Abstract
Combining single molecule atomic force microscopy (AFM) and protein engineering techniques, here we demonstrate that we can use recombination-based techniques to engineer novel elastomeric proteins by recombining protein fragments from structurally homologous parent proteins. Using I27 and I32 domains from the muscle protein titin as parent template proteins, we systematically shuffled the secondary structural elements of the two parent proteins and engineered 13 hybrid daughter proteins. Although I27 and I32 are highly homologous, and homology modeling predicted that the hybrid daughter proteins fold into structures that are similar to that of parent protein, we found that only eight of the 13 daughter proteins showed β-sheet dominated structures that are similar to parent proteins, and the other five recombined proteins showed signatures of the formation of significant α-helical or random coil-like structure. Single molecule AFM revealed that six recombined daughter proteins are mechanically stable and exhibit mechanical properties that are different from the parent proteins. In contrast, another four of the hybrid proteins were found to be mechanically labile and unfold at forces that are lower than the ∼20 pN, as we could not detect any unfolding force peaks. The last three hybrid proteins showed interesting duality in their mechanical unfolding behaviors. These results demonstrate the great potential of using recombination-based approaches to engineer novel elastomeric protein domains of diverse mechanical properties. Moreover, our results also revealed the challenges and complexity of developing a recombination-based approach into a laboratory-based directed evolution approach to engineer novel elastomeric proteins.
Keywords: single-molecule force spectroscopy, elastomeric protein, recombination, mechanical stability, mechanical unfolding
Elastomeric proteins are an important class of mechanical proteins that are subject to stretching force under physiological conditions (Labeit and Kolmerer 1995; Kellermayer et al. 1997; Rief et al. 1997; Tskhovrebova et al. 1997; Oberhauser et al. 1998; Tatham and Shewry 2000; Gosline et al. 2002; Li et al. 2002; Bao and Suresh 2003). They function as molecular springs to provide tissues with extensibility, elasticity, and mechanical strength. Elastomeric proteins can also function as biomaterials of superb mechanical properties. For example, spider dragline silk is the best-known fibrous material that outperforms any manmade high-performance fibrous materials (Gosline et al. 1999; Becker et al. 2003). Understanding the molecular details of the design of elastomeric proteins is not only important for elucidating biophysical principles underlying a wide variety of biological processes, but also may pave the way to use bottom-up approaches to design and engineer novel elastomeric proteins with well-defined mechanical properties for applications ranging from material sciences to nanobiotechnology (Goodsell 2004; Elvin et al. 2005).
Using single-molecule force spectroscopy techniques, it is possible to stretch individual protein molecules to measure their mechanical properties at the single molecule level (Rief et al. 1997). In combination with protein engineering techniques, it has become feasible to investigate the molecular determinants of the mechanical stability of proteins (Fisher et al. 1999). Insights into the design of mechanically stable elastomeric proteins have significantly expanded the scope of elastomeric proteins by including nonmechanical proteins, which by nature are not subject to stretching force under their normal working conditions, and yet are mechanically stable (Yang et al. 2000; Best et al. 2001; Dietz and Rief 2004; Brockwell et al. 2005; Cecconi et al. 2005; Cao et al. 2006; Sharma et al. 2007). It has been recognized that there exists a key region in the protein that is critical for mechanical stability (Li et al. 2000a; Lu and Schulten 2000), and can be considered as the mechano-active site, similar to the active site in enzymes (Li 2007). The mechanical stability of proteins is not only governed by the mechano-active site and the noncovalent interactions involved in the active site, but also by interactions mediated by amino acid residues adjacent to the mechano-active site. Despite the significant progress in understanding the elastomeric proteins over the last decade, thorough understanding of the molecular determinants for the mechanical stability of proteins remains elusive and largely unestablished. Rational engineering of proteins with well-defined mechanical stability in a systematic fashion currently is still out of the reach of current experimental approaches.
Analogous to the engineering of enzymes of well-defined enzymatic activity, engineering proteins of well-defined mechanical stability is to large extent about how to design and engineer the mechano-active site to produce the desired mechanical properties. Efforts along two different directions have been pursued in enzyme engineering: one is rational design and engineering-based methods (Kortemme and Baker 2004; Ashworth et al. 2006; Jiang et al. 2008), and the other one is a laboratory-based directed evolution approach (Arnold and Georgiou 2003). Inspired by the successful approaches of directed evolution of enzymes, especially those involving recombination (Voigt et al. 2002; Otey et al. 2004; Bloom et al. 2005; Griswold et al. 2005; Carbone and Arnold 2007), we have started to explore a recombination-based approach to engineer elastomeric proteins with novel mechanical properties (Sharma et al. 2006) in parallel to our ongoing efforts using rational design and engineering approaches.
DNA shuffling-based recombination is an important mechanism used by nature to improve protein traits such as enzymatic activity and confer new properties to proteins during evolution. Recombination offers the advantage of combining beneficial mutations from multiple parents into a single offspring. This approach has also been used and developed extensively in directed evolution of proteins in laboratory settings, and has become one of the most important strategies in enzyme engineering (Voigt et al. 2002; Bittker et al. 2004; Otey et al. 2004; Griswold et al. 2005). Recombination is based on DNA shuffling of proteins sharing high-sequence homology and identity. Recent developments have extended this method to proteins that are distantly related and share low-sequence homology (Bittker et al. 2004; Griswold et al. 2005). In a previous paper (Sharma et al. 2006), we reported the proof of principle of using DNA shuffling-based recombination technique to engineer elastomeric proteins with novel mechanical properties. Here, we report our much expanded experimental efforts in developing DNA shuffling-based recombination into a versatile approach in engineering proteins of well-defined mechanical properties. Using I27 and I32 from the muscle protein titin as model parent proteins (Carrion-Vazquez et al. 1999b; Li et al. 2002), we systematically shuffled the secondary structural elements between the two parent proteins and generated a family of hybrid proteins. Using single-molecule AFM techniques and conventional stopped-flow spectrofluorimetry methods, we characterized their mechanical properties as well as their chemical unfolding kinetics in detail. Our results not only demonstrate the great potential of recombination-based methodology in engineering proteins of novel mechanical properties, but also illustrate the direction for future experimental efforts.
Results
Many naturally occurring elastomeric proteins are tandem modular proteins that consist of many individually folded homologous globular domains (Tatham and Shewry 2000). The giant muscle protein titin is an ideal example in this regard (Labeit and Kolmerer 1995). Titin contains hundreds of individually folded, homologous immunoglobulin (Ig) domains. These homologous Ig domains exhibit distinct mechanical stability and constitute ideal target for recombination (Carrion-Vazquez et al. 1999b; Li et al. 2002). Building upon previous single-molecule AFM work (Carrion-Vazquez et al. 1999b; Li et al. 2002), we employ the 27th and 32nd Ig domains from the giant muscle protein titin as parent templates to generate hybrid daughter proteins by recombining fragments from the two parent proteins. I27 and I32 share a high sequence homology (Fig. 1A, identity 42%, similarity 57%), but displays sharply different mechanical stability (Li et al. 2002). I27 has a characteristic β-sandwich structure with seven β-strands packed into two β-sheets (Pfuhl and Pastore 1995). The A′G region of I27 has been shown to be the mechano-active site of I27 and offers the major mechanical resistance to mechanical unfolding of I27 (Lu et al. 1998; Carrion-Vazquez et al. 1999a; Li et al. 2000a; Lu and Schulten 2000). Homology modeling predicts that I32 folds into a three-dimensional structure that is very similar to that of I27, and single-molecule AFM studies suggest that the mechano-active site for I32 is also located at the A′G region of the protein (unpublished data). Thus, I27 and I32 constitute ideal parent template proteins for recombination.
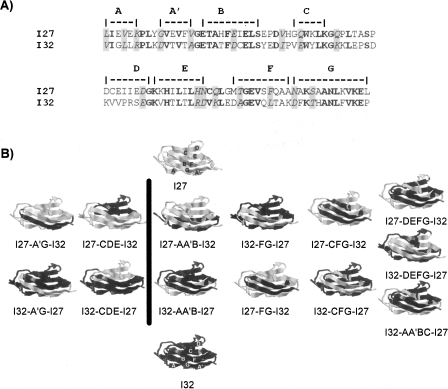
Figure 1.
Hybrid Ig proteins engineered by shuffling fragments between the two parent proteins, I27 and I32. (A) Sequence alignment of I27 and I32. The secondary structural elements are indicated above the sequences. Residues in block indicate those residues that are identical in both I27 and I32, and residues shaded in gray indicate those that are similar in both parent proteins. (B) Three-dimensional structures of parent proteins as well as the engineered hybrid Ig domains. By interchanging the various secondary structural elements between the two parent proteins, we engineered 13 hybrid proteins. In the hybrid proteins, the fragments coming from parent protein I27 are shown in gray, while those from I32 are shown in black. Left column shows the hybrid daughter proteins whose A′G strands are kept intact during recombination, while the hybrid ones in the right column contain A′G strands from two different parent proteins.
Construction of hybrid proteins by shuffling secondary structural elements between I27 and I32
In our previous communication (Sharma et al. 2006) we shuffled the force-bearing strands (A′G strands) and non-force-bearing β-strands (CDE strands) between the two parent proteins. In these four hybrid proteins, the force-bearing strands, which encode the mechano-active site for the two Ig domains, were kept intact. To fully explore the possibility of recombination and the potential benefits in creating proteins of hybrid force-bearing β-strands, here we attempt to systematically shuffle the secondary structural elements between the two parent proteins. In this study, the crossover sites were chosen at various flexible loop regions of I27 and I32, and AA′B β hairpin, AA′BC strands, FG hairpin, and CFG β-strands were shuffled between the two parent proteins. In all of these hybrid proteins, the original force-bearing β-strands from a given parent protein were separated and distributed into two hybrid daughter proteins. For example, for hybrid protein I32-FG-I27, the FG β-hairpin was shuffled from I27 into I32. In the hybrid daughter protein, the force-bearing β-strand AA′ comes from I32, while force-bearing strand G comes from I27. In this way, we will examine whether force-bearing β-strands coming from two different parent proteins can reconstitute a fully functional mechano-active site. Together with the previously reported shuffling of A′G and CDE β-strands, these hybrid proteins will enable us to investigate the effect of the mechano-active site on the mechanical stability of Ig domains. Figure 1B shows the three-dimensional structures of the hybrid proteins predicted from homology modeling using a web-based package Swiss-Model by First Approach Mode (http://swissmodel.expasy.org/). It is evident that homology modeling predicts that all hybrid daughter proteins will fold into three-dimensional structures that are similar to those of the wild-type parent proteins.
Secondary structures of the hybrid proteins generated by recombination
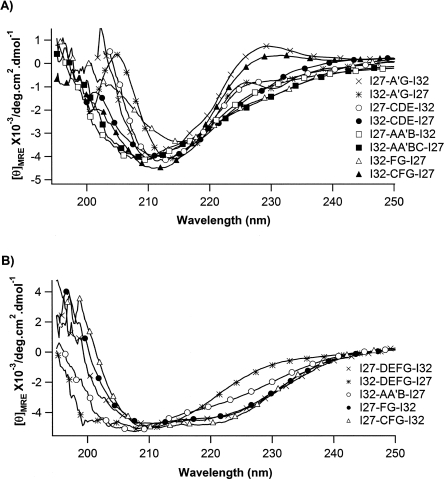
In order to experimentally examine whether the engineered hybrid proteins fold into three-dimensional structures that are similar to those of parent proteins, we carried out far-ultraviolet circular dichroism (far-UV CD) spectroscopy measurements on the monomeric hybrid proteins. The CD spectra of the 13 engineered hybrid proteins are shown in Figure 2. Different from homology modeling prediction results, CD spectra clearly reveal the changes in the secondary structures of some of the hybrid proteins. The CD spectra of the hybrid proteins show diverse signatures for the formation of different secondary structures, and can be classified into two categories: The first category of proteins (consisting of I27-A′G-I32, I32-A′G-I27, I27-CDE-I32, I32-CDE-I27, I27-AA′B-I32, I32-AA′BC-I27, I32-FG-I27, and I32-CFG-I27) show single minima between 210 nm and 218 nm in their CD spectra, which is indicative of β-sheet structure, suggesting that these hybrid proteins fold into structures that are predominantly β-sheets, which resemble those of the parent proteins I27 and I32 (Politou et al. 1995). In contrast, the second category of hybrid proteins exhibits clear signatures of the formation of non-β-sheet secondary structures. The CD spectra of hybrid proteins I27-DEFG-I32, I32-AA′B-I27, I27-FG-I32, and I27-CFG-I32 show a broad negative peak between 208 nm and 222 nm, with two minima at ∼208 nm and ∼222 nm, respectively, indicating the formation of various amounts of α-helical structures in these hybrid proteins together with the β-sheet structure, suggesting that the structures of hybrid proteins deviate from those predicted by homology modeling. Moreover, for proteins I32-DEFG-I27 and I32-AA′B-I27, the minima at 208 nm show a significant shift toward lower wavelength, suggestive of the increasing content of random coil-like sequences in these two hybrid proteins. These results suggest that the secondary structure of the sequences in parent proteins is context dependent: Shuffling secondary structures between highly homologous proteins does not guarantee the formation of the correct secondary structure, and the secondary structure of the structural elements being shuffled depends on the new environment in the hybrid proteins.
Figure 2.
Far-UV CD spectra for the recombined hybrid Ig domains. (A) CD spectra of eight engineered hybrid Ig proteins that show predominantly β-sheet structure. These spectra are characterized by single minima between 210 and 218 nm. (B) CD spectra of engineered hybrid Ig domains that show secondary structures deviating from typical β-sheet structure. The CD spectra of these proteins show a broad negative peak between 208 nm and 222 nm, with two minima at ∼208 nm and ∼222 nm, suggesting the formation of an α-helical structure.
Mechanical properties of recombined hybrid proteins
The mechanical properties of I27-A′G-I32, I32-A′G-I27, I27-CDE-I32, and I32-CDE-I27 were investigated in detail in our previous single-molecule AFM studies (Sharma et al. 2006) using (GB1)4–hybrid–(GB1)4 constructs, which allowed us to screen the mechanical properties of hybrid proteins relatively quickly. To investigate the mechanical stability of the hybrid proteins in greater detail and obtain much improved statistics, here we constructed polyproteins (GB1–hybrid)4, in which the GB1 alternates with the hybrid proteins, for our single-molecule AFM experiments. In the hybrid proteins, the GB1 domains, which are characterized by contour length increment ΔLc of ∼18 nm and unfolding forces of ∼180 pN (Cao et al. 2006; Cao and Li 2007), serve as internal fingerprints, allowing us to identify single-molecule stretching events and discern the mechanical signature of the hybrid proteins in an unambiguous way (Li et al. 2001). Since the hybrid Ig domains alternate with GB1 domains in the heteropolyprotein, we can be certain that the force–extension curve must contain the mechanical signature of the unfolding of at least one hybrid domain if there are two unfolding events of GB1. If the hybrid proteins are properly folded and of significant mechanical stability, the unfolding of such hybrid proteins will result in unfolding events with ΔLc that are similar to that of wild-type parent proteins I27 and I32, which is ∼28 nm. If the hybrid proteins are not properly folded or mechanically labile, we will not observe unfolding events of ΔLc of ∼28 nm. Instead, the stretching and unfolding of such hybrid domains will give rise to random coil-like elastic behaviors or irregular low unfolding force peaks. Indeed, in our constructed hybrid proteins, we observed both types of unfolding behaviors. For clarity, the mechanical unfolding of hybrid proteins are classified into three categories according to their distinct mechanical properties: One category corresponds to mechanically stable hybrid Ig domains; the second one corresponds to mechanically labile ones, and the third one exhibits dual mechanical features of the first two categories.
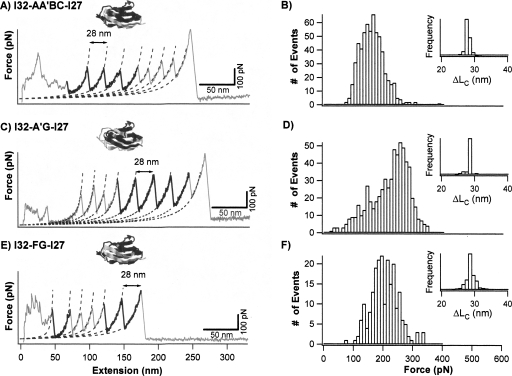
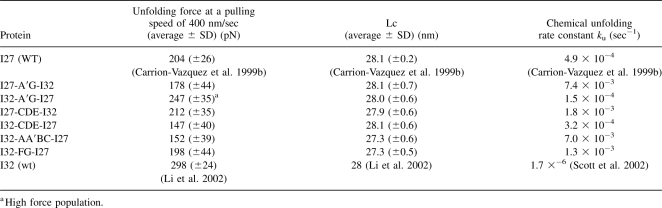
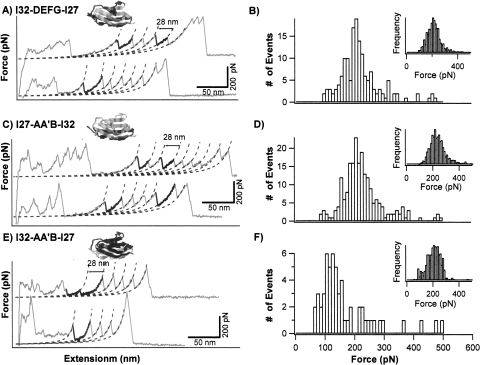
The first category consists of hybrid Ig domains I27-A′G-I32, I32-A′G-I27, I27-CDE-I32, I32-CDE-I27, I32-AA′BC-I27, and I32-FG-I27. Stretching heteropolyprotein (GB1-I32-AA′BC-I27)4 results in force–extension curves showing unfolding force peaks with two apparent ΔLc: one is 18 nm and the other is 28 nm (Fig. 3A). The unfolding events of ΔLc of ∼18 nm correspond to the mechanical unraveling of the GB1 domains in the polyprotein (Cao et al. 2006; Cao and Li 2007), while the unfolding events of ΔLc of 28 nm correspond to the complete mechanical unfolding of the properly folded I32-AA′BC-I27 domains (inset of Fig. 3B). The mechanical properties of this category of hybrid Ig domains were studied using similar approaches, and the force–extension curves of I32-A′G-I27 and I32-FG-I27 are shown in Figure 3C and E, and unfolding force histograms of these hybrid Ig domains are shown in Figure 3B, D, and F, respectively. It is clear that the unfolding force distribution of I32-AA′BC-I27 and I32-FG-I27 have similar width to that of the parent proteins as well as that of the previously studied I27-A′G-I32, I27-CDE-I32, and I32-CDE-I27. The average unfolding forces are 152 ± 39 pN for I32-AA′BC-I27 and 198 ± 44 pN for I32-FG-I27. In contrast, the unfolding force distribution of I32-A′G-I27 is much broader and shows a bimodal distribution, suggestive of the existence of conformational heterogeneity in the native states of I32-A′G-I27. The bimodal distribution of I32-A′G-I27 shows one peak at ∼150 pN and the other one at ∼250 pN. The average unfolding forces for all the recombined hybrid proteins are shown in Table 1. It is clear that recombination of protein fragments can lead to proteins of diverse mechanical properties, with mechanical stability ranging from 130 pN to >250 pN.
Figure 3.
Mechanical properties of mechanically stable hybrid Ig domains I32-AA′BC-I27, I32-A′G-I27, and I32-FG-I27. (A,C, and E) Typical force–extension curves of stretching heteropolyproteins (GB1-I32-AA′BC-I27)4, (GB1-I32-A′G-I27)4, and (GB1-I32-FG-I27)4, respectively. The unfolding events of GB1 are characterized by ΔLc of ∼18 nm and unfolding force of ∼180 pN, and are colored in gray. The unfolding events with ΔLc of ∼28 nm correspond to the unfolding of well-folded engineered hybrid Ig domains and are in black. Dotted lines correspond to WLC fits. (B,D, and F) Unfolding force histograms for hybrid Ig domains I32-AA′BC-I27, I32-A′G-I27, and I32-FG-I27, respectively. The unfolding forces of I32-AA′BC-I27 and I32-FG-I27 show unimodal distribution with average unfolding forces of 152 ± 39 pN (n = 602) and 198 ± 44 pN (n = 215), respectively. The unfolding forces of I32-A′G-I27 show a bimodal distribution, with the first peak centered at 150 pN and the second peak centered around 250 pN, respectively. Insets are histograms for ΔLc of individual hybrid Ig domains.
Table 1.
Mechanical and chemical unfolding properties of hybrid Ig domains
The width of the unfolding force distribution for hybrid proteins is similar among them (except I32-A′G-I27) and slightly broader than that of the two parent proteins. The width of the unfolding force distribution carries important information about the unfolding distance Δx u between the folded state and mechanical unfolding transition state. As shown by Evans (2001), the unfolding force distribution, even in the absence of experimental uncertainty, is set by the thermo force, k BT/Δx u, where k B is the Boltzmann constant, T is temperature, and Δx u is the unfolding distance between the native state and mechanical unfolding transition state. A similar width of unfolding force distributions observed for different hybrid proteins suggests that the unfolding distance Δx u is similar for different hybrid proteins, and the difference in mechanical stability originates from the difference in barrier height for the mechanical unfolding reaction.
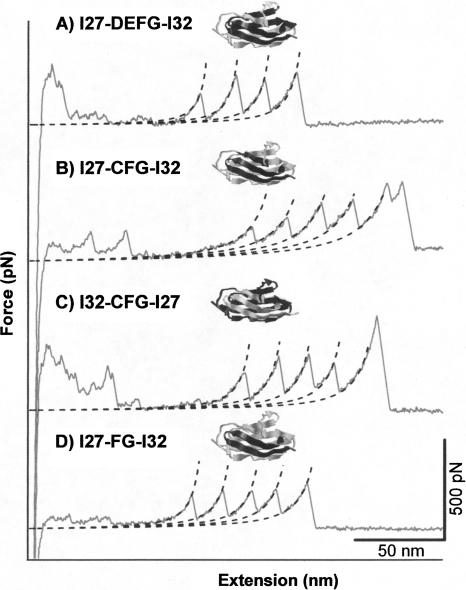
The second category consists of hybrid Ig domains I27-DEFG-I32, I27-CFG-I32, I32-CFG-I27, and I27-FG-I32. This category of hybrid Ig domains does not exhibit any unfolding force peaks with ΔLc of ∼28 nm. Instead, their force–extension curves are characterized by a long featureless spacer preceding the unfolding of GB1 domains. For example, Figure 4A shows a typical force–extension curve of (GB1-I27-DEFG-I32)4, which is characterized by a long featureless spacer followed by three GB1 unfolding events of ΔLc of ∼18 nm. The long featureless spacers in force–extension curves are mechanical signatures of random coil-like polymers. Since GB1 alternates with I27-DEFG-I32 in the heteropolyprotein, the long featureless spacer must correspond to the stretching and unfolding of I27-DEFG-I32 domains. The lack of unfolding events of ΔLc of 28 nm strongly suggests that I27-DEFG-I32 unfolds at very low forces, implying that the hybrid protein is either folded but mechanically labile or unstructured.
Figure 4.
Typical force–extension curves of heteropolyprotein (A–D) (GB1-I27-DEFG-I32)4, (GB1- I27-CFG-I32)4, (GB1- I32-CFG-I27)4, and (GB1-I27-FG-I32)4. The force–extension curves of these heteropolyproteins are characterized by long featureless spacers followed by the unfolding events of GB1 domains, which corresponds to the stretching of hybrid Ig domains that unfold at low force (below our detection limit). Dotted lines are WLC fits to the experimental data.
The third category consists of hybrid Ig domains I32-DEFG-I27, I27-AA′B-I32, and I32-AA′B-I27. These three proteins exhibit dual “personality” in their mechanical properties. In the polyprotein chimeras (GB1-hybrid)4, hybrid Ig domains alternate with GB1 domains. Therefore, the ratio between the numbers of the observed unfolding events for hybrid Ig domains and for GB1 domains should be close to 1. For example, if one observed three GB1 unfolding events in a force–extension curve, one should observe two or three or four unfolding events of the hybrid Ig domains (Li et al. 2001). Indeed, for I32-A′G-I27, I32-AA′BC-I27, and I32-FG-I27 heteropolyproteins, the ratio of the number of unfolding events for GB1 and hybrid proteins are smaller than 2, close to the theoretically predicted ratio. However, the force–extension curves of the third category of hybrid Ig domains are characterized by significantly more GB1 unfolding events than Ig unfolding events. For example, Figure 5A shows two typical force–extension curves of (GB1-I32-DEFG-I27)4. In the top curve, three GB1 unfolding events and two I32-DEFG-I27 unfolding events were observed, consistent with the polyprotein construction. In contrast, the number of GB1 unfolding events in the bottom curve is much more than the number of I32-DEFG-I27 unfolding events. This force–extension curve contains five unfolding events of GB1, which result from the stretching of a dimer of the heteropolyprotein formed by the air oxidation of the C-terminal cysteine residues in the two different heteropolyprotein molecules. Therefore, this force–extension curve should contain the signature of stretching and unfolding of at least three I32-DEFG-I27 domains, yet only one unfolding force peak was observed to match the complete unfolding of I32-DEFG-I27, as evidenced by its ΔLc of 28 nm. This result indicates that only one I32-DEFG-I27 domain unfolded at high forces while the other two hybrid Ig domains unfolded at low forces that are below our detection limit. Therefore, the same protein I32-DEFG-I27 exhibits two distinct mechanical stabilities. These results suggest that this category of hybrid Ig domains exist in two distinct states that give rise to the observed duality of the mechanical stability. The distribution of the hybrid domains between these two mechanically distinct states can be best described by R hybrid/GB1, the ratio between the number of unfolding events for hybrid Ig domains versus the number of GB1 unfolding events. The observed R hybrid/GB1 is 0.27 for I32-DEFG-I27, 0.18 for I27-AA′B-I32, and 0.15 for I32-AA′B-I27, respectively, suggesting that only a small fraction of these three hybrid proteins are mechanically stable and the majority of them are mechanically labile and unfold at low force that is below our detection limit. It is worth noting that the ratio R hybrid/GB1 can be effected by the relative stability between GB1 domains and hybrid Ig domains, as well as the strength of the adhesion bonds between the polyprotein and the AFM tip or substrate. If the adhesion force is not very strong and hybrid Ig domains were of much higher mechanical stability than GB1 domains, it is possible that all the weaker GB1 domains in the polyprotein would have unfolded, but stronger domains would remain folded when the adhesion bond ruptures. In such cases, the force–extension curves will have more unfolding events of the weaker GB1 domains than the stronger hybrid Ig domains. Thus, although unlikely, we cannot completely rule out the possibility that some of the hybrid Ig domains in the third category might indeed unfold at forces that are much higher than that of GB1.
Figure 5.
Hybrid Ig domains I32-DEFG-I27, I27-AA′B-I32, and I32-AA′B-I27 exhibit duality in their mechanical stability. (A,C, and E) Typical force–extension curves of (GB1-I32-DEFG-I27)4, (GB1-I27-AA′B-I32)4, and (GB1-I32-AA′B-I27)4, respectively. The unfolding events of GB1 are colored in gray. The unfolding events with ΔLc of ∼28 nm correspond to the unfolding of hybrid Ig domains and are in black. It is of note that the number of GB1 unfolding events in the force–extension curves are much greater than the number of the unfolding events of hybrid Ig domains. (B,D, and F) Unfolding force histograms of I32-DEFG, I27-AA′B-I32, and I32-AA′B-I27 domains in the heteropolyproteins. The number of events in these histograms is 154, 211, and 57, respectively. Insets in B, D, and F are unfolding force histograms of GB1 domains in the heteropolyproteins, respectively. The number of events is 570, 1199, and 377, respectively.
It is important to note that all the six mechanically stable hybrid daughter proteins (the first category of hybrid proteins) exhibit secondary structural features that are similar to that of wild-type (WT) parent proteins. It seems that the mechano-active site can be reconstituted by recombining AA′ and G β-strands from the two parent proteins. However, it is important to point out that folding does not necessarily entail mechanical stability, at least from the point of view of secondary structures of the hybrid proteins. For example, hybrid protein I32-CFG-I27 displays secondary structure that is similar to that of parent proteins, yet is mechanically weak and unfolds at forces that are below the detection limit of our AFM.
Correlation of mechanical stability with chemical unfolding rate constant?
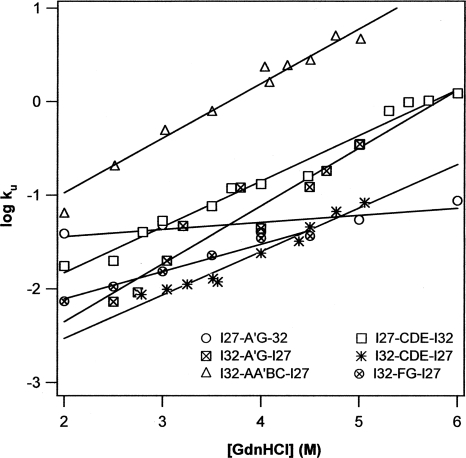
Mechanical stability of proteins is determined by the kinetic energy barrier for mechanical unfolding as well as the distance between the native state and transition state. Hence, the mechanical stability directly correlates with the mechanical unfolding rate constant α0 at zero force. It was observed that the mechanical unfolding rate constant and chemical unfolding rate constant coincide for distal Ig domains from titin (Li et al. 2000b, 2002; Scott et al. 2002), leading to the strong correlation between mechanical stability and chemical unfolding rate constant. I27 and I32 are two examples for such coincidences. In order to investigate whether such correlation is preserved in hybrid proteins after shuffling structural elements between the two parent proteins, we carried out stopped-flow spectrofluorimetry experiments to directly characterize the chemical unfolding kinetics of the six hybrid proteins that are well-folded and mechanically stable. The chemical unfolding kinetics experiments were carried out with the monomers of the recombined hybrids in the presence of denaturant GdnHCl. The unfolding arms of the Chevron plots are plotted in Figure 6. It is clear that the logarithmic of the unfolding rate constants of the hybrid proteins increases linearly as a function of the denaturant concentration, but with different slopes for different hybrid proteins, indicating that the chemical unfolding energy barrier for different hybrid proteins respond differently to the increasing denaturant concentration. Extrapolation of the unfolding arms to zero denaturant concentration provides estimates of the spontaneous unfolding rate constant k u of hybrid proteins in water, which are tabulated in Table 1.
Figure 6.
Plot of logarithmics of chemical unfolding rate constant k u versus the concentration of guanidinium hydrochloride for the engineered mechanically stable hybrid Ig domains. The spontaneous unfolding rate constants in water k u (H2O) for these hybrid Ig domains, which were obtained by extrapolating the unfolding arm to zero denaturant concentration, are tabulated in Table 1.
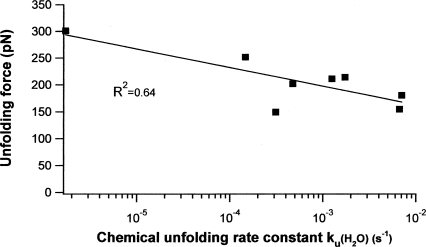
By plotting the mechanical stability of hybrid proteins versus the chemical unfolding rate constant, we notice that there is a weak correlation (R 2 = 0.64) between the chemical unfolding rate constant and mechanical unfolding forces (Fig. 7): The higher the chemical unfolding rate constant is, the lower the unfolding force is. However, this correlation is much weaker than that observed for WT Ig domains from the I-band of titin. This result suggests that within this family of highly homologous hybrid proteins, the chemical unfolding rate constant can only serve as a very rough guide for screening hybrid proteins that are of significant mechanical stability.
Figure 7.
The relationship between the mechanical unfolding force and the spontaneous chemical unfolding for Ig domains. There is a weak correlation between the mechanical unfolding force and the spontaneous chemical unfolding rate constant for the six mechanically stable hybrid daughter proteins, as well as the two parent proteins.
Discussion
Recombination is a promising strategy to engineer proteins of diverse mechanical stability
Using two structurally homologous proteins, I27 and I32, as parent proteins, we have demonstrated that shuffling protein fragments between them can generate hybrid proteins of diverse mechanical stability. Although A′ and G β-strands function as mechano-active sites that are critical to the mechanical stability of I27-like proteins, interactions in other regions can also affect the mechano-active site and change the mechanical stability. Here, we demonstrate that shuffling the A′G β-strands between I27 and I32 does not lead to the transfer of the mechanical stability between the two proteins, suggesting that the existence of the mechano-active site is context dependent. Furthermore, our results showed that dissecting the mechano-active sites and then reconstituting new ones could result in well-folded and mechanically stable hybrid proteins. These results corroborate that not only the mechano-active site plays critical roles in determining the mechanical stability of proteins, the noncovalent interactions neighboring the mechano-active sites are also important in fine tuning the mechanical resistance of proteins. Therefore, these results highlight the global attributes of mechanical stability and suggest that changing the context of the mechano-active site and reconstituting mechano-active sites are feasible ways to engineer proteins of diverse mechanical stability. Considering that the molecular determinants of the mechanical stability of proteins remain largely unclear, DNA shuffling-based recombination is thus a promising strategy to fulfill such requirements, and to engineer proteins of diverse mechanical stability.
Although some hybrid daughter proteins are mechanically comparable to or even more stable than one of the parent proteins, I27, the mechanical stability of the hybrid proteins is yet to surpass that of I32. In order to fully take advantage of the recombination-based approach to engineer protein of tailored mechanical stability (such as improved mechanical stability), one will have to build and examine a library of hybrid proteins that is large enough to carry beneficial mutations. Such a large library of highly homologous hybrid proteins with diverse mechanical stability will also offer an unprecedented training data set for machine learning-based approach to use a statistical analysis method to analyze the correlation between the observed mechanical stability and primary sequence and three-dimensional structures, and identify features that are important for the mechanical stability of proteins. The potential of such approaches has been elegantly demonstrated recently by Arnold and coworkers in the chemical recombination and engineering of cytochrome P450s (Li et al. 2007). We anticipate that similar experiments in engineering elastomeric proteins will pave the way to develop the recombination method into a directed-evolution based tool to tailor the mechanical stability of proteins, which will complement the rational design approach.
Challenges in protein recombination to novel mechanical properties
Although our results demonstrated the great potential of shuffling protein fragments among homologous parent proteins to engineer novel mechanically stable proteins, there exist some challenges to fully realize the potential of recombination-based approaches to engineer novel mechanical proteins.
Folding is the basis for mechanical stability
Acquiring desirable three-dimensional structures and proper folding are the structural basis for mechanical stability. Despite the high sequence homology and identity, almost half of the hybrid daughter proteins we generated exhibit secondary structural signatures that differ from the parent proteins. It becomes evident that hybrid proteins resulting from shuffling structural elements between parent proteins does not guarantee the formation of correctly folded proteins. Although structural elements, such as β-hairpins, were interchanged between two parent proteins at identical sites of the parent proteins, the context in which the structural elements formed has been changed. Hence, the particular structural elements do not necessarily form the same secondary structural elements as they do in original parent protein, leading to misfolding and/or conformational heterogeneity in the native conformations of the hybrid proteins. For example, in hybrid proteins I27-DEFG-I32 and I32-DEFG-I27, four β-strands were interchanged between I27 and I32. The interchange results in the formation of α-helical structures and the increasing random coil content, leading to the disruption of the folded structure of the resulted hybrid proteins. These observations are consistent with early studies on context-dependent secondary structure formation (Minor Jr. and Kim 1996).
Moreover, our current approach does not take into account the nonlocal interactions when we shuffle the secondary structural element, such as the β-hairpin, between parent proteins. It was shown that the folding of I27 involves a diffused folding nucleus involving non-adjacent residues (Fowler and Clarke 2001). It is thus likely that such shuffling may disrupt long-range interactions that are critical for folding, leading to misfolding. Therefore, it is important to design recombined hybrid proteins with the least disruption of such interactions that are critical for folding. Toward such a designed “shuffling” approach, computational algorithms, such as SCHEMA (Voigt et al. 2002; Meyer et al. 2003, 2006), which was developed for such purposes by Arnold and Georgiou (2003), will be of particular importance for our future endeavors.
In addition to correctly folded native structures, folding kinetics is also an important aspect for designing new elastomeric proteins. Although the folding rate constant, which is determined by the folding energy barrier, does not correlate with the mechanical stability of proteins, efficient folding is important for elastomeric proteins to regain their mechanical stability after mechanical unfolding during multiple stretching–relaxation cycles. Optimizing folding properties of tandem modular proteins will be important tasks for future endeavors.
Developing methodologies to efficiently screen proteins that are correctly folded and mechanically resistant
To build a large library of hybrid proteins to screen proteins of desirable mechanical stability, it is also of paramount importance to establish a methodology to enable efficient screening of hybrid daughter proteins that are correctly folded and are likely to retain functional mechano-active site. Generally speaking, well-folded three-dimensional structures should be the foundation for the mechanical stability of proteins. Our current study achieved an ∼50% success rate for constructing proteins that are well-folded and mechanically stable. However, our method used here will not be sufficient to deal with a large library of hybrid proteins. Utilizing high-throughput screening methods to screen correctly folded proteins is key to significantly improving the efficiency of constructing novel hybrid proteins with significant mechanical stability.
Furthermore, developing efficient alternative methods to screen proteins of desirable mechanical stability will be another important task for future endeavors. Currently, the examination of the mechanical properties of the hybrid proteins relies on single-molecule AFM and the construction of polyproteins. With a large library of hybrid proteins, it will become impractical to use single-molecule AFM to screen proteins of desirable mechanical stability. Developing new alternative high-throughput screening methods will be critical to using a recombination method to engineer proteins with defined mechanical properties. This will help to implement a methodology, which is analogous to a directed evolution approach, to purposefully evolve mechanical proteins with desired mechanical properties.
Materials and Methods
Engineering of hybrid proteins by shuffling fragment from I27 and I32
WT I27 and I32 were PCR amplified from the plasmid Ig8-GFP (Dietz and Rief 2004) encoding I27 to I34 genes of human cardiac titin and GFP (a kind gift from Professor Matthias Rief). I27 and I32 were then subcloned into the pUC19 vector with the help of the BamH I site at the 5′ end and Bgl II and Kpn I sites at the 3′ end, to obtain pUC19-I27 and pUC19-I32, respectively. I27-A′G-I32 was generated by the megaprimer method. Residues 15 to 78 were PCR amplified using the standard PCR protocol. The primers encoding the A′ strand (11–14) and G strand (79–88) of I32 were used as the forward and reverse primers flanked with one to four residues of I27, respectively. The amplified PCR product was then used as the megaprimer to generate pUC19I27-A′G-I32 using pUC19I27 as the template. pUC19I32-A′G-I27 was also generated in a similar fashion.
For the generation of pUC19I27-CDE-I32 and pUC19I32-CDE-I27, Apa I and Age I were introduced, respectively, in I27 and I32 after the residues 28 and 67 by site-directed mutagenesis, to generate the plasmids pUC19I27(a-a) and pUC19I32(a-a). The residues 29 to 66 codes the CDE region of I27 along with its adjacent loop, and was obtained by digestion with Apa I and Age I restriction enzymes and ligated into pUC19I32(a-a) linearized with the same set of enzymes to obtain pUC19I32-CDE-I27. pUC19I27-CDE-I32 was obtained in a similar fashion.
In order to generate the other hybrids, the Ava I site was introduced between E and F strands of pUC19I17(a-a) and pUC19I32(a-a). For the generation of pUC19I27-DEFG-I32 and pUC19I32-DEFG-I27, pUC19I27(a-a) and pUC19I32(a-a) were digested with Ava I and Kpn I, to get the insert DEFG, and they were shuffled between pUC19I27(a-a) and pUC19I32(a-a). Similarly, for the generation of pUC19I27-AA′B-I32, and pUC19I32-AA'B-I27, BamH I and Apa I sites were utilized, and for the case of pUC19I32-FG-I27 and pUC19I27-FG-I32, BamH I and Age I sites were utilized.
For the generation of I27-CFG-I32 and I32-CFG-I27, Apa I, Ava I, and Age I sites were introduced, respectively, between the strands B–C, C–D, and E–F in I27 and I32, respectively. In the first step, I27-C-I32 and I32-C-I27 were generated utilizing the Apa I and Ava I sites. In the second step, the Age I site was utilized to generate I27-CFG-I32 and I32-CFG-I27.
I32-AA′BC-I27 was generated using overlap PCR. First, DNA fragment encoding residues 1–38 were PCR amplified. The forward primer contains the restriction site BamHI at the 5′ end and the reverse primer encodes residues 31–38 of I27 and also contains 5′ DNA overhang encoding residues 39–44 of I32. Similarly, DNA encoding amino acids 31–89 of I32 were PCR amplified. The forward primer encodes amino acids 39–45 of I32 and a 5′ overhang of residues 32–38 of I27. The reverse primer contains restriction site BglII, followed by kpnI, respectively. The above two PCR products were gel purified and mixed together and re-amplified using forward and reverse primers containing restriction sites BamHI and BglII, followed by KpnI, respectively. The amplified product was gel purified and digested with restriction enzymes BamHI and KpnI, and ligated into plasmid pQE80 digested with similar restriction enzymes. All the generated hybrids were confirmed by DNA sequencing.
The protein expression was carried out in pQE80 vector. For the generation of pQE80(GB1)4I27-A′G-I32, pQE80(GB1)4I32-A′G-I27, pQE80(GB1)4I27-CDE-I32, and pQE80(GB1)4I32-CDE-I27, the hybrid genes were digested with BamH I and Kpn I and subcloned into the pQE80(GB1)4 vector linearized with Bgl II and Kpn I restriction enzymes to obtain pQE80(GB1)4–hybrid. In order to get pQE80(GB1)4–hybrid–(GB1)4, pQE80(GB1)4 was digested with BamH I and Kpn I to get the insert (GB1)4, and ligated with Bgl II and Kpn I linearized pQE80(GB1)4–hybrid. But in the case of pQE80(GB1-I32-AA′BC-I27)4, pQE80(GB1-I27-AA′B-I32)4, and pQE80(GB1-I32-FG-I27)4, the hybrid insert was obtained by digestion with BamH I and Kpn I and ligated with pQE80GB1, then linearized with Bgl II and Kpn I. The then obtained heterodimer was then digested with BamH I and Kpn I to get the insert and with Bgl II and Kpn I to get the linearized vector. The proteins were expressed in DH5α. The six histidine residues in the expression vector made the purification process easier with the help of NiNTA resin. The purified protein (Cao et al. 2006) was stored in 1× PBS containing 200 mM imidazole and 5 mM DTT to prevent the air oxidation of the two cystine residues that were introduced at the 3′ end of the gene. The protein was stored at 4°C.
Single-molecule AFM experiments
Single-molecule AFM experiments were carried out as described previously (Cao et al. 2006).
Stopped-flow spectrofluorimetry and far-UV CD measurements
The chemical unfolding experiments were carried out on a Bio Logic SFM-300 stopped-flow module, using 7 M guanidinium hydrochloride in 1× PBS buffer, pH 7.4, as the stock denaturing agent. Monomeric hybrid proteins of the concentration of about 1 mg/mL were used in the study. The unfolding kinetics curves were fitted to a single exponential curve to obtain the unfolding rate constants. The far-UV CD spectra were recorded on a JASCO-J810 spectropolarimeter flushed with nitrogen gas. A 0.2-cm path length cuvette was used as the sample container. The data reported here is an average of three scans, with a scan rate of 20 nm min−1. The data are expressed as mean residue ellipticity. For fluorescence and CD studies the hybrid proteins were dialyzed extensively against 0.5× PBS to get rid of imidazole, before carrying out the experiments.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada, Canada Research Chairs Program, and Canada Foundation for Innovation.
Footnotes
Reprint requests to: Hongbin Li, Department of Chemistry, The University of British Columbia, Vancouver, British Columbia V6T 1Z1, Canada; e-mail: hongbin@chem.ubc.ca; fax: (604) 822-2847.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036376.108.
References
- Arnold, F.H., Georgiou, G. Directed enzyme evolution. Humana Press Inc; Clifton, NJ: 2003. [Google Scholar]
- Ashworth, J., Havranek, J.J., Duarte, C.M., Sussman, D., Monnat R.J., Jr, Stoddard, B.L., Baker, D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, G., Suresh, S. Cell and molecular mechanics of biological materials. Nat. Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- Becker, N., Oroudjev, E., Mutz, S., Cleveland, J.P., Hansma, P.K., Hayashi, C.Y., Makarov, D.E., Hansma, H.G. Molecular nanosprings in spider capture-silk threads. Nat. Mater. 2003;2:278–283. doi: 10.1038/nmat858. [DOI] [PubMed] [Google Scholar]
- Best, R.B., Li, B., Steward, A., Daggett, V., Clarke, J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 2001;81:2344–2356. doi: 10.1016/S0006-3495(01)75881-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittker, J.A., Le, B.V., Liu, J.M., Liu, D.R. Directed evolution of protein enzymes using nonhomologous random recombination. Proc. Natl. Acad. Sci. 2004;101:7011–7016. doi: 10.1073/pnas.0402202101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, J.D., Meyer, M.M., Meinhold, P., Otey, C.R., MacMillan, D., Arnold, F.H. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005;15:447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Brockwell, D.J., Beddard, G.S., Paci, E., West, D.K., Olmsted, P.D., Smith, D.A., Radford, S.E. Mechanically unfolding the small, topologically simple protein L. Biophys. J. 2005;89:506–519. doi: 10.1529/biophysj.105.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., Li, H. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat. Mater. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- Cao, Y., Lam, C., Wang, M., Li, H. Nonmechanical protein can have significant mechanical stability. Angew. Chem. Int. Ed. Engl. 2006;45:642–645. doi: 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- Carbone, M.N., Arnold, F.H. Engineering by homologous recombination: Exploring sequence and function within a conserved fold. Curr. Opin. Struct. Biol. 2007;17:454–459. doi: 10.1016/j.sbi.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., Marszalek, P.E., Oberhauser, A.F., Fernandez, J.M. Atomic force microscopy captures length phenotypes in single proteins. Proc. Natl. Acad. Sci. 1999a;96:11288–11292. doi: 10.1073/pnas.96.20.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., Oberhauser, A.F., Fowler, S.B., Marszalek, P.E., Broedel, S.E., Clarke, J., Fernandez, J.M. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. 1999b;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi, C., Shank, E.A., Bustamante, C., Marqusee, S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Dietz, H., Rief, M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin, C.M., Carr, A.G., Huson, M.G., Maxwell, J.M., Pearson, R.D., Vuocolo, T., Liyou, N.E., Wong, D.C., Merritt, D.J., Dixon, N.E. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- Evans, E. Probing the relation between force–lifetime and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- Fisher, T.E., Oberhauser, A.F., Carrion-Vazquez, M., Marszalek, P.E., Fernandez, J.M. The study of protein mechanics with the atomic force microscope. Trends Biochem. Sci. 1999;24:379–384. doi: 10.1016/s0968-0004(99)01453-x. [DOI] [PubMed] [Google Scholar]
- Fowler, S.B., Clarke, J. Mapping the folding pathway of an immunoglobulin domain: Structural detail from φ value analysis and movement of the transition state. Structure. 2001;9:355–366. doi: 10.1016/s0969-2126(01)00596-2. [DOI] [PubMed] [Google Scholar]
- Goodsell, D.S. Bionanotechnology. Wiley-Liss; Hoboken, NJ: 2004. [Google Scholar]
- Gosline, J.M., Guerette, P.A., Ortlepp, C.S., Savage, K.N. The mechanical design of spider silks: From fibroin sequence to mechanical function. J. Exp. Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- Gosline, J., Lillie, M., Carrington, E., Guerette, P., Ortlepp, C., Savage, K. Elastic proteins: Biological roles and mechanical properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:121–132. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold, K.E., Kawarasaki, Y., Ghoneim, N., Benkovic, S.J., Iverson, B.L., Georgiou, G. Evolution of highly active enzymes by homology-independent recombination. Proc. Natl. Acad. Sci. 2005;102:10082–10087. doi: 10.1073/pnas.0504556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L., Althoff, E.A., Clemente, F.R., Doyle, L., Rothlisberger, D., Zanghellini, A., Gallaher, J.L., Betker, J.L., Tanaka, F., Barbas C.F., III, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer, M.S., Smith, S.B., Granzier, H.L., Bustamante, C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- Kortemme, T., Baker, D. Computational design of protein–protein interactions. Curr. Opin. Chem. Biol. 2004;8:91–97. doi: 10.1016/j.cbpa.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Labeit, S., Kolmerer, B. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Li, H. Engineering proteins with tailored nanomechanical properties: A single molecule approach. Org. Biomol. Chem. 2007;5:3399–3406. doi: 10.1039/b710321m. [DOI] [PubMed] [Google Scholar]
- Li, H., Carrion-Vazquez, M., Oberhauser, A.F., Marszalek, P.E., Fernandez, J.M. Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 2000a;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- Li, H., Oberhauser, A.F., Fowler, S.B., Clarke, J., Fernandez, J.M. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. 2000b;97:6527–6531. doi: 10.1073/pnas.120048697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Oberhauser, A.F., Redick, S.D., Carrion-Vazquez, M., Erickson, H.P., Fernandez, J.M. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. 2001;98:10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Linke, W.A., Oberhauser, A.F., Carrion-Vazquez, M., Kerkvliet, J.G., Lu, H., Marszalek, P.E., Fernandez, J.M. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Li, Y., Drummond, D.A., Sawayama, A.M., Snow, C.D., Bloom, J.D., Arnold, F.H. A diverse family of thermostable cytochrome P450s created by recombination of stabilizing fragments. Nat. Biotechnol. 2007;25:1051–1056. doi: 10.1038/nbt1333. [DOI] [PubMed] [Google Scholar]
- Lu, H., Isralewitz, B., Krammer, A., Vogel, V., Schulten, K. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 1998;75:662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Schulten, K. The key event in force-induced unfolding of Titin's immunoglobulin domains. Biophys. J. 2000;79:51–65. doi: 10.1016/S0006-3495(00)76273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M.M., Silberg, J.J., Voigt, C.A., Endelman, J.B., Mayo, S.L., Wang, Z.G., Arnold, F.H. Library analysis of SCHEMA-guided protein recombination. Protein Sci. 2003;12:1686–1693. doi: 10.1110/ps.0306603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M.M., Hochrein, L., Arnold, F.H. Structure-guided SCHEMA recombination of distantly related β-lactamases. Protein Eng. Des. Sel. 2006;19:563–570. doi: 10.1093/protein/gzl045. [DOI] [PubMed] [Google Scholar]
- Minor D.L., Jr, Kim, P.S. Context-dependent secondary structure formation of a designed protein sequence. Nature. 1996;380:730–734. doi: 10.1038/380730a0. [DOI] [PubMed] [Google Scholar]
- Oberhauser, A.F., Marszalek, P.E., Erickson, H.P., Fernandez, J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- Otey, C.R., Silberg, J.J., Voigt, C.A., Endelman, J.B., Bandara, G., Arnold, F.H. Functional evolution and structural conservation in chimeric cytochromes p450: Calibrating a structure-guided approach. Chem. Biol. 2004;11:309–318. doi: 10.1016/j.chembiol.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Pfuhl, M., Pastore, A. Tertiary structure of an immunoglobulin-like domain from the giant muscle protein titin: A new member of the I set. Structure. 1995;3:391–401. doi: 10.1016/s0969-2126(01)00170-8. [DOI] [PubMed] [Google Scholar]
- Politou, A.S., Thomas, D.J., Pastore, A. The folding and stability of titin immunoglobulin-like modules, with implications for the mechanism of elasticity. Biophys. J. 1995;69:2601–2610. doi: 10.1016/S0006-3495(95)80131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief, M., Gautel, M., Oesterhelt, F., Fernandez, J.M., Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Scott, K.A., Steward, A., Fowler, S.B., Clarke, J. Titin; a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 2002;315:819–829. doi: 10.1006/jmbi.2001.5260. [DOI] [PubMed] [Google Scholar]
- Sharma, D., Cao, Y., Li, H. Engineering proteins with novel mechanical properties by recombination of protein fragments. Angew. Chem. Int. Ed. Engl. 2006;45:5633–5638. doi: 10.1002/anie.200600382. [DOI] [PubMed] [Google Scholar]
- Sharma, D., Perisic, O., Peng, Q., Cao, Y., Lam, C., Lu, H., Li, H. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc. Natl. Acad. Sci. 2007;104:9278–9283. doi: 10.1073/pnas.0700351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham, A.S., Shewry, P.R. Elastomeric proteins: Biological roles, structures and mechanisms. Trends Biochem. Sci. 2000;25:567–571. doi: 10.1016/s0968-0004(00)01670-4. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova, L., Trinick, J., Sleep, J.A., Simmons, R.M. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- Voigt, C.A., Martinez, C., Wang, Z.G., Mayo, S.L., Arnold, F.H. Protein building blocks preserved by recombination. Nat. Struct. Biol. 2002;9:553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- Yang, G., Cecconi, C., Baase, W.A., Vetter, I.R., Breyer, W.A., Haack, J.A., Matthews, B.W., Dahlquist, F.W., Bustamante, C. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc. Natl. Acad. Sci. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]