Abstract
G protein-coupled receptors (GPCRs) are notoriously difficult to express, particularly in microbial systems. Using GPCR fusions with the green fluorescent protein (GFP), we conducted studies to identify bacterial host effector genes that result in a general and significant enhancement in the amount of membrane-integrated human GPCRs that can be produced in Escherichia coli. We show that coexpression of the membrane-bound AAA+ protease FtsH greatly enhances the expression yield of four different class I GPCRs, irrespective of the presence of GFP. Using this new expression system, we produced 0.5 and 2 mg/L of detergent-solubilized and purified full-length central cannabinoid receptor (CB1) and bradykinin receptor 2 (BR2) in shake flask cultures, respectively, two proteins that had previously eluded expression in microbial systems.
Keywords: G protein-coupled receptor, Escherichia coli, membrane protein, FtsH, genetic engineering, cannabinoid receptor, bradykinin receptor
The array of functions that membrane proteins (MPs) perform in cells is very broad, ranging from the maintenance of cell structure and protection against toxins in the environment to the production of energy and respiration. Consistent with these vital roles, genes for MPs typically account for 15%–30% of an organism's genome. However, <0.5% of the protein structures contained in the Protein Data Bank are of MPs. Furthermore, the great majority of extant MP structures are of prokaryotic proteins, with only about two dozen distinct eukaryotic MP structures currently available to the public (see http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html for a well-curated database of known MP structures). The reason for this relative lag in structural biology efforts toward MPs is twofold: The first bottleneck is the determination of suitable crystallization conditions (Loll 2003). The second limitation is the production of sufficient amounts of MP suitable for initiation/optimization of crystallization trials. The latter limitation is particularly pertinent to mammalian MPs, which may be present in only miniscule quantities in native tissues. The most promising avenue for addressing the protein production bottleneck is the development of robust heterologous protein expression systems in well-understood organisms (Grisshammer and Tate 1995; Link and Georgiou 2007).
One class of mammalian MPs of particular interest is the G protein-coupled receptor (GPCR) family. These proteins are characterized by a seven-transmembrane helical topology, and represent the most abundant class of MPs in animals with ∼1100 genes (5% of the genome) in the model organism Caernohabditis elegans (Bargmann 1998). In addition to their prevalence, GPCRs are of outstanding pharmacological interest, and it has been estimated that as many as 65% of currently marketed small-molecule drugs target these receptors (Bartfai et al. 2004). Despite their importance from both a basic biological standpoint and as an avenue for human health, structural biology efforts have yielded only two distinct GPCR structures: those of bovine and squid rhodopsin (Palczewski et al. 2000; Murakami and Kouyama 2008) and of the human β2 adrenergic receptor (Cherezov et al. 2007; Rasmussen et al. 2007). The determination of the rhodopsin structures was facilitated by the natural abundance of this protein in the eye, while material for crystallization of the β2 adrenergic receptor was obtained by heterologous expression of the protein in an insect cell line.
Microbial organisms, such as the bacterium Escherichia coli, have been extensively used for the expression of MPs for structural studies (Wagner et al. 2006) given their ease of growth and manipulation, and ability to be propagated in chemically defined medium, allowing for substitution with selenomethionine for X-ray crystal structure determination (Hendrickson et al. 1990) or with isotopically labeled amino acids for NMR structural studies. However, when expressed in E. coli, GPCRs are often toxic to the host cell, are subject to degradation, or accumulate in inclusion bodies that, with some exceptions (Busuttil et al. 2001; Baneres et al. 2003; 2005; Tian et al. 2005), are difficult to solubilize and refold (Lundstrom et al. 2006). Only a very limited number of human class I GPCR, such as the peripheral cannabinoid receptor and the neurotensin receptor, fused to polypeptides that prevent aggregation or enhance stability, have been expressed in membrane-integrated form in E. coli (Tucker and Grisshammer 1996; Calandra et al. 1997).
In this work, we have examined the effects of the overexpression of a panel of translocon components, molecular chaperones, and other proteins involved in E. coli MP biogenesis on the expression of four human class I GPCRs (Bockaert and Pin 1999) in E. coli: the central and peripheral cannabinoid receptors (CB1 and CB2, respectively), the bradykinin receptor 2 (BR2), and the neurokinin (Substance P) receptor 1 (NKR1). In order to rapidly and quantitatively assess the effect of these factors, we constructed GPCR–GFP fusions and used GFP fluorescence as a reporter of GPCR expression. The fluorescence of MP–GFP fusions has been previously found to be a good indicator of the amount of membrane-localized MPs (Drew et al. 2005). We found that coexpression of the membrane-anchored AAA+ protease FtsH significantly enhanced the expression of all the GPCRs tested, as well as the amount of recombinant CB1 and BR2 that can be solubilized by detergents and isolated. The availability of a system for the expression of GPCRs in bacteria that can produce large amounts of membrane-bound protein constitutes the first step toward the ultimate goal of obtaining large quantities of ligand-binding receptors for structural studies.
Results
We investigated the expression of four human class I GPCRs in E. coli: CB1, CB2, BR2, and NKR1. Each of these receptors is of therapeutic interest with the cannabinoid receptors being targets for pain, nausea, anxiety, glaucoma, and wasting diseases (Porter and Felder 2001); BR2 a target for inflammation and pain (Marceau and Regoli 2004); whereas NKR1 antagonists are potential antidepressants and antiemetics for cancer patients who receive chemotherapy (Brain and Cox 2006). In earlier studies, CB1 could be expressed only at trace levels in E. coli (Calandra et al. 1997). BR2 has not been expressed in microbial systems to date, and it expresses only with poor yields in mammalian cell cultures (Camponova et al. 2007). Finally, NKR1 expressed in bacterial or yeast cell hosts could not be properly localized in the membrane (Butz et al. 2003; Bane et al. 2007).
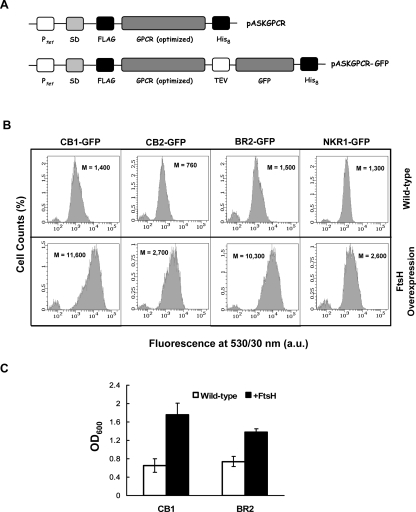
Fully E. coli codon-optimized genes were synthesized and fused to sequences encoding an N-terminal FLAG octapeptide and C-terminal octahistidine, and placed under the control of the tet promoter of the vector pASK75 (Skerra 1994) to minimize leaky expression, generating the plasmid family pASKGPCR (Fig. 1A). The motivation for using fully codon-optimized genes is the presence of several rare codons in the sequences of some of the GPCRs studied. For example, the human CB1 includes nine of the rare AGG codon for arginine in its cDNA sequence. We also constructed vectors expressing C-terminal fusions of the codon-optimized GPCR genes with GFP (pASKGPCR-GFP plasmid family; Fig. 1A). Previous work on GFP fusions of bacterial MPs indicated that the total cell GFP fluorescence is a good indicator of the amount of properly targeted and folded MP (Drew et al. 2005; 2006).
Figure 1.
(A) Constructs used in this study: (top) The pASKGPCR family of expression vectors includes a codon-optimized GPCR gene, flanked by affinity tags (N-terminal FLAG octapeptide and C-terminal octahistidine tag), under the control of the tet promoter with an optimized Shine-Dalgarno (SD) sequence. (Bottom) The pASKGPCR–GFP plasmid family encodes GPCRs with C-terminally fused GFP and a tobacco etch virus (TEV) cleavage site between the GPCR and GFP. (B) Fluorescence histograms of cells expressing GPCR–GFP fusions either without (top row) or with (bottom row) FtsH coexpression. FtsH coexpression results in at least a twofold, and up to an eightfold, increase in the mean cell fluorescence (M) of all GPCRs tested. Fluorescence histograms correspond to a population of 10,000 cells. a.u.: arbitrary units. (C) FtsH coexpression results in a growth enhancement of cultures expressing GPCRs. Cell densities at saturation after 36 h of expression at 12°C for CB1 and BR2 are reported. Data represent the average value of three independent experiments, and the error bars represent one SD from the mean value. All cultures were induced at OD600 between 0.5 and 0.7. OD600: optical density at 600 nm.
Initially, we monitored the effect of coexpression of a panel of select E. coli proteins known to affect bacterial protein translocation, folding, degradation, or membrane insertion on the fluorescence of cells expressing CB1–GFP (Table 1). As the majority of MPs in E. coli are targeted to the membrane by the signal recognition particle (SRP) and inserted into the plasma membrane by the Sec translocon (Luirink and Sinning 2004), the effects of overproduction of the protein and RNA components of the SRP (Ffh and the 4.5S RNA), the SRP receptor FtsY, and translocon components SecY and SecE were investigated. Similarly, we examined the effect of the membrane integrase YidC (Samuelson et al. 2000), which had been implicated in Sec-independent translocation of certain MPs (van Bloois et al. 2005). We also investigated the role of the cytoplasmic molecular chaperones/co-chaperones DnaK/DnaJ, SecB, and GroEL/GroES, as well as the peptidyl-prolyl isomerase trigger factor (Tig). Finally, we also investigated the effects of overexpressing the membrane-anchored AAA+ protease FtsH (also called HflB), which appears to play an important role in MP degradation and quality control (Akiyama et al. 1996; Ito and Akiyama 2005).
Table 1.
Effects of various helper proteins on the expression of CB1–GFP as assessed by flow cytometry
Coexpression of Ffh, DnaK/DnaJ, or Tig resulted in a modest but reproducible (two- to threefold) increase in CB1–GFP fluorescence (Table 1). More importantly, a large (eightfold) increase in fluorescence was observed in cells coexpressing the membrane-anchored AAA+ protease FtsH (Fig. 1B). In addition, coexpression of FtsH enhanced bacterial growth, resulting in nearly twofold higher final cell density (Fig. 1C). Similarly large increases in cell fluorescence and enhancements in growth were observed with all four GPCR-GFP fusions tested (Fig. 1B,C, and data not shown). Plating experiments on the cells following protein expression confirmed that the increased growth observed in FtsH coexpression cultures was not due to increased plasmid loss.
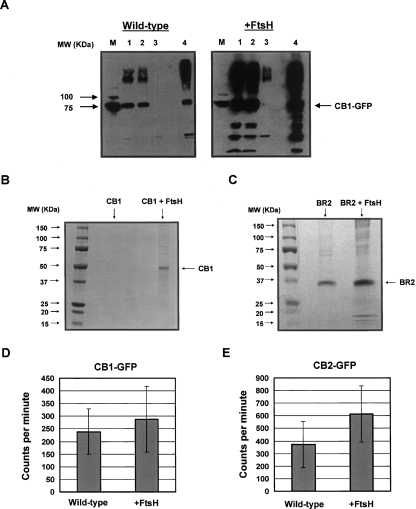
Western blotting of cell lysates and of membrane fractions from cells expressing CB1–GFP revealed that FtsH coexpression results in a large increase in expression of intact and membrane-targeted CB1–GFP fusion (Fig. 2A).
Figure 2.
(A) Western blot analysis (antipoly-His) on MC4100A cells expressing CB1–GFP with either empty pBAD33 (left) or pBAD–FtsH (right), indicating an enhancement of CB1–GFP expression in both total cell lysates and total membrane fractions. A sample corresponding to an equal number of cells was loaded in each lane. M, molecular weight marker; 1, total cell lysate; 2, insoluble fraction; 3, supernatant from membrane pellet; 4, total membranes. (B) SDS-PAGE analysis of purified CB1 either without or with FtsH coexpression. The expected molecular weight of full-length CB1 is ∼53 kDa. (C) SDS-PAGE analysis of purified BR2 either without or with FtsH coexpression. The expected molecular weight of full-length BR2 is ∼45 kDa. The increased electrophoretic mobility observed here occurs frequently with MPs, and it has been previously observed with BR2 expressed in human cell culture (Camponova et al. 2007). The intactness of purified BR2 was verified with Western blotting using anti-His and anti-FLAG antibodies (data not shown). (D) Specific CP 55,940 binding of cells expressing CB1–GFP in the presence and absence of FtsH overexpression. Error bars represent the standard error of the mean from four replicates. Data were taken at the saturating ligand concentration of 10 nM. (E) Specific CP 55,940 binding of cells expressing CB2–GFP in the presence or absence of FtsH overexpression. Error bars are as described above in D, and data were taken at a saturating ligand concentration of 10 nM.
As described in previous work, the GFP moiety simplifies the selection of a detergent suitable for MP solubilization (Drew et al. 2005). Optimal detergents for solubilizing CB1–GFP and BR2–GFP were identified by performing a detergent screen (see Materials and Methods). CB1 and BR2 lacking the GFP fusion partner were expressed in the presence and absence of FtsH at 12°C for ∼36 h in order to minimize receptor proteolysis that was observed at higher temperatures, although increased GPCR–GFP fluorescence and suppression of growth arrest associated with FtsH coexpression were observed at all expression temperatures tested up to 25°C. Following solubilization of the proteins with the detergents Cymal-7 and LMPC, respectively, and purification via immobilized metal affinity chromatography, protein yields were measured using the bicinchoninic acid (BCA) assay. Overexpression of FtsH led to a sevenfold increase in the yield of purified CB1 under these conditions (500 μg/L with FtsH overexpression vs. 70 μg/L without; Fig. 2B). A similar large increase in the yield of purified receptor was also observed with BR2. In this case, FtsH overexpression led to an eightfold increase in the production of purified BR2, resulting in 2 mg/L of isolated receptor (Fig. 2C). For comparison, mammalian expression of BR2 in human embryonic kidney-293S cells yielded only 140 μg/L of purified protein (Camponova et al. 2007).
GPCRs expressed in microbial systems are typically inactive, with only a fraction of the protein present in a ligand-binding conformation (Sarramegna et al. 2003; White et al. 2004). Grishammer and coworkers reported that CB1 expressed in E. coli is essentially inactive, presumably due to heavy proteolysis, whereas bacterially expressed CB2 retains ligand-binding activity (Calandra et al. 1997). In order to investigate whether coexpression of FtsH affects the amount of active protein in addition to increasing the overall GPCR yield, we performed saturation binding assays with the synthetic radioligand CP 55,940 on whole cells expressing either CB1–GFP or CB2–GFP. These experiments revealed that both CB1 and CB2 receptors are produced in active form (Fig. 2D,E). The data for specific CP 55,940 binding to cells expressing CB1–GFP could be fit to a single-site saturation model and indicate that ∼60 active receptors per cell are produced (Supplemental Fig. 1A). However, coexpression of FtsH does not appear to result in a significant enhancement in the amount of active receptors (Fig. 2D; Supplemental Fig. 1B). It should be noted that the inherent “stickiness” of the CP 55,940 ligand and the accompanying large errors in the measurements interferes with the quantitative determination of the differences in the levels of active protein. For cells expressing CB2–GFP at a saturating concentration of CP 55,940 (10 nM), the specific radioligand binding of cells expressing both CB2–GFP and FtsH was nearly twofold higher than that of cells expressing CB2–GFP alone (Fig. 2E; Supplemental Fig. 2), indicating a modest increase in the production of active receptors upon FtsH coexpression.
Discussion
In this work, we used GPCR–GFP fusions and flow cytometry to investigate the effect of the overexpression of a number of bacterial effector genes on GPCR production in E. coli. We found that coexpression of the membrane-bound AAA+ protease FtsH results in a marked increase in the bacterial expression yield of several GPCRs, including CB1 and BR2. BR2 has not been previously expressed in microbial hosts, and CB1 was expressed in E. coli with vanishingly low yields due to extensive proteolysis (Calandra et al. 1997). It must also be noted that CB1 and BR2 production is low even when these receptors are expressed in higher animal cell cultures (Andersson et al. 2003; Camponova et al. 2007). We have found that using codon-optimized genes and expression at a very low temperature results in the production of a small but significant number of active CB1 receptor molecules, as determined by ligand-binding assays. To our knowledge, CB1 had not been previously expressed in active form in bacteria, even as part of a fusion with the generally stabilizing maltose-binding protein (Calandra et al. 1997). FtsH coexpression, however, does not increase the amount of active CB1 significantly, while it appears to enhance the production of ligand-binding-competent CB2 only modestly. Nonetheless, having established a system for the production of markedly enhanced amounts of membrane-bound GPCRs in E. coli, additional studies can be undertaken to maximize the yield of active receptors. Ultimately, the use of a receptor-specific ligand column (White et al. 2004) to separate active receptors from inactive, soluble receptors may be needed in order to prepare material suitable for biophysical studies.
Given that FtsH is a protease with a putative role in MP quality control, our observed results were unexpected. It had been previously suggested that abrogation of the proteolytic activity of FtsH might lead to an increase in the yield of MPs overexpressed in E. coli (Wagner et al. 2007). The results presented here point to the opposite conclusion, namely, that overexpression of FtsH is a positive effector for MP production in E. coli. Preliminary results have shown that FtsH overexpression also increases the bacterial production of native E. coli MPs such as YidC.
The proteolytic activity of FtsH appears to be necessary for the observed increase in GPCR expression, as a deletion mutant of FtsH in which the protease domain (amino acids 399–644) of the protein was completely removed, failed to improve expression of CB1–GFP as judged by flow cytometry, or suppress the growth arrest that accompanies CB1 production (unpublished results). Furthermore, it appears that FtsH is unique among similar proteins in its ability to promote GPCR expression. Overexpression of the membrane-associated, stress-induced AAA+ protease HtpX (Sakoh et al. 2005) and the putative membrane-associated AAA+ proteases YcaL, YfgC, and YggG on CB1–GFP expression was tested, but none of these proteins led to increased GPCR expression or suppression of growth arrest (data not shown). Preliminary transcriptome analysis indicated a marked upregulation of genes involved in glycerol metabolism, indicating possible changes in the lipid composition of the cytoplasmic membrane (Flower 2001).
This work on the genetic optimization of the expression host represents a key step toward the “rationalization” of MP expression advocated by Wagner et al. (2006). While our work here has identified a novel positive effector of MP expression in E. coli, the use of GPCR–GFP or other MP–GFP fusions is particularly well suited for more global, library-based approaches (Link et al. 2007). The techniques described herein may prove invaluable for the discovery of other overexpressed genes or, alternatively, of chromosomal lesions that improve the MP expression capacity of the host. Studies along these lines are under way in our laboratory.
Materials and Methods
Strains and plasmids
The E. coli strain MC4100A (MC4100 ara+) (Santini et al. 2001) was used for all experiments. The construction of the plasmid families pASKGPCR and pASKGPCR–GFP is described in the Supplemental material.
Protein expression
MC4100A cells were freshly transformed with the appropriate GPCR encoding-plasmid and with either a second plasmid encoding a putative effector of GPCR expression or empty pBAD33. Following incubation in SOC medium for 1 h, the cells were plated on LB plates with 100 mg/L ampicillin and 25 mg/L chloramphenicol. Single colonies were inoculated into LB with antibiotics. Following ∼10 h of growth at 37°C, the culture was diluted 500-fold into fresh media with antibiotics and containing 0.2% arabinose. The cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.5–0.7, at which point the culture was shifted to 12°C. Following a short temperature equilibration step, protein synthesis was induced by the addition of anhydrotetracycline at a concentration of 0.2 mg/L.
SDS-PAGE and Western blotting
SDS-PAGE was performed on 4%–20% gradient precast gels (NuSep). Detection of proteins with Western blots was performed using horseradish peroxidase-conjugated antipolyhistidine or anti-FLAG antibodies (Sigma) following the manufacturer's recommendations.
Flow cytometry
Cell fluorescence was analyzed using a BD FACSAria flow cytometer and FACSDiva software.
Detergent screening and protein purification
Detergent screening and protein purification procedures are described in the Supplemental material.
Radioligand-binding assays
Assays were performed on CB1 and CB2 essentially as described elsewhere (Felder et al. 1992), except that Millipore 96-well HTS plates were used. Each well contained 5 × 107 cells in assay buffer (50 mM Tris, 5 mM MgCl2, 2.5 mM EDTA, 5 mg/mL bovine serum albumin, pH 7.4) with concentrations of 3H-CP 55,940 (Perkin-Elmer) ranging from 0 to 10 nM. To measure nonspecific binding, non-radioactive CP 55,940 (Sigma) was added at a concentration of 5 μM. This control was critical as ∼80% of the observed binding of CP 55,940 was nonspecific. Wells were washed three times with wash buffer (50 mM Tris, 0.5 mg/mL bovine serum albumin, pH 7.4) before removal of the membrane and scintillation counting. Each measurement was repeated four times. Data were fit to a single-site saturation-binding model using Sigmaplot software (SPSS).
Acknowledgments
This work was supported by the Welch Foundation (grant Welch F-1629) and by the National Institutes of Health (grant NIH GM 55090). A.J.L. was supported by an NIH Kirschstein/NRSA postdoctoral fellowship.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: George Georgiou, Department of Chemical Engineering, University of Texas at Austin, 1 University Station C0400, Austin, TX 78712-0231, USA; e-mail: gg@che.utexas.edu; fax: (512) 471-7963.
Abbreviations: GPCR, G protein-coupled receptor; MP, membrane protein; GFP, green fluorescent protein; CB1, central cannabinoid receptor; CB2, peripheral cannabinoid receptor; BR2, bradykinin receptor 2; NKR1, neurokinin (substance P) receptor 1; SRP, signal recognition particle; LMPC, 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035980.108.
References
- Akiyama, Y., Kihara, A., Tokuda, H., Ito, K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- Andersson, H., D'Antona, A.M., Kendall, D.A., Von Heijne, G., Chin, C.N. Membrane assembly of the cannabinoid receptor 1: Impact of a long N-terminal tail. Mol. Pharmacol. 2003;64:570–577. doi: 10.1124/mol.64.3.570. [DOI] [PubMed] [Google Scholar]
- Bane, S.E., Velasquez, J.E., Robinson, A.S. Expression and purification of milligram levels of inactive G protein-coupled receptors in E. coli . Protein Expr. Purif. 2007;52:348–355. doi: 10.1016/j.pep.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneres, J.L., Martin, A., Hullot, P., Girard, J.P., Rossi, J.C., Parello, J. Structure-based analysis of GPCR function: Conformational adaptation of both agonist and receptor upon leukotriene B-4 binding to recombinant BLT1. J. Mol. Biol. 2003;329:801–814. doi: 10.1016/s0022-2836(03)00438-8. [DOI] [PubMed] [Google Scholar]
- Baneres, J.L., Mesnier, D., Martin, A., Joubert, L., Dumuis, A., Bockaert, J. Molecular characterization of a purified 5-HT4 receptor—a structural basis for drug efficacy. J. Biol. Chem. 2005;280:20253–20260. doi: 10.1074/jbc.M412009200. [DOI] [PubMed] [Google Scholar]
- Bargmann, C.I. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- Bartfai, T., Benovic, J.L., Bockaert, J., Bond, R.A., Bouvier, M., Christopoulos, A., Civelli, O., Devi, L.A., George, S.R., Inui, A., et al. The state of GPCR research in 2004. Nat. Rev. Drug Discov. 2004;3:574–626. doi: 10.1038/nrd1458. [DOI] [PubMed] [Google Scholar]
- Bockaert, J., Pin, J.P. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain, S.D., Cox, H.M. Neuropeptides and their receptors: Innovative science providing novel therapeutic targets. Br. J. Pharmacol. 2006;147:S202–S211. doi: 10.1038/sj.bjp.0706461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil, B.E., Turney, K.L., Frauman, A.G. The expression of soluble, full-length, recombinant human TSH receptor in a prokaryotic system. Protein Expr. Purif. 2001;23:369–373. doi: 10.1006/prep.2001.1519. [DOI] [PubMed] [Google Scholar]
- Butz, J.A., Niebauer, R.T., Robinson, A.S. Co-expression of molecular chaperones does not improve the heterologous expression of mammalian G protein-coupled receptor expression in yeast. Biotechnol. Bioeng. 2003;84:292–304. doi: 10.1002/bit.10771. [DOI] [PubMed] [Google Scholar]
- Calandra, B., Tucker, J., Shire, D., Grisshammer, R. Expression in Escherichia coli and characterisation of the human central CB1 and peripheral CB2 cannabinoid receptors. Biotechnol. Lett. 1997;19:425–428. [Google Scholar]
- Camponova, P., Baud, S., Mattras, H., Duroux-Richard, I., Bonnafous, J.C., Marie, J. High-level expression and purification of the human bradykinin B2 receptor in a tetracycline-inducible stable HEK293S cell line. Protein Expr. Purif. 2007;55:300–311. doi: 10.1016/j.pep.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Cherezov, V., Rosenbaum, D.M., Hanson, M.A., Rasmussen, S.G.F., Thian, F.S., Kobilka, T.S., Choi, H.J., Kuhn, P., Weis, W.I., Kobilka, B.K., et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, D., Slotboom, D.J., Friso, G., Reda, T., Genevaux, P., Rapp, M., Meindl-Beinker, N.M., Lambert, W., Lerch, M., Daley, D.O., et al. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci. 2005;14:2011–2017. doi: 10.1110/ps.051466205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, D., Lerch, M., Kunji, E., Slotboom, D.J., de Gier, J.W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- Felder, C.C., Veluz, J.S., Williams, H.L., Briley, E.M., Matsuda, L.A. Cannabinoid agonists stimulate both receptor-mediated and nonreceptor-mediated signal transduction pathways in cells transfected with and expressing cannabinoid receptor clones. Mol. Pharmacol. 1992;42:838–845. [PubMed] [Google Scholar]
- Flower, A.M. SecG function and phospholipid metabolism in Escherichia coli . J. Bacteriol. 2001;183:2006–2012. doi: 10.1128/JB.183.6.2006-2012.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisshammer, R., Tate, C.G. Overexpression of integral membrane-proteins for structural studies. Q. Rev. Biophys. 1995;28:315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A., Horton, J.R., Lemaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD)—a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Akiyama, Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- Link, A.J., Georgiou, G. Advances and challenges in membrane protein expression. AIChE J. 2007;53:752–756. [Google Scholar]
- Link, A.J., Jeong, K.J., Georgiou, G. Beyond toothpicks: New methods for isolating mutant bacteria. Nat. Rev. Microbiol. 2007;5:680–688. doi: 10.1038/nrmicro1715. [DOI] [PubMed] [Google Scholar]
- Loll, P.J. Membrane protein structural biology: The high-throughput challenge. J. Struct. Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Luirink, J., Sinning, I. SRP-mediated protein targeting: Structure and function revisited. Bichim. Biophys. Acta. 2004;1694:17–35. doi: 10.1016/j.bbamcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Lundstrom, K., Wagner, R., Reinhart, C., Desmyter, A., Cherouati, N., Magnin, T., Zeder-Lutz, G., Courtot, M., Prual, C., Andre, N., et al. Structural genomics on membrane proteins: Comparison of more than 100 GPCRs in 3 expression systems. J. Struct. Funct. Genomics. 2006;7:77–91. doi: 10.1007/s10969-006-9011-2. [DOI] [PubMed] [Google Scholar]
- Marceau, F., Regoli, D. Bradykinin receptor ligands: Therapeutic perspectives. Nat. Rev. Drug Discov. 2004;3:845–852. doi: 10.1038/nrd1522. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Kouyama, T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- Palczewski, K., Kumasaka, T., Hori, T., Behnke, C.A., Motoshima, H., Fox, B.A., Le Trong, I., Teller, D.C., Okada, T., Stenkamp, R.E., et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Porter, A.C., Felder, C.C. The endocannabinoid nervous system: Unique opportunities for therapeutic intervention. Pharmacol. Ther. 2001;90:45–60. doi: 10.1016/s0163-7258(01)00130-9. [DOI] [PubMed] [Google Scholar]
- Rasmussen, S.G.F., Choi, H.J., Rosenbaum, D.M., Kobilka, T.S., Thian, F.S., Edwards, P.C., Burghammer, M., Ratnala, V.R.P., Sanishvili, R., Fischetti, R.F., et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Sakoh, M., Ito, K., Akiyama, Y. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli . J. Biol. Chem. 2005;280:33305–33310. doi: 10.1074/jbc.M506180200. [DOI] [PubMed] [Google Scholar]
- Samuelson, J.C., Chen, M.Y., Jiang, F.L., Moller, I., Wiedmann, M., Kuhn, A., Phillips, G.J., Dalbey, R.E. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- Santini, C.L., Bernadac, A., Zhang, M., Chanal, A., Ize, B., Blanco, C., Wu, L.F. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 2001;276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- Sarramegna, V., Talmont, F., Demange, P., Milon, A. Heterologous expression of G protein-coupled receptors: Comparison of expression systems fron the standpoint of large-scale production and purification. Cell. Mol. Life Sci. 2003;60:1529–1546. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra, A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli . Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- Tian, C.L., Breyer, R.M., Kim, H.J., Karra, M.D., Friedman, D.B., Karpay, A., Sanders, C.R. Solution NMR spectroscopy of the human vasopressin V2 receptor, a G protein-coupled receptor. J. Am. Chem. Soc. 2005;127:8010–8011. doi: 10.1021/ja051161b. [DOI] [PubMed] [Google Scholar]
- Tucker, J., Grisshammer, R. Purification of a rat neurotensin receptor expressed in Escherichia coli . Biochem. J. 1996;317:891–899. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bloois, E., Nagamori, S., Koningstein, G., Ullers, R.S., Preuss, M., Oudega, B., Harms, N., Kaback, H.R., Herrmann, J.M., Luirink, J. The Sec-independent function of Escherichia coli YidC is evolutionarily-conserved and essential. J. Biol. Chem. 2005;280:12996–13003. doi: 10.1074/jbc.M414094200. [DOI] [PubMed] [Google Scholar]
- Wagner, S., Bader, M.L., Drew, D., de Gier, J.W. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wagner, S., Baars, L., Ytterberg, A.J., Klussmeier, A., Wagner, C.S., Nord, O., Nygren, P.A., van Wijk, K.J., de Gier, J.W. Consequences of membrane protein overexpression in Escherichia coli . Mol. Cell. Proteomics. 2007;6:1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- White, J.F., Trinh, L.B., Shiloach, J., Grisshammer, R. Automated large-scale purification of a G protein-coupled receptor for neurotensin. FEBS Lett. 2004;564:289–293. doi: 10.1016/S0014-5793(04)00195-4. [DOI] [PubMed] [Google Scholar]