez, L.E., Urquiza, M., Ocampo, M., Suarez, J., Curtidor, H., Guzman, F., Vargas, L.E., Trivinos, M., Rosas, M., Patarroyo, M.E. Plasmodium falciparum EBA-175 kDa protein peptides which bind to human red blood cells. Parasitology. 2000;120:225–235. doi: 10.1017/s003118209900551x. [DOI] [PubMed] [Google Scholar]
- Rodriguez, L.E., Ocampo, M., Vera, R., Puentes, A., Lopez, R., Garcia, J., Curtidor, H., Valbuena, J., Suarez, J., Rosas, J., et al. Plasmodium falciparum EBA-140 kDa protein peptides that bind to human red blood cells. J. Pept. Res. 2003;62:175–184. doi: 10.1034/j.1399-3011.2003.00079.x. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y. Plasmodium falciparum: Analysis of protein–protein interactions of the 140/130/110-kDa rhoptry protein complex using antibody and mouse erythrocyte binding assays. Exp. Parasitol. 1993;77:179–194. doi: 10.1006/expr.1993.1075. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y., Ndengele, M.M. Monoclonal antibody epitope mapping of Plasmodium falciparum rhoptry proteins. Exp. Parasitol. 1993;76:46–58. doi: 10.1006/expr.1993.1006. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y., Perkins, M.E. Binding of Plasmodium falciparum rhoptry proteins to mouse erythrocytes and their possible role in invasion. Mol. Biochem. Parasitol. 1990;39:91–100. doi: 10.1016/0166-6851(90)90011-a. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y., Perkins, M.E. Interaction of the 140/130/110 kDa rhoptry protein complex of Plasmodium falciparum with the erythrocyte membrane and liposomes. Exp. Parasitol. 1991;73:161–171. doi: 10.1016/0014-4894(91)90019-s. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y., Shio, H., Perkins, M.E. Secretion of Plasmodium falciparum rhoptry protein into the plasma membrane of host erythrocytes. J. Cell Biol. 1988;106:1507–1513. doi: 10.1083/jcb.106.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y., Wang, T., Fujioka, H., Drazba, J.A., Aikawa, M., Brochak, W. Sequence analysis of the Rhop-3 gene of Plasmodium berghei and P. chabaudi, reactivity of Rhop-3 protein within isolated rhoptries and binding of Rhop-3 to mouse erythrocytes. J. Protozool. Res. 2000;10:71–89. [Google Scholar]
- Sanders, P.R., Gilson, P.R., Cantin, G.T., Greenbaum, D.C., Nebl, T., Carucci, D.J., McConville, M.J., Schofield, L., Hodder, A.N., Yates J.R., III, et al. Distinct protein classes including novel merozoite surface antigens in raft-like membranes of Plasmodium falciparum . J. Biol. Chem. 2005;280:40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- Shirano, M., Tsuboi, T., Kaneko, O., Tachibana, M., Adams, J.H., Torii, M. Conserved regions of the Plasmodium yoelii rhoptry protein RhopH3 revealed by comparison with the P. falciparum homologue. Mol. Biochem. Parasitol. 2001;112:297–299. doi: 10.1016/s0166-6851(00)00366-2. [DOI] [PubMed] [Google Scholar]
- Siddiqui, W.A., Tam, L.Q., Kramer, K.J., Hui, G.S., Case, S.E., Yamaga, K.M., Chang, S.P., Chan, E.B., Kan, S.C. Merozoite surface coat precursor protein completely protects Aotus monkeys against Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. 1987;84:3014–3018. doi: 10.1073/pnas.84.9.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, R.J. Radioisotopes in biology. Oxford University Press; New York: 1990. pp. 192–199. [Google Scholar]
- Sreerama, N., Venyaminov, S.Y., Woody, R.W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama, N., Venyaminov, S.Y., Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Summer, A.P., Stauffer, W.M., Fischer, P.R. Pediatric malaria in the developing world. Semin. Pediatr. Infect. Dis. 2005;16:105–115. doi: 10.1053/j.spid.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Tam, J.P., Heath, W.F., Merrifield, R.B. SN 1 and SN 2 mechanisms for the deprotection of synthetic peptides by hydrogen fluoride. Studies to minimize the tyrosine alkylation side reaction. Int. J. Pept. Protein Res. 1983;21:57–65. doi: 10.1111/j.1399-3011.1983.tb03078.x. [DOI] [PubMed] [Google Scholar]
- Thwing, J., Skarbinski, J., Newman, R.D., Barber, A.M., Mali, S., Roberts, J.M., Slutsker, L., Arguin, P.M. Malaria surveillance—United States, 2005. MMWR Surveill. Summ. 2007;56:23–40. [PubMed] [Google Scholar]
- Topolska, A.E., Lidgett, A., Truman, D., Fujioka, H., Coppel, R.L. Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum . J. Biol. Chem. 2004;279:4648–4656. doi: 10.1074/jbc.M307859200. [DOI] [PubMed] [Google Scholar]
- Urquiza, M., Rodriguez, L.E., Suarez, J.E., Guzman, F., Ocampo, M., Curtidor, H., Segura, C., Trujillo, E., Patarroyo, M.E. Identification of Plasmodium falciparum MSP-1 peptides able to bind to human red blood cells. Parasite Immunol. 1996;18:515–526. doi: 10.1046/j.1365-3024.1996.d01-15.x. [DOI] [PubMed] [Google Scholar]
- Wang, L., Mohandas, N., Thomas, A., Coppel, R.L. Detection of detergent-resistant membranes in asexual blood-stage parasites of Plasmodium falciparum . Mol. Biochem. Parasitol. 2003;130:149–153. doi: 10.1016/s0166-6851(03)00165-8. [DOI] [PubMed] [Google Scholar]
- Wang, T., Fujioka, H., Drazba, J.A., Sam-Yellowe, T.Y. Rhop-3 protein conservation among Plasmodium species and induced protection against lethal P. yoelii and P. berghei challenge. Parasitol. Res. 2006;99:238–252. doi: 10.1007/s00436-006-0136-9. [DOI] [PubMed] [Google Scholar]
- WHO. Expert committee on malaria technical report. World Health Organization; Geneva: 2005. [Google Scholar]
- Wyatt, C.R., Goff, W., Davis, W.C. A flow cytometric method for assessing viability of intraerythrocytic hemoparasites. J. Immunol. Methods. 1991;140:23–30. doi: 10.1016/0022-1759(91)90122-v. [DOI] [PubMed] [Google Scholar]
- Yamamura, H.I., Enna, S.J., Kuhar, M.J. Neurotransmitter receptor binding. Raven Press; New York: 1978. [Google Scholar]
- Yang, J.C., Blanton, R.E., King, C.L., Fujioka, H., Aikawa, M., Sam-Yellowe, T.Y. Seroprevalence and specificity of human responses to the Plasmodium falciparum rhoptry protein Rhop-3 determined by using a C-terminal recombinant protein. Infect. Immun. 1996;64:3584–3591. doi: 10.1128/iai.64.9.3584-3591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]