Abstract
VX-680, also known as MK-0457, is an ATP-competitive small molecule inhibitor of the Aurora kinases that has entered phase II clinical trials for the treatment of cancer. We have solved the cocrystal structure of AurA/TPX2/VX-680 at 2.3 Å resolution. In the crystal structure, VX-680 binds to the active conformation of AurA. The glycine-rich loop in AurA adopts a unique bent conformation, forming a π–π interaction with the phenyl group of VX-680. In contrast, in the published AurA/VX-680 structure, VX-680 binds to AurA in the inactive conformation, interacting with a hydrophobic pocket only present in the inactive conformation. These data suggest that TPX2, a protein cofactor, can alter the binding mode of VX-680 with AurA. More generally, the presence of physiologically relevant cofactor proteins can alter the kinetics, binding interactions, and inhibition of enzymes, and studies with these multiprotein complexes may be beneficial to the discovery and optimization of enzyme inhibitors as therapeutic agents.
Keywords: Aurora kinase, TPX2, crystal structure, cofactor, SAR
The Aurora kinases (AurA, B, and C) are Ser/Thr kinases critical for mitosis and cytokinesis (Bischoff and Plowman 1999; Carmena and Earnshaw 2003; Andrews 2005). AurA mainly associates with the centrosome and the spindle microtubules during mitosis and functions in centrosome maturation, spindle assembly, maintenance of spindle bipolarity, and mitotic checkpoint control. Activity and localization of AurA is regulated by multiple binding partners, such as the target protein of XKlp2 (TPX2), an important regulator of AurA (Kufer et al. 2002; Bayliss et al. 2003, 2004; Eyers and Maller 2004; Anderson et al. 2007). Crystal structures of AurA and TPX2, as well as biochemical analysis, suggested that TPX2 enhances the kinase activity of AurA by stabilizing the active conformation of the kinase, facilitating substrate binding, and preventing dephosphorylation of Thr288 in the activation loop of AurA (Bayliss et al. 2003; Anderson et al. 2007).
AurA, frequently amplified and overexpressed in human tumors, is oncogenic since ectopical expression of AurA leads to colony formation in cell culture and tumors in nude animals (Bischoff et al. 1998). A number of potent inhibitors of the Aurora kinases have been reported to inhibit tumor growth in xenograft models and have progressed into human clinical trials (Harrington et al. 2004; Mortlock et al. 2005; Gautschi et al. 2006; Heron et al. 2006).
VX-680, also known as MK-0457, is an ATP-competitive small molecule inhibitor of the Aurora kinases that has entered phase II clinical trials for the treatment of cancer (Harrington et al. 2004). The crystal structure of VX-680 bound to AurA has been published recently. As described by Cheetham et al. (2007), VX-680 binds to a hydrophobic pocket within the active site that is only present in the inactive or “closed” conformation. The authors have hence proposed that VX-680 would have weak affinity for the active conformation of AurA and probably binds to the closed conformation of its other kinases targets (including Flt-3 and Abl).
We have reported previously that the presence of TPX2 alters the affinity of VX-680 toward AurA (Anderson et al. 2007). Our biochemical data suggested that the inhibition constant of VX-680 toward AurA is only 4.2-fold lower when TPX2 is present (Ki = 5.9 nM for AurA/TPX2). To understand the molecular basis of the potent inhibition of AurA/TPX2 by VX-680, we have solved the cocrystal structure of AurA/TPX2/VX-680 at 2.3 Å resolution. In this crystal structure, VX-680 binds to the ATP site of AurA, which adopts an active conformation in the presence of TPX2. The glycine-rich loop has a unique bent conformation compared to other AurA-inhibitor structures and forms a π–π interaction with the phenyl group of VX-680. Therefore, binding modes of compounds like VX-680 are very different in the presence and absence of TPX2. Because of the physiologically relevant role of TPX2 in AurA function, we propose that it is important to obtain crystal structures of small molecule/AurA complexes in the presence of TPX2. Our structural observations, as well as our biochemical analysis, also have broad implications for enzyme catalysis and inhibition in general. Physiologically relevant cofactors need to be considered for enzyme assays, since they can significantly impact enzyme catalysis and alter inhibitor interactions.
Results
Crystallography was carried out to investigate the effect of TPX2 on inhibitor binding to AurA. The final model of the structure includes the catalytic core of the kinase (residues 123–387AUR) and two segments of TPX2 (residues 6–21TPX and 26–43TPX), as well as VX-680. No ordered electron density is present for the four intervening residues of TPX2 (22–25TPX), and therefore that section of TPX2 was not modeled.
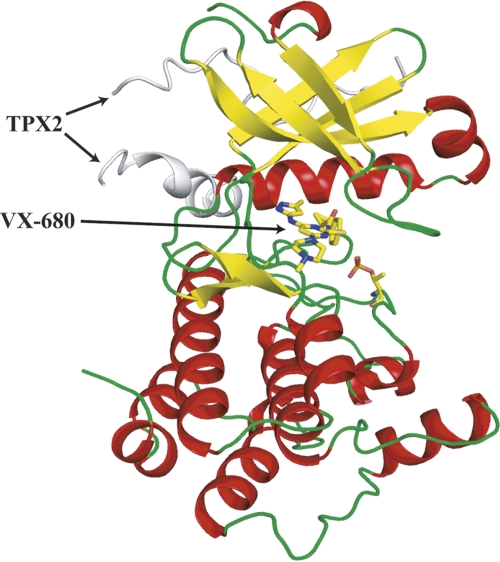
The overall structure of AurA/TPX2/VX-680 (Fig. 1) is similar to the crystal structure of AurA/TPX2/ADP (pdb: 1ol5) described previously by Bayliss et al. (2003). Briefly, the Aurora A catalytic core has the typical bilobal kinase fold (Hanks et al. 1988; Bossemeyer 1995; Nolen et al. 2004; Liao 2007), comprised of an N-terminal lobe (residues 123–210) and a large C-terminal lobe (residues 217–387). The N-terminal lobe predominantly consists of β-strands, as well as two α-helices, including the αC helix. The C lobe is mainly α-helical. The N and C lobes are linked together by a hinge region that forms an important part of the catalytic active site. The active site is situated at the interface between the lobes and includes the ATP binding site, the catalytic base (Asp256AUR), and the kinase activation segment (residues 274–299AUR). Due to autoactivation, the AurA used for crystallization is phosphorylated on Thr288AUR (80% based on LC/MS analysis after trypsin digestion), the key residue required for AurA activation. Both Thr287AUR and Thr288 AUR are phosphorylated in AurA/TPX2/ADP structure (Bayliss et al. 2003). Despite the lack of phosphorylation of Thr287AUR in the AurA/TPX2/VX-680 structure, the activation segment of Aurora A is well ordered, with the phosphorylated Thr288AUR clearly identified in the electron density, and the activation loop adopts an active conformation similar to that in the AurA/TPX2/ADP structure. The activation segment conformation is similar to other Ser/Thr kinases in their active state (Nolen et al. 2004). Structural comparison shows that all the conserved residues at the active site have the same orientation as the AurA/TPX2/ADP structure (Bayliss et al. 2003), including the positively charged residue Lys162AUR (which aligns the phosphates of ATP for catalysis), the negatively charged residue Asp274AUR (which coordinates the magnesium ion bound to the nucleotide), and the catalytic base Asp256AUR (which is involved in transferring the γ-phosphate of ATP to the hydroxyl group of a substrate serine or threonine residue).
Figure 1.
A ribbon diagram of AurA kinase domain in complex with TPX2 and VX-680, colored by secondary structure. VX-680 is shown in a ball-and-stick presentation: yellow for carbon, orange for sulfur, red for oxygen, and blue for nitrogen atoms. The phosphorylated Thr288AUR residue is also shown in the ball-and-stick presentation.
As in the AurA/TPX2/ADP structure, TPX2 binds AurA with two separate stretches. The upstream stretch (residues 6–21TPX), binds to the N-terminal lobe of Aurora A in a mostly extended conformation. The downstream stretch (residues 26–43TPX) adopts an α-helical conformation that interacts with both helix αC and the activation segment of Aurora A. The two Aurora A binding motifs of TPX2 are connected by a flexible linker (disordered in the structure). The conserved residues, Tyr8TPX, Tyr10TPX, Ala12TPX, Phe16TPX, Ile17TPX, Phe19TPX, Trp34TPX, Phe35TPX, and Ala39TPX, interact closely with the AurA catalytic core. However, in contrast to the AurA/TPX2/ADP structure, where the side chain of Ser14TPX points to the solvent (Bayliss et al. 2003), the side chain of Ser14TPX now points to the core of AurA and forms an interaction with the main chain carbonyl oxygen of Tyr199AUR in AurA/TPX2/VX-680.
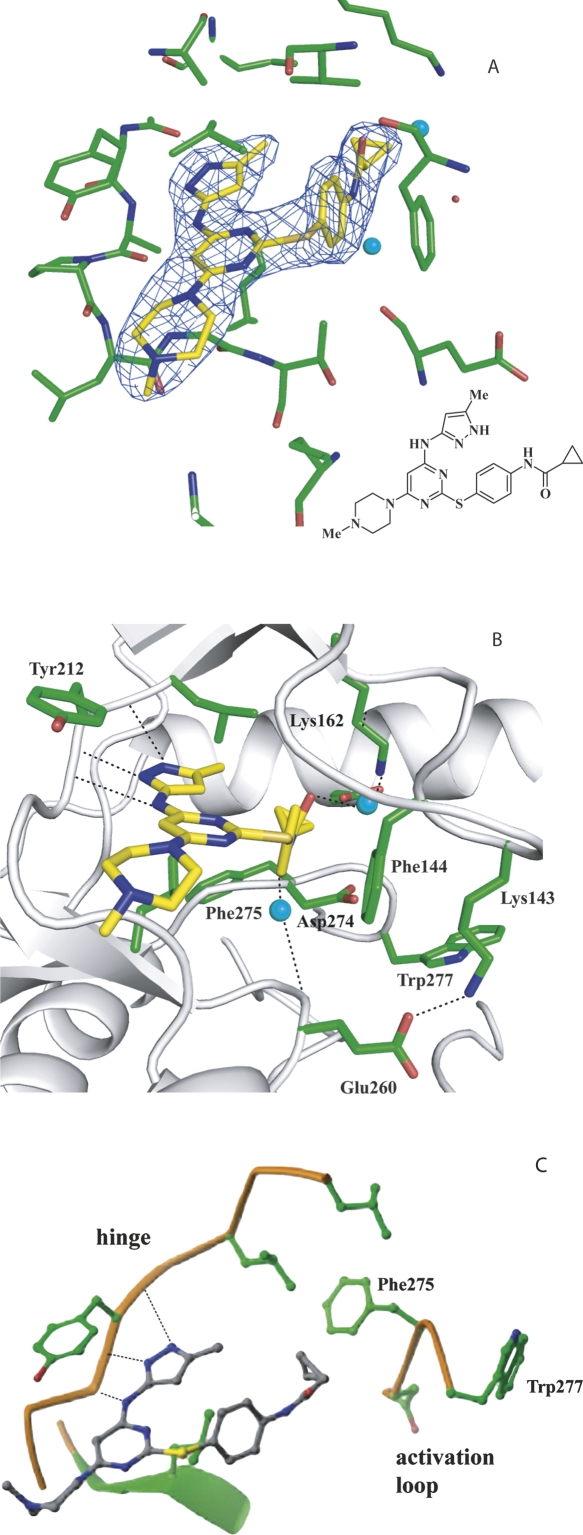
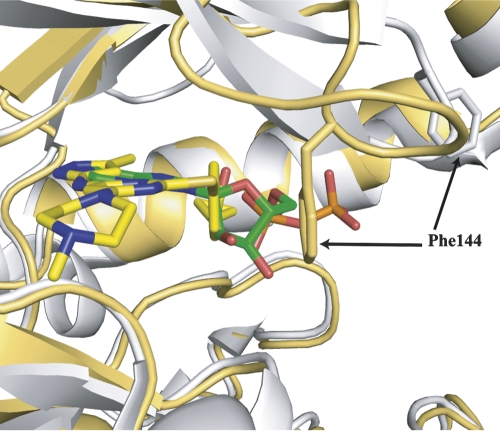
VX-680 is located in the ATP binding site of Aurora A and is clearly identified by electron density (Fig. 2A,B). VX-680 is a Y-shaped molecule with one arm consisting of the aminopyrazole group, which is packed against the hinge region. The other arm of the Y shape, consisting of a substituted phenyl group, inserts into the back pocket of the active site, with its cyclopropyl group surrounded by Val147AUR, Lys162AUR, Ala273AUR, and Asp274AUR. Three hydrogen bonds are formed between VX-680 and the main chain carbonyl and amine groups of Glu211AUR and Ala213AUR in the hinge region. The methylpiperazine group, forming the base of the Y, stretches out from the active site into the solvent. The central pyrimidine group is located in a hydrophobic pocket, surrounded by Leu139AUR, Tyr212AUR, Gly216AUR, Thr217AUR, and Leu263AUR. Even though the crystal structure of AurA/TPX2/VX-680 is very similar to the AurA/TPX2/ADP structure, there are significant differences in the conformation of the glycine-rich loop of AurA (Fig. 3). In the AurA/TPX2/VX-680 structure, the glycine-rich loop is bent with the Cα atoms of residues Lys143AUR and Phe144AUR translated by >4 and 8 Å, respectively, from their extended-loop position in the AurA/TPX2/ADP structure. The side chain of Phe144AUR in the glycine-rich loops forms a π–π interaction with the phenyl group of VX-680. With the movement of the glycine-rich loop, residue Lys143AUR is able to interact with Glu260AUR in the C lobe to form a charge–charge interaction. In contrast, in the AurA/TPX2/ADP structure, the glycine-rich loop is in an extended conformation, with the side chain of Lys143AUR pointing toward the solvent. We also noted the presence of two water molecules in our structure (Fig. 2B), which may further enhance the binding of VX-680 to AurA/TPX2 by forming hydrogen bonds from the carbonyl and amide groups in VX-680, to the ε–amino group of Lys162AUR, and to the main chain carbonyl group of Glu260AUR, respectively.
Figure 2.
Binding of VX-680 in the active site of AurA/TPX2. (A) The electron density of VX-680 contoured at 1.5σ from the final 2fo-fc map. The AurA carbons are shown in green, inhibitor carbons in yellow, oxygens in red, nitrogens in blue, sulfur in orange, and water molecules in light blue. (B) Interactions of VX-680 with the active site of AurA when TPX2 is bound. The backbone tracing is in gray and hydrogen bonds are shown as dotted black lines. (C) Interactions of VX-680 with the active site of AurA without TPX2 bound (adapted from Cheetham et al. 2007 and reprinted with permission from Elsevier ©2007).
Figure 3.
Superposition of the backbones of AurA/TPX2/VX-680 (gold) and AurA/TPX2/ADP (gray, pdb: 1ol5) structures focusing on the different conformations of the glycine-rich loop. The Cα of Phe144AUR in the AurA/TPX2/VX-680 structure has moved by >8 Å. The VX-680 carbons are in yellow, the ADP carbons in green, oxygens in red, nitrogens in blue, sulfur and phosphorus in orange. The side chain of Phe144 is shown in stick form.
Discussion
In order to understand how the presence of the protein cofactor TPX2 affects the molecular interaction of inhibitors with AurA, we solved the crystal structure of AurA/TPX2/VX-680 at 2.3 Å resolution. Even though structures of multiple AurA-small molecular inhibitors have been published, our structure is the first one with a small molecule inhibitor (other than ADP) bound to AurA in the presence of TPX2. Since the coordinates of AurA/VX-680 structure (Cheetham et al. 2007) are not yet available, we have been unable to directly overlay the two structures and compare them at atomic detail. However, compared to the AurA/VX-680 structure described by Cheetham et al. (2007) we have observed important differences in binding modes in the presence and absence of TPX2.
In the AurA/VX-680 crystal structure described by Cheetham et al. (2007) the activation loop of AurA exhibits large movement from the AurA/TPX2/ADP structure (pdb: 1ol5) (7 Å shift of Asp275) and converts the Mg2+ binding pocket in the active conformation into a hydrophobic pocket (Fig. 2C). VX-680 forms multiple hydrogen bonds with the hinge region of AurA and hydrophobic interactions through the pyrimidine core. In addition, the cyclopropyl group of VX-680 makes extensive hydrophobic interactions with residues that form this unique hydrophobic pocket only present in the inactive conformation, and such interactions were proposed to be important for high-affinity binding. Based on this observation, one would expect that VX-680 would bind to the active conformation of AurA with much lower affinity.
In our previous biochemical studies, we reported that the presence of TPX2 alters the inhibitor affinity toward AurA (Anderson et al. 2007). We hypothesized that TPX2 binding decreases the size and accessibility of the hydrophobic back pocket, adjacent to the ATP site, to inhibitors. Since VX-680 does not reach deeply into this back pocket, we predicted that it would still be a potent inhibitor of AurA/TPX2. We showed that even though VX-680 is 4.2-fold less potent against AurA in the presence of TPX2, it is still a potent inhibitor of AurA/TPX2 with a Ki = 5.9 nM (Anderson et al. 2007), contradictory to the predictions by Cheetham et al. (2007).
Our crystal structure of AurA/TPX2/VX-680 has shed some light on this apparent paradox. Comparing our structure with the AurA/VX-680 structure (Cheetham et al. 2007), the interaction between VX-680 and the hinge region of AurA appears to be the same or similar, since both structures contain the same three hydrogen bonds between inhibitor and protein. However, the cyclopropyl and the phenyl groups of VX-680 form very different sets of interactions with AurA depending on whether or not TPX2 is present. In the AurA/VX-680 structure, where AurA is unphosphorylated and inactive, the activation loop adopts a closed conformation (Cheetham et al. 2007). The orientation of the catalytic residues Asp274AUR, Lys162AUR, and Glu181AUR are largely disrupted for catalysis. The Asp274AUR residue is rotated by 180° and shifted by 7 Å to form part of a unique hydrophobic pocket that facilitates VX-680 binding. In the AurA/TPX2/VX-680 structure, however, VX-680 binds to the active conformation of AurA. As a result, the inhibitor is unable to interact with the hydrophobic pocket present only in the inactive conformation observed in the AurA/VX-680 structure. However, the glycine-rich loop exhibits significant movement upon VX-680 binding, collapsing into the ATP site. As a result, the side chain of Phe144AUR in the glycine-rich loop moves by 10 Å to form a π–π interaction with the phenyl group in VX-680. Presumably, this interaction serves as an anchor to stabilize this unique conformation of the glycine-rich loop and contribute to the binding affinity of VX-680 toward AurA/TPX2. We also noted the presence of two water molecules in our structure, which may further enhance the binding of VX-680 to AurA/TPX2 by forming hydrogen bonds from the carbonyl and amide groups in VX-680, to the ε–amino group of Lys162AUR, and to the main chain carbonyl group of Glu260AUR, respectively.
Similar to other kinases such as Abl, the AurA kinase is structurally flexible and can exist in multiple conformational states. The equilibrium between these conformational states can be altered by the binding of small molecule inhibitors or macromolecule cofactors such as TPX2. One such example through our current study is the glycine-rich loop, which undergoes a dramatic conformational change due to TPX2 and VX-680 binding. The glycine-rich loop of kinases is known to be very flexible and can adopt different conformations that facilitate the binding of small molecule inhibitors (Mohammadi et al. 1997; Schindler et al. 2000; Nagar et al. 2002). In fact, the glycine-rich loop of abl kinase adopts a similarly bent confirmation upon binding to Gleevec (Schindler et al. 2000; Nagar et al. 2002). Another example is the conformation of the activation loop of AurA, which is altered by both Thr288AUR phosphorylation and TPX2 binding. Without phosphorylation of Thr288, the activation loop is disordered (Cheetham et al. 2002) except when a phosphate ion is bound (Nowakowski et al. 2002). Even when Thr288AUR is phosphorylated, Bayliss et al. (2003) noted that the activation loop of AurA adopts an inactive conformation in the AurA/ADP structure (pdb: 1ol7), due to conformational flexibility. On the other hand, our biochemical analysis and previous reports indicate that AurA does possess kinase activity in the absence of TPX2. Taken together, these results suggest that the activation loop of AurA is flexible and can adopt both active (for catalysis) and inactive conformation (as observed in crystal structures) in the absence of TPX2. TPX2 binding to the phosphorylated AurA limits the movement of the activation loop, stabilizes its active conformation, and holds it in place for catalysis.
It is important to point out that while AurA is phosphorylated on Thr288 in our AurA/TPX2/VX-680 structure, it is unphosphorylated in the AurA/VX-680 structure. Therefore, to illustrate the effect of TPX2, it would have been best to compare our structure with the structure of AurA/VX-680 where Thr288 is phosphorylated. Such a structure is not available to date. As mentioned previously, the activation loop adopts an inactive conformation even when Thr288 is phosphorylated in the AurA/ADP structure (pdb: 1ol7) (Bayliss et al. 2003) due to conformational flexibility. We speculate that TPX2 binding is the key factor that determines the differential binding modes for VX-680 to AurA in the AurA/TPX2/VX-680 and AurA/VX-680 structures, rather than Thr288 phosphorylation alone.
Cheetham et al. (2007) have proposed that binding of VX-680 to an inactive kinase conformation explains its potency and selectivity profile, especially toward Abl and its Gleevec-resistant mutants like T315I and H396P. However, in the crystal structure of VX-680 in complex with the H396P Abl kinase mutant (Young et al. 2006), the Abl enzyme adopts an open and active conformation, casting doubts on the hypothesis of preferential binding to the inactive conformation by VX-680. Our AurA/TPX2/VX-680 structure demonstrates that, in fact, VX-680 can interact with AurA in both the active and the inactive conformations. It possesses different set of interactions with active and inactive AurA depending on the phosphorylation state and the presence of TPX2.
Many inhibitors are reported to have affinity for both the phosphorylated and the unphosphorylated forms of kinases. For example, Gleevec is reported to inhibit the phosphorylated form of Abl with >20-fold less potency than the unphosphorylated form (Schindler et al. 2000). When it was crystallized with Abl, the activation loop of Abl adopted the inactive conformation. In contrast, compounds like PD173955 (Nagar et al. 2002) and Dasatinib (Tokarski et al. 2006) preferentially bind to the active conformation even when Abl is unphosphorylated. While phosphorylation usually shifts the equilibrium toward the active kinase conformation, compound binding can in fact induce the active conformation in unphosphorylated kinases or the inactive conformation in phosphorylated kinases. It seems to be very difficult to obtain crystal structures of a compound binding to both the active and the inactive conformation of kinases. In the case of VX-680 binding to AurA, it is only possible to capture compound binding to the active versus the inactive AurA conformation using TPX2 as the conformational switch that “freezes” the active conformation.
Significance
Many enzymes form multiprotein complexes in the cellular context that are critical for their biological activity, localization, and degradation. We report here an example where an enzyme inhibitor against Aurora A, an important cancer target, has different binding modes and inhibition potency against the target in the presence of the cofactor protein than in its absence. The purified AurA/TPX2 protein complex may provide a more physiologically relevant enzyme form for the characterization of inhibitors. More generally, the presence of physiologically relevant cofactor proteins may alter the characteristics of enzymes, and studies with these multiprotein complexes may be beneficial to discovery and optimization of enzyme inhibitors as therapeutic agents.
Materials and Methods
AurA kinase domain expression and purification
His-tagged AurA 125-391 construct with a cleavable Tev (Tobacco Etch Virus) sequence (His6-Tev-AurA 125-391) was expressed in Escherichia coli and purified using Ni-NTA resin. Formation of the AurA/TPX2 complex was carried out by immobilizing purified GST-Tev-TPX2 onto a glutathione Sepharose 4B column followed by passing the purified His6-Tev-AurA 125–391 through the column. The complex was treated with the Tev protease, and the cleaved complex was eluted from the column and further purified according to Bayliss et al. (2003). Purified complex was incubated with VX-680, with molar ratio of compound versus protein of 5:1 for 16 h at 4°C and concentrated by spin-filter (Vivascience) to 15 mg/mL for crystallization.
Crystallization and structure determination
Crystals of AurA in complex with TPX2 and VX-680 were grown by sitting-drop vapor diffusion from a solution of 16% polyethylene glycol 3350 and 0.2 M lithium sulfate buffered with 100 mM Bis-Tris, pH 6.9. The drops were formed by mixing 2 μL of the above solution with 2 μL of protein complex. The best crystals were obtained after several rounds of seeding at 20°C. Crystals were allowed to equilibrate in a similar crystallization solution, in the presence of 18% ethylene glycol, prior to freezing in liquid nitrogen. X-ray diffraction data to 2.3 Å resolution were collected using the MARCCD165 detector at the Industrial Macromolecular Crystallography Association beamline 17-ID in the Advanced Photon Source at Argonne National Laboratory. Diffraction data were indexed and scaled using the HKL2000 software package (see Table 1; Otwinowski and Minor 1997). The crystal belongs to the orthorhombic space group C2221, with a single AurA/TPX2/VX-680 complex per asymmetric unit. The structure of the triplex was solved by molecular replacement method based on published coordinates of AurA/TPX2/ADP (pdb: 1ol5, crystallized with ATP-γS, nucleotide assigned as ADP since only the adenosine and two ordered phosphates were visible) (Bayliss et al. 2003) using the program Phaser (McCoy 2007). Clear density was observed for VX-680 in the initial difference-Fourier maps. The structure was refined to 2.3 Å resolution with a crystallographic R-value of 0.20 and a free R-value of 0.25. The final structure includes Aurora A residues from 125 to 389, TPX2 residues from 6 to 21 and from 26 to 42, VX-680, one sulfate, and 58 water molecules (Table 1).
Table 1.
Data collection and refinement statistics
Our AurA/TPX2/VX-680 structure was compared to the crystal structure of AurA/TPX2/ADP (pdb: 1ol5). The superposition of the different AurA crystal structures was achieved by aligning the hinge loop and two β-strands at the bottom of the ATP pocket (residues 209–214, 262–264, and 270–272).
Protein Data Bank deposition
The coordinates have been deposited in the Protein Data Bank (pdb: 3e5a).
Acknowledgments
We thank Drs. Lisa Shewchuk and Luhua Lai for insightful comments on the manuscript. We gratefully acknowledge Drs. Peter J. Tummino, Robert A. Copeland, Domingos Silva, Jerry Adams, Mary Ann Hardwicke, and Denis Patrick for support and encouragement during the course of this work.
Footnotes
Reprint requests to: Zhihong Lai, Enzymology and Mechanistic Pharmacology, GlaxoSmithKline, 1250 South Collegeville Road, Collegeville, PA 19426, USA; e-mail: zhihong.v.lai@gsk.com; fax: (610) 917-7901; or Baoguang Zhao, Department of Computational and Structural Chemistry, GlaxoSmithKline, 709 Swedeland Road, King of Prussia, PA 19406, USA; e-mail: baoguang.zhao@gsk.com; fax: (610) 270-5093.
Abbreviations: AurA, Aurora A; AurB, Aurora B; AurC, Aurora C; TPX2, targeting protein for Xenopus kinesin-like protein 2.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036590.108.
References
- Anderson, K., Yang, J., Koretke, K., Nurse, K., Calamari, A., Kirkpatrick, R.B., Patrick, D., Silva, D., Tummino, P.J., Copeland, R.A., et al. Binding of TPX2 to Aurora A alters substrate and inhibitor interactions. Biochemistry. 2007;46:10287–10295. doi: 10.1021/bi7011355. [DOI] [PubMed] [Google Scholar]
- Andrews, P.D. Aurora kinases: Shining lights on the therapeutic horizon? Oncogene. 2005;24:5005–5015. doi: 10.1038/sj.onc.1208752. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., Sardon, T., Vernos, I., Conti, E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., Sardon, T., Ebert, J., Lindner, D., Vernos, I., Conti, E. Determinants for Aurora-A activation and Aurora-B discrimination by TPX2. Cell Cycle. 2004;3:404–407. [PubMed] [Google Scholar]
- Bischoff, J.R., Anderson, L., Zhu, Y., Mossie, K., Ng, L., Souza, B., Schryver, B., Flanagan, P., Clairvoyant, F., Ginther, C., et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., Plowman, G.D. The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- Bossemeyer, D. Protein kinases–structure and function. FEBS Lett. 1995;369:57–61. doi: 10.1016/0014-5793(95)00580-3. [DOI] [PubMed] [Google Scholar]
- Carmena, M., Earnshaw, W.C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Cheetham, G.M., Knegtel, R.M., Coll, J.T., Renwick, S.B., Swenson, L., Weber, P., Lippke, J.A., Austen, D.A. Crystal structure of Aurora-2, an oncogenic serine/threonine kinase. J. Biol. Chem. 2002;277:42419–42422. doi: 10.1074/jbc.C200426200. [DOI] [PubMed] [Google Scholar]
- Cheetham, G.M., Charlton, P.A., Golec, J.M., Pollard, J.R. Structural basis for potent inhibition of the Aurora kinases and a T315I multi-drug resistant mutant form of Abl kinase by VX-680. Cancer Lett. 2007;251:323–329. doi: 10.1016/j.canlet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Eyers, P.A., Maller, J.L. Regulation of Xenopus Aurora A activation by TPX2. J. Biol. Chem. 2004;279:9008–9015. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- Gautschi, O., Mack, P.C., Davies, A.M., Lara P.N., Jr, Gandara, D.R. Aurora kinase inhibitors: A new class of targeted drugs in cancer. Clin. Lung Cancer. 2006;8:93–98. doi: 10.3816/CLC.2006.n.036. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., Quinn, A.M., Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Harrington, E.A., Bebbington, D., Moore, J., Rasmussen, R.K., Ajose-Adeogun, A.O., Nakayama, T., Graham, J.A., Demur, C., Hercend, T., Diu-Hercend, A., et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Heron, N.M., Anderson, M., Blowers, D.P., Breed, J., Eden, J.M., Green, S., Hill, G.B., Johnson, T., Jung, F.H., McMiken, H.H., et al. SAR and inhibitor complex structure determination of a novel class of potent and specific Aurora kinase inhibitors. Bioorg. Med. Chem. Lett. 2006;16:1320–1323. doi: 10.1016/j.bmcl.2005.11.053. [DOI] [PubMed] [Google Scholar]
- Kufer, T.A., Sillje, H.H., Korner, R., Gruss, O.J., Meraldi, P., Nigg, E.A. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, J.J. Molecular recognition of protein kinase binding pockets for design of potent and selective kinase inhibitors. J. Med. Chem. 2007;50:409–424. doi: 10.1021/jm0608107. [DOI] [PubMed] [Google Scholar]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, M., McMahon, G., Sun, L., Tang, C., Hirth, P., Yeh, B.K., Hubbard, S.R., Schlessinger, J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Mortlock, A., Keen, N.J., Jung, F.H., Heron, N.M., Foote, K.M., Wilkinson, R., Green, S. Progress in the development of selective inhibitors of Aurora kinases. Curr. Top. Med. Chem. 2005;5:199–213. doi: 10.2174/1568026053507651. [DOI] [PubMed] [Google Scholar]
- Nagar, B., Bornmann, W.G., Pellicena, P., Schindler, T., Veach, D.R., Miller, W.T., Clarkson, B., Kuriyan, J. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- Nolen, B., Taylor, S., Ghosh, G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Nowakowski, J., Cronin, C.N., McRee, D.E., Knuth, M.W., Nelson, C.G., Pavletich, N.P., Rogers, J., Sang, B.C., Scheibe, D.N., Swanson, R.V., et al. Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure. 2002;10:1659–1667. doi: 10.1016/s0969-2126(02)00907-3. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Schindler, T., Bornmann, W., Pellicena, P., Miller, W.T., Clarkson, B., Kuriyan, J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Tokarski, J.S., Newitt, J.A., Chang, C.Y., Cheng, J.D., Wittekind, M., Kiefer, S.E., Kish, K., Lee, F.Y., Borzillerri, R., Lombardo, L.J., et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- Young, M.A., Shah, N.P., Chao, L.H., Seeliger, M., Milanov, Z.V., Biggs W.H., III, Treiber, D.K., Patel, H.K., Zarrinkar, P.P., Lockhart, D.J., et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006;66:1007–1014. doi: 10.1158/0008-5472.CAN-05-2788. [DOI] [PubMed] [Google Scholar]