Abstract
The fibroblast growth factor receptor (FGFR) can be activated through direct interaction with the neural cell adhesion molecule (NCAM). The extracellular part of the FGFR consists of three immunoglobulin-like (Ig) modules, and that of the NCAM consists of five Ig and two fibronectin type III (F3) modules. NCAM–FGFR interactions are mediated by the third FGFR Ig module and the second NCAM F3 module. Using surface plasmon resonance and nuclear magnetic resonance analyses, the present study demonstrates that the second Ig module of FGFR also is involved in binding to the NCAM. The second Ig module residues involved in binding were identified and shown to be localized on the “opposite sides” of the module, indicating that when NCAMs are clustered (e.g., due to homophilic binding), high-affinity FGFR binding sites may be formed by the neighboring NCAMs.
Keywords: FGF receptor, NCAM, F3 module, NMR spectroscopy, surface plasmon resonance spectroscopy
The fibroblast growth factor (FGF) receptors (FGFR) 1 through 4 can be activated by their cognate ligands, the FGFs (Mc Keehan et al. 1998; Itoh and Ornitz 2004), and by cell adhesion molecules (CAMs) such as the neural cell adhesion molecule (NCAM), L1-CAM, and N-cadherin (Doherty and Walsh 1996; Kiselyov et al. 2003). Whereas the basic principles of FGFR activation by FGFs are well understood (Plotnikov et al. 1999; Pellegrini et al. 2000), the mechanism of FGFR activation by CAMs is not clear. In the absence of FGF, FGFR exists in the cell membrane as a monomeric protein that dimerizes upon binding to FGF. FGFR dimerization brings the tyrosine kinase domains of two receptor molecules into close proximity to each other, followed by autophosphorylation of the kinase domains, and thus receptor activation. In contrast to transiently expressed FGFs, CAMs are expressed constitutatively, and are thought to activate FGFR only when they are involved in cell–cell adhesion (Kiselyov et al. 2005).
The NCAM is a cell-surface glycoprotein belonging to the Ig superfamily (Berezin et al. 2000; Kiselyov et al. 2005). The NCAM can be expressed as three major isoforms (A, B, and C), with differences in the cytoplasmic domain. The extracellular region of the NCAM is identical for the three isoforms, and consists of five Ig-like and two fibronectin type III (F3) modules. The NCAM plays a major role during nervous system development, mediating adhesion between neural cells, stimulating neurite outgrowth and fasciculation, and promoting cell survival and synaptic plasticity (Cremer et al. 1997; Berezin et al. 2000; Bruses and Rutishauser 2001; Rougon and Hobert 2003; Walmod et al. 2004). The NCAM mediates cell–cell adhesion through homophilic binding and regulates neurite outgrowth via FGFR (Doherty and Walsh 1996; Kiselyov et al. 2003). The mechanism of the NCAM homophilic binding, although extensively studied, is still controversial (for a thorough review of the available structural data, see Kiselyov et al. 2005). The Ig1 and Ig2 modules of the NCAM were demonstrated to bind to each other, which indicates that these modules are involved in a symmetrical double-reciprocal interaction (Kiselyov et al. 1997; Atkins et al. 1999; Jensen et al. 1999; Kasper et al. 2000). Recently, the crystal structure of the first three N-terminal NCAM modules has been determined (Soroka et al. 2003), and based on this structure, a model of the NCAM homophilic binding has been suggested. According to this model, interactions between the Ig1 and Ig2 modules lead to formation of the NCAM cis-dimers on the surface of the same cell. The cis-dimers from two opposing cells can form two kinds of one-dimensional “zippers.” When combined, the two types of “zippers” may form a two-dimensional “zipper.”
The extracellular region of the prototypical FGFR consists of three Ig modules of the intermediate subtype (Plotnikov et al. 1999; Pellegrini et al. 2000; Kiselyov et al. 2006a). The Ig2 and Ig3 modules mediate binding to FGF and heparin, whereas the Ig1 module has an autoinhibitory function (Wang et al. 1995; Olsen et al. 2004) through direct binding to Ig2 (Kiselyov et al. 2006b). The FGFR site involved in binding to the NCAM has been mapped to the Ig3 module, and the corresponding NCAM site to the F3(2) module (Kiselyov et al. 2003). Affinity of interaction between the FGFR Ig2–3 modules and the F3(1–2) modules of the NCAM is much higher than affinity of interaction between the single Ig3 and the F3(2) module. Therefore, it seems likely that Ig2 and the F3(1) module also are involved in this interaction. Thus, it is of interest to test whether the FGFR Ig2 module is involved in the FGFR–NCAM interaction.
Results
To study the possible binding between the FGFR Ig2 module and the NCAM, recombinant proteins of the NCAM F3(1–2) modules and the FGFR Ig2 module expressed in a yeast expression system of Pichia pastoris were used. Both proteins were properly folded as judged by one-dimensional NMR.
The FGFR Ig2 module binds to the NCAM
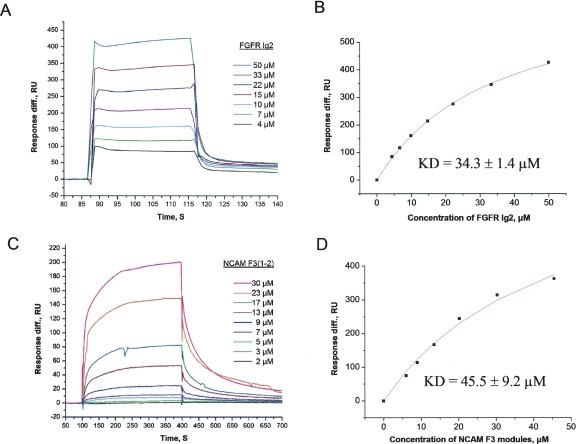
It has previously been shown by SPR that the double-module construct of the F3(1–2) modules of the NCAM binds to the double Ig2–3 FGFR module construct with a dissociation constant (Kd) of 10 μM (Kiselyov et al. 2003). However, interactions between the individual modules were minimally detected by SPR, indicating that both NCAM modules as well as both FGFR modules are involved in binding or are necessary for maximal binding. By using a method more sensitive to weak binding, NMR, an interaction between the F3(2) module of the NCAM and the FGFR Ig3 module was detected (Kiselyov et al. 2003). The binding site in the F3(2) module of the NCAM was mapped to the FG-loop region of the module, which is located in the N-terminal part of the module (Kiselyov et al. 2003). This indicates that the C-terminal part of the F3(1) module of the NCAM, together with the N-terminal part of the F3(2) module, might form a single binding site for FGFR, and this site may be destroyed when the modules are separated. Binding of the FGFR Ig2 module to the F3(1–2) modules of the NCAM was therefore tested. Using SPR, binding of the soluble FGFR Ig2 module to the immobilized F3(1–2) modules of the NCAM was detected (Fig. 1A). The Kd value for this interaction was estimated to be 34.3 ± 1.4 μM by fitting the plot of the maximum binding versus ligand concentration with the theoretical curve (Fig. 1B). When the FGFR Ig2 module was immobilized, binding of the soluble F3(1–2) modules of the NCAM to the immobilized Ig2 module could also be demonstrated (Fig. 1C). The Kd value for this binding was estimated to be 45.5 ± 9.2 μM by fitting the plot of the maximum binding versus ligand concentration with the theoretical curve (Fig. 1D). These data indicate that the Ig2 FGFR module is also involved in binding to the NCAM F3 modules.
Figure 1.
Surface plasmon resonance analysis of the binding between the FGFR Ig2 module and the NCAM F3 modules. (A) Binding of the soluble FGFR Ig2 module to the immobilized NCAM F3 modules (with a fitting of the saturation plot shown in B). (C) Binding of the soluble NCAM F3 modules to the immobilized FGFR Ig2 module (with a fitting of the saturation plot shown in D). The experiment was repeated nine times.
Mapping of the FGFR Ig2 module residues involved in binding to the NCAM
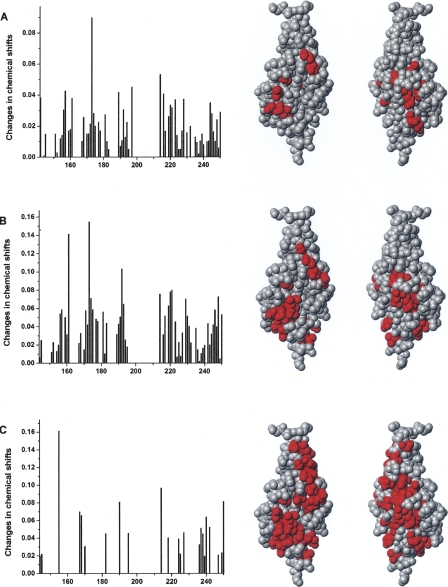
Since the FGFR Ig2 module binds to the F3(1–2) modules of the NCAM, it was of interest to identify the residues of the Ig2 module in the vicinity of the binding site using NMR spectroscopy. The 15N-heteronuclear single quantum correlation (HSQC) spectrum of a 15N-labeled protein records the one-bond coupling of an H–N bond, and is therefore a useful tool for monitoring site-specific perturbations. The assignment of the 15N-HSQC spectrum of a 15N-labeled Ig2 FGFR module has been reported recently (Kiselyov et al. 2006b). The chemical shift changes of the signals provide a means to identify the amino acid residues whose NMR signals are perturbed by the binding of another molecule. 15N-HSQC spectra of a 40 μM Ig2 module was recorded in the presence of 0, 13, 25, or 40 μM F3(1–2) modules of NCAM. Addition of F3(1–2) modules of the NCAM led to either line-broadening, chemical shift changes or, for certain residues, disappearance of the NMR signals. The residues with NMR signal undergoing significant chemical shift changes, >0.03 ppm for the 13 μM F3(1–2) modules and 0.05 ppm for the 25 and 40 μM F3(1–2) modules, or disappearing completely, were considered to be specifically perturbed by the binding. The recorded changes of chemical shifts and mapping of the significantly perturbed residues onto the structure of the Ig2 module (Plotnikov et al. 1999) are shown in Figure 2. As can be seen from Figure 2C, addition of 40 μM F3(1–2) modules perturbed most of the Ig2 module's residues, and most of the signals disappeared. This indicates that the exchange between the bound and unbound form of the Ig2 module is intermediate on the NMR timescale. Since addition of 13 μM F3(1–2) modules led to perturbation of only a few residues (Fig. 2A), 40 μM F3(1–2) modules perturbed most of the residues (Fig. 2C), whereas 25 μM F3(1–2) modules perturbed 23 residues (Fig. 2B), the latter were decided to represent the residues of the Ig2 module perturbed by the F3(1–2) modules of the NCAM. As can be seen from Figure 2B, the perturbed residues appear to consist of two clusters. The first cluster involves residues T156, S157, E159, A171, T173, V174, K175, S181, S214, M217, D218, S219, and V220, while the second cluster involves residues M161, L191, K192, N193, F197, V221, T229, C230, D246, and V248. Perturbation of these residues shows that the presence of the NCAM F3(1–2) modules close to the FGFR Ig2 module alters the chemical environment at the perturbed residues, indicating that the perturbed residues are either a part of, or in the vicinity of, the binding site for the interaction between the NCAM and FGFR.
Figure 2.
Mapping of the FGFR Ig2 module's residues involved in the NCAM binding. Changes in chemical shifts of the 10 μM 15N-labeled Ig2 module after addition of (A) 13 μM, (B) 25 μM, and (C) 40 μM unlabeled NCAM F3 modules and mapping of the significantly perturbed residues onto the FGFR Ig2 module structure (right panels). The crystal structure of human FGFR Ig2 module (PDB code: 1EVT) was used for mapping. MolMol software version 2K.2 was used for creating graphical representations of molecular structures. Origin software version 6.1 was used to create diagrams.
Validation of the binding sites in the FGFR Ig2 module identified by NMR
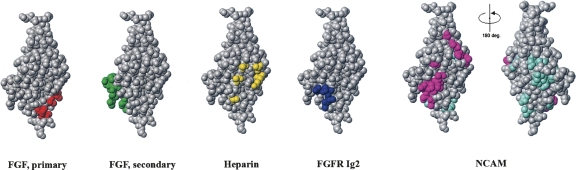
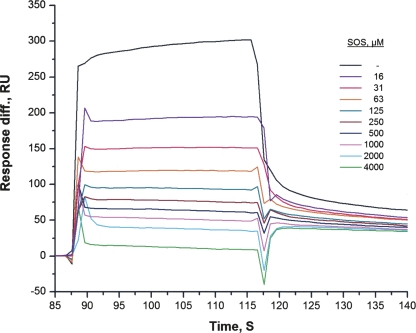
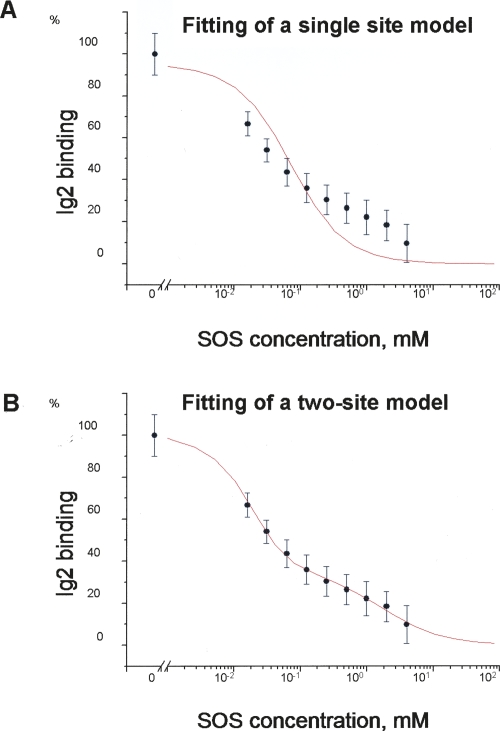
The first cluster is located in close proximity to the binding sites for FGF, heparin, and Ig2 itself (Fig. 3; Plotnikov et al. 1999; Pellegrini et al. 2000). From Figure 3, it appears that heparin may inhibit the binding of the FGFR Ig2 module to NCAM. To test this assumption, the ability of sucrose octasulphate (SOS), a well-known heparin analog, to inhibit binding of the soluble Ig2 module to the immobilized F3(1–2) modules of NCAM was investigated. As shown in Figure 4, SOS was indeed capable of inhibiting the Ig2–NCAM binding, as expected based on the fact that the first cluster is located close to the heparin-binding site of the FGFR Ig2 module. Since there is no suitable ligand binding to the FGFR Ig2 module in the vicinity of the second cluster, we attempted to demonstrate the presence of the second binding site in the FGFR Ig2 module for the NCAM by analysis of a binding isotherm (measured by SPR) of the NCAM F3(1–2) modules (using a concentration range from 1 to 500 μM) to the immobilized Ig2 module. It should be noted that this method can demonstrate the presence of two binding sites only if their Kd values differ by >100-fold. The recorded binding isotherm failed to show the presence of the second site (data not shown), which could be due to the fact that the Kd values of the two sites are approximately the same (or not sufficiently different). However, analysis of the competition isotherm, in which the binding level of Ig2 (from Fig. 4) versus SOS concentration is shown on a logarithmic scale (see Fig. 5A), reveals the presence of the second site. One can clearly see that the competition isotherm is not monophasic, and fitting of the data with a single site model produces a very bad fit (see Fig. 5A). Fitting of the data with a two-site model produces an excellent fit (see Fig. 5B) in which SOS inhibits 67 ± 2% of the Ig2 binding with a Kd of 7.5 ± 1.4 μM and 33 ± 2% of the binding with a Kd of 1.9 ± 0.4 mM. Since the Kd of the heparin–FGFR binding is in the μM range, it is reasonable to assume that the 7.5 μM Kd corresponds to an interaction of SOS with the heparin-binding site of Ig2, and thus 67% of the Ig2 binding occurs via the first site identified by NMR (which is in the vicinity of the heparin-binding site). The remaining 33% of the binding occurs via a different site. This interaction can also be inhibited by SOS (binding, probably unspecifically, either to the FGFR or NCAM), but affinity of the SOS binding to this site is extremely low (1.9 mM Kd). The fact that there is only a twofold difference between the binding levels of the two sites (67% and 33% of the total binding) indicates that affinities for the two sites are similar. Another way of demonstrating the presence of two binding sites is to estimate the Kd values for the two sites from the concentration dependence of the chemical shifts changes. Unfortunately, this method is not applicable in our case due to the intermediate exchange (see above) between the bound and unbound forms of the protein.
Figure 3.
Mapping of the various FGFR Ig2 module's binding sites onto the module's structure. For the NCAM binding site, the first and second clusters are shown in magenta and aquamarine, respectively. The crystal structure of the human FGFR Ig2 module (PDB code: 1EVT) was used for mapping. MolMol software version 2K.2 was used for creating graphical representations of molecular structures.
Figure 4.
Surface plasmon resonance analysis of the inhibitory effect of sucrose octasulphate (SOS) on binding of the FGFR Ig2 module to the NCAM F3 modules. The experimental setup was the same as described in Figure 1. The binding of 20 μM FGFR Ig2 module to the immobilized NCAM F3 modules was studied in the presence of the indicated concentrations of SOS. The experiment was repeated nine times.
Figure 5.
Competition isotherm (by SOS) of the FGFR Ig2 module binding to the NCAM. The isotherm shows the maximum binding level of Ig2 at 20 μM concentration versus SOS concentration. The data were fitted with a single site model in A and a two-site model in B. The data are shown as averages of six replicates, with error bars corresponding to standard deviations.
Discussion
The present study demonstrates for the first time that the FGFR Ig2 module is involved in direct binding to the F3(1–2) modules of the NCAM. The binding affinity for the Ig2 module and the F3(1–2) modules (Kd value of ∼34–46 μM) was three to four times lower than affinity for the double-module constructs of the FGFR Ig2–3 modules and the F3(1–2) modules of the NCAM (Kd value of ∼10 μM), which may be expected because the Ig3 module is also involved in this binding (Kiselyov et al. 2003). Since the Ig3 module is known to bind to the F3(2) module of the NCAM (Kiselyov et al. 2003), the Ig2 module is expected to bind to the F3(1) module. Surprisingly, the Ig2 module was found by NMR to have two possible binding sites for the NCAM, which are located on the “opposite sides” of the module. It should be noted that it is not possible to distinguish by NMR whether an identified site corresponds to a real binding event or the perturbation occurs due to slight structural changes in the module caused by binding at another site. Fortunately, one of the identified sites is located in the vicinity of the Ig2 module's site for heparin, and the presence of this site was confirmed by inhibition of the Ig2–NCAM binding by a heparin analog, SOS. Analysis of the binding isotherm has not given any information about whether or not the second site corresponds to a real binding event. However, analysis of the competition isotherm (by SOS) reveals the presence of two sites in the FGFR Ig2 module with similar affinities for the NCAM (which is probably the reason why the two sites are indistinguishable in the binding isotherm), but very different affinities for SOS. The fact that SOS can inhibit the two sites with binding constants differing by almost 300-fold makes the two sites easily identifiable in the competition isotherm. SOS inhibits one of the sites with 7.5 μM Kd (which corresponds to binding of SOS to the heparin binding site of FGFR) and another one, with 1.9 mM Kd. The latter interaction (either with the NCAM or FGFR) is probably unspecific, and is very unlikely to have any physiological relevance, but it turned out to be quite useful in showing the presence of the second site. It should be noted that one cannot be certain that this site corresponds to the second cluster of perturbed residues identified by NMR. However, the fact that the competition isotherm clearly demonstrates the presence of two binding sites in the FGFR Ig2 module makes it quite plausible that the two clusters of perturbed residues (identified by NMR) correspond to real binding events.
The mechanism of the FGFR activation by the NCAM is not well understood. It has been hypothesized that most FGFR molecules are involved in a transient interaction with the NCAM (Kiselyov et al. 2003, 2005), and when the NCAM is not involved in cell–cell adhesion, the NCAMs are uniformly spread on the cell surface. However, when the NCAM is involved in cell–cell adhesion (via homophilic binding), NCAMs may arrange themselves into the so-called “zipper” formations (Soroka et al. 2003), presumably leading to clustering of the NCAMs, and as a result, to subsequent clustering of the FGFR molecules. The increase in local concentration of the FGFR molecules may be expected to increase the number of the FGFR molecules involved in the direct FGFR–FGFR binding, which could result in the FGFR activation (Kiselyov et al. 2005).
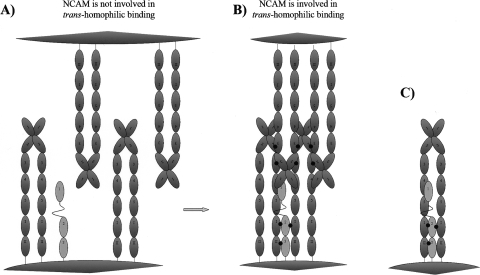
The possible FGFR binding simultaneously to two NCAMs, found in this study, permits modification of the aforementioned model of the FGFR activation by the NCAM. We hypothesize that in the absence of the NCAM-mediated cell–cell adhesion (i.e., when the NCAM is not involved in trans-homophilic binding), the FGFR molecules do not bind substantially to the NCAM (see Fig. 6A). However, when the NCAM is involved in trans-homophilic binding, the NCAMs become clustered due to formation of “zipper”-like structures (Soroka et al. 2003), thus allowing an FGFR molecule to bind simultaneously to two neighboring NCAMs with a much higher affinity than to individual NCAMs, ensuring an efficient NCAM–FGFR interaction (Fig. 6B). Thus, FGFR is expected to bind to, and become activated by, NCAMs only when the NCAM is clustered through a trans-homophilic binding mechanism. It should be noted that FGFR may also be bound to the neighboring NCAMs belonging to the same NCAM cis-dimer, as shown in Figure 6C, which appears to be nonphysiological, since it would lead to a constitutatively strong interaction between the NCAM and FGFR regardless of whether the NCAM is involved in the trans-homophilic binding or not. However, this model cannot be excluded based on our data alone.
Figure 6.
Schematic representation of the proposed model for the interaction between the FGFR and NCAM. (A) The NCAM is not involved in the trans-homophilic binding, and there is no substantial binding between the FGFR and NCAM. (B) The NCAM is involved in trans-homophilic binding and the FGFR binds to two neighboring NCAM molecules belonging to the different cis-dimers. (C) The FGFR is bound to two NCAM molecules belonging to the same cis-dimer.
Thus, we have demonstrated for the first time a direct binding between the FGFR Ig2 module and the NCAM. The Ig2 module turned out to have two putative binding sites for the NCAM, which allowed us to formulate a speculative model for the molecular mechanism of the NCAM interaction with the FGFR. Whether or not the model is true requires further investigation.
Materials and Methods
Production of recombinant proteins
The mouse FGFR1 Ig2 module (3c isoform) and the rat double-module construct of the F3(1–2) modules of the NCAM were produced as previously described (Kiselyov et al. 2003, 2006b).
Surface plasmon resonance analysis
The binding experiments were performed at 25°C using a CM5 sensor chip. Proteins were immobilized on the chip surface by amine coupling. PBS (pH 7.40) was used as running buffer. Immobilization and binding analysis were performed using a BIAcore2000 instrument (Biosensor Applications AB) as previously described (Kiselyov et al. 2003). The Kd values were determined by fitting the saturation data to the theoretical binding curve.
NMR measurements
The following samples were used for recording of NMR spectra: 40 μM 15N-FGFR Ig2 module with or without 13, 25, and 40 μM F3 modules of the NCAM. The buffer was 10 mM sodium phosphate containing 150 mM NaCl, pH 7.4. The samples were analyzed by recording 15N-HSQC spectra using the standard setup provided by ProteinPack. The spectra were processed by NMRPipe (Delaglio et al. 1995) and analyzed by Pronto3D (Kjaer et al. 1994). Changes of chemical shifts were calculated as [(5 · ΔH)2 + ΔN2]0.5, where ΔH is the change of the 1H chemical shift, and ΔN is the change of the 15N chemical shift. The NMR experiments were performed using Varian Unity Inova 750 and 800 MHz spectrometers. All spectra were recorded at 25°C.
Acknowledgments
The Structural Biology and NMR Laboratory (SBiN-Lab) were established through funding from The John and Birthe Meyer Foundation to F.M.P. This work was financially supported by grants from the Danish Medical and Natural Science Research Council, the Lundbeck Foundation, the Danish Cancer Society, and by the European Commission (FP6 frameprogram-integrated project PROMEMORIA-LSHM-CT-2005-512012).
Footnotes
Reprint requests to: Flemming M. Poulsen, Structural Biology and NMR Laboratory (SBiNLab), Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark; e-mail: fmp@bio.ku.dk; fax: 45-35-360116; or Arthur Kochoyan, University of Copenhagen, Bledamsvej 3C, Building 24.2, DK-2200 Copenhagen N, Denmark; e-mail: Arthur@sund.ku.dk; fax: 45-35-360116.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035964.108.
References
- Atkins, A.R., Osborne, M.J., Lashuel, H.A., Edelman, G.M., Wright, P.E., Cunningham, B.A., Dyson, H.J. Association between the first two immunoglobulin-like domains of the neural cell adhesion molecule N-CAM. FEBS Lett. 1999;451:162–168. doi: 10.1016/s0014-5793(99)00554-2. [DOI] [PubMed] [Google Scholar]
- Berezin, V., Bock, E., Poulsen, F.M. The neural cell adhesion molecule. Curr. Opin. Drug Discov. Dev. 2000;3:605–609. [PubMed] [Google Scholar]
- Bruses, J.L., Rutishauser, U. Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie. 2001;83:635–643. doi: 10.1016/s0300-9084(01)01293-7. [DOI] [PubMed] [Google Scholar]
- Cremer, H., Chazal, G., Goridis, C., Represa, A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol. Cell. Neurosci. 1997;8:323–335. doi: 10.1006/mcne.1996.0588. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Doherty, P., Walsh, F.S. CAM-FGF receptor interactions: A model for axonal growth. Mol. Cell. Neurosci. 1996;8:99–111. doi: 10.1006/mcne.1996.0049. [DOI] [PubMed] [Google Scholar]
- Itoh, N., Ornitz, D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Jensen, P.H., Soroka, V., Thomsen, N.K., Ralets, I., Berezin, V., Bock, E., Poulsen, F.M. Structure and interactions of NCAM modules 1 and 2, basic elements in neural cell adhesion. Nat. Struct. Biol. 1999;6:486–493. doi: 10.1038/8292. [DOI] [PubMed] [Google Scholar]
- Kasper, C., Rasmussen, H., Kastrup, J.S., Ikemizu, S., Jones, E.Y., Berezin, V., Bock, E., Larsen, I.K. Structural basis of cell–cell adhesion by NCAM. Nat. Struct. Biol. 2000;7:389–393. doi: 10.1038/75165. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Berezin, V., Maar, T.E., Soroka, V., Edvardsen, K., Schousboe, A., Bock, E. The first immunoglobulin-like neural cell adhesion molecule (NCAM) domain is involved in double-reciprocal interaction with the second immunoglobulin-like NCAM domain and in heparin binding. J. Biol. Chem. 1997;272:10125–10134. doi: 10.1074/jbc.272.15.10125. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Skladchikova, G., Hinsby, A.M., Jensen, P.H., Kulahin, N., Soroka, V., Pedersen, N., Tsetlin, V., Poulsen, F.M., Berezin, V., et al. Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure. 2003;11:691–701. doi: 10.1016/s0969-2126(03)00096-0. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Soroka, V., Berezin, V., Bock, E. Structural biology of NCAM homophilic binding and activation of FGFR. J. Neurochem. 2005;94:1169–1179. doi: 10.1111/j.1471-4159.2005.03284.x. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Bock, E., Berezin, V., Poulsen, F.M. NMR structure of the first Ig module of mouse FGFR1. Protein Sci. 2006a;15:1512–1515. doi: 10.1110/ps.062207906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov, V.V., Kochoyan, A., Poulsen, F.M., Bock, E., Berezin, V. Elucidation of the mechanism of the regulatory function of the Ig1 module of the fibroblast growth factor receptor 1. Protein Sci. 2006b;15:2318–2322. doi: 10.1110/ps.062206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaer, M., Andersen, K.V., Poulsen, F.M. Automated and semiautomated analysis of homo- and heteronuclear multidimensional nuclear magnetic resonance spectra of proteins: The program Pronto. Methods Enzymol. 1994;239:288–307. doi: 10.1016/s0076-6879(94)39010-x. [DOI] [PubMed] [Google Scholar]
- McKeehan, W.L., Wang, F., Kan, M. The heparan sulfate-fibroblast growth factor family: Diversity of structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- Olsen, S.K., Ibrahimi, O.A., Raucci, A., Zhang, F., Eliseenkova, A.V., Yayon, A., Basilico, C., Linhardt, R.J., Schlessinger, J., Mohammadi, M. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, L., Burke, D.F., von Delft, F., Mulloy, B., Blundell, T.L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- Plotnikov, A.N., Schlessinger, J., Hubbard, S.R., Mohammadi, M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- Rougon, G., Hobert, O. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu. Rev. Neurosci. 2003;26:207–238. doi: 10.1146/annurev.neuro.26.041002.131014. [DOI] [PubMed] [Google Scholar]
- Soroka, V., Kolkova, K., Kastrup, J.S., Diederichs, K., Breed, J., Kiselyov, V.V., Poulsen, F.M., Larsen, I.K., Welte, W., Berezin, V., et al. Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion. Structure. 2003;11:1291–1301. doi: 10.1016/j.str.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Walmod, P.S., Kolkova, K., Berezin, V., Bock, E. Zippers make signals: NCAM-mediated molecular interactions and signal transduction. Neurochem. Res. 2004;29:2015–2035. doi: 10.1007/s11064-004-6875-z. [DOI] [PubMed] [Google Scholar]
- Wang, F., Kan, M., Yan, G., Xu, J., McKeehan, W.L. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 1995;270:10231–10235. doi: 10.1074/jbc.270.17.10231. [DOI] [PubMed] [Google Scholar]