Abstract
Urate oxidase catalyzes the oxidation of uric acid with poor solubility to produce 5-hydroxyisourate and allantoin. Since allantoin is excreted in vivo, urate oxidase has the potential to be a therapeutic target for the treatment of gout. However, its severe immunogenicity limits its clinical application. Furthermore, studies on the structure–function relationships of urate oxidase have proven difficult. We developed a method for genetically incorporating p-azido-L-phenylalanine into target protein in Escherichia coli in a site-specific manner utilizing a tyrosyl suppressor tRNA/aminoacyl-tRNA synthetase system. We substituted p-azido-L-phenylalanine for Phe170 or Phe281 in urate oxidase. The products were purified and their enzyme activities were analyzed. In addition, we optimized the system by adding a “Shine-Dalgarno (SD) sequence” and tandem suppressor tRNA. This method has the benefit of site-specifically modifying urate oxidase with homogeneous glycosyl and PEG derivates, which can provide new insights into structure–function relationships and improve pharmacological properties of urate oxidase.
Keywords: urate oxidase, site-directed mutagenesis, tyrosyl suppressor tRNA/aminoacyl-tRNA synthetase system, p-azido-L-phenylalanine, amber nonsense codon
Uric acid is the ultimate product of purine degradation. Animals use urate oxidase (uricase) to convert uric acid to allantoin, which is more soluble and easily excreted. High levels of uric acid in the blood lead to renal failure and a number of diseases, such as gout. As uricase can rapidly decrease the plasma urate levels, it is an attractive target for the treatment of gout (Coiffier et al. 2003). However, repeated treatment with the native uricase is accompanied by strong immunoreaction, which not only attenuates its bioavailability but also causes severe side effects (Bardin 2004). Uricase exists in monomeric and multimeric forms; its active form is a tetramer (Cammalleri and Malaguarnera 2007). Therefore, structure–function relationships of uricase are essential for exploiting its therapeutic potential.
Peter G. Schultz and his research team developed a method for site-specific incorporation of unnatural amino acids into proteins in vivo (Wang and Schultz 2005). These unnatural amino acids carry reactive groups that are lacking in natural amino acids (Wang and Schultz 2005). Proteins containing unnatural amino acids can be site-specifically modified with biophysical probes, cytotoxic agents, and cross-linking agents, and are thus convenient for analyzing the structure and function of proteins. These proteins can also be site-specifically modified with other agents such as homogeneous glycosyl or PEG derivates for improving their pharmacological properties (Dougherty 2000; Deiters et al. 2004). Recently, Dougherty and coworkers have found optimal suppressor tRNAs to optimize site-specific modification of the unnatural amino acids in target proteins (Rodriguez et al. 2007a,b).
Here, we established an orthogonal tyrosyl suppressor tRNA/aminoacyl-tRNA synthetase system that can selectively incorporate p-azido-L-phenylalanine into the amber nonsense codon TAG of uricase in Escherichia coli. We optimized this system by adding a “Shine-Dalgarno (SD) sequence” and tandem suppressor tRNA in order to increase the expression levels of tyrosyl suppressor tRNA and aminoacyl-tRNA synthetase. Moreover, we substituted p-azido-L-phenylalanine for uricase Phe170 or Phe281 with good efficiencies and high selectivities, while the products retained enzyme activities. These uricases containing p-azido-L-phenylalanine can be used for site-specific PEGylation with alkynyl-PEG derivates in order to reduce their antigenicity and immunogenicity, or to carry out site-specific modification with biophysical probes to study the structure–function relationships of uricase.
Results
Results of positive selection and negative screen
To test the orthogonality of the system, especially the fidelity of the mutant aminoacyl-tRNA synthetase in E. coli, an amber codon was introduced at a permissive site (Asp112) in the chloramphenicol acetyltransferase gene. The mutants of MjTyrRS were obtained through PCR-based random mutagenesis at translation active sites based on the crystal structure and dynamics simulations of MjTyrRS (Zhang et al. 2005; Turner et al. 2006). The activities of the MjTyrRS mutants were measured by the survivability of E. coli cells in cultures with various chloramphenicol concentrations. pAC-tRNA encoding the suppressor tRNA was cotransformed with the mutants of MjTyrRS in E. coli DH10B cells, and positive selections and negative screens were carried out with and without pAzpa. Any mutant of MjTyrRS that could just aminoacylate endogenous tRNA was screened and eliminated. E. coli that survived in the presence of pAzpa and died in the absence of pAzpa were isolated, and the mutant genes of MjTyrRS were isolated from these cells and sequenced.
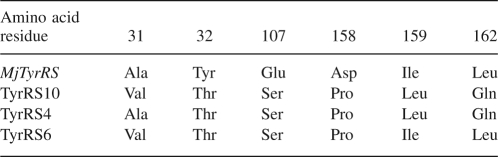
DNA sequencing of the mutant MjTyrRSs revealed that the major sites of enzyme activity were at 31Ala, 32Tyr, 107Glu, 158Asp, and 159Ile. We selected three mutants of MjTyrRS–TyrRS10, TyrRS4, and TyrRS6. The mutations in TyrRS10, TyrRS4, and TyrRS6 are shown in Table 1. The positive colonies were selected according to the suppression of TAG at Asp112 inside the chloramphenicol acetyltransferase gene. The chloramphenicol tolerance of DH10B containing TyrRS10 in the positive selection reached 60 μg/mL, while the chloramphenicol tolerance was lower than 15 μg/mL in the negative screen, as shown in Figure 1.
Table 1.
Amino acid residues of active sites in the wt MjTyrRS and the mutant MjTyrRSs with specificities for p-azido-L-phenylalanine
Figure 1.
Positive selection and negative screen based on the suppression of Asp112TAG in the chloramphenicol acetyltransferase gene. pBR-TyrRS encoding the mutant MjTyrRS and pAC-tRNA encoding the suppressor tRNA were cotransformed in DH10B cells. (A) Positive selection was carried out in GMML liquid medium with various concentrations of chloramphenicol and p-azido-L-phenylalanine. (B) Negative screen was carried out in GMML liquid medium with various concentrations of chloramphenicol in the absence of p-azido-L-phenylalanine.
Optimization results
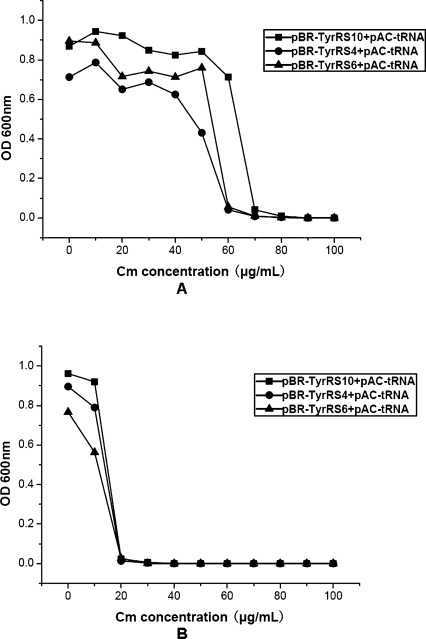
There are SD sequences in bacterial mRNAs, which are in charge of initiating translation from the initiation codon AUG. We added an SD sequence (AGGAA) in front of the gene of mutant MjTyrRS to increase the expression levels of the MjTyrRS gene. As shown in Figure 2, after inserting this SD sequence before the start codon for the plasmid-encoded mutant MjTyrRS, the chloramphenicol tolerance of DH10B containing TyrRS10 in the positive selection increased from 60 to 110 μg/mL.
Figure 2.
After placement of a SD sequence before the start codon for the plasmid-encoded mutant MjTyrRS, positive selection and negative screen were carried out. These selections were based on the suppression of Asp112TAG in the chloramphenicol acetyltransferase gene. pBR-SD-TyrRS encoded the mutant MjTyrRS that was cotransformed with pAC-tRNA in DH10B cells. (A) Positive selection was carried out in GMML liquid medium with various concentrations of chloramphenicol and p-azido-L-phenylalanine. (B) Negative screen was carried out in GMML liquid medium with various concentrations of chloramphenicol in the absence of p-azido-L-phenylalanine.
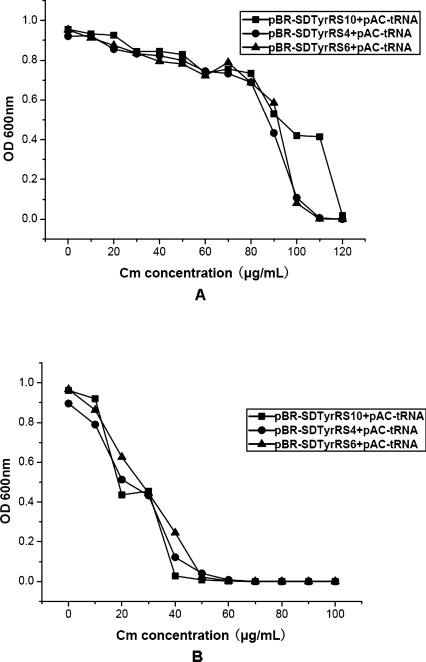
Codon usage has been identified as the most important factor during the expression of prokaryotic genes. This is because the larger the abundance of cognate tRNAs available within the cell, the more preferred the codons will be (Gustafsson et al. 2004). Similarly, high expression levels of proteins with amber nonsense codons are correlated with the amount of the corresponding tyrosyl suppressor tRNA. Since tyrosyl suppressor tRNA lies in the stringent plasmid pACYC184, we hoped to increase the expression level of suppressor tRNA through inserting four copies of the suppressor tRNA gene into pAC-tRNA at the same site.
In prokaryote chromosomes, DNA fragments encoding tRNAs are often multicistronic. However, studies have shown that multicistronic expression vectors can decrease the expression level of the second gene relative to that of the first gene, which is adjacent to the promoter. This problem can be solved by inserting an additional promoter in front of the second gene. These corresponding two-promoter vectors yielded four to nine times more protein than multicistronic expression vectors (Kim et al. 2004). Therefore, we designed (lpp-tRNA-rrnC)4 to improve the expression of tyrosyl suppressor tRNA.
As expected, we increased the expression levels of tyrosyl suppressor tRNA, as proven by the survivability in cultures with various chloramphenicol concentrations. As shown in Figure 3, cells cotransformed with pAC-tRNA4/pBR-SD-TyrRS grew at a faster rate and the chloramphenicol tolerance in the positive selection increased from 60 to 130 μg/mL.
Figure 3.
After incorporation of multiple copies of plasmid-encoded Methanococcus jannaschii-derived suppressor tRNA, a series of selections based on the suppression of Asp112TAG in the chloramphenicol acetyltransferase gene were carried out. (A) In order to assess the efficiency of the multiple copies of suppressor tRNA, pBR-SD-TyrRS10 encoding the mutant MjTyrRS was cotransformed with pAC-tRNA and pAC-tRNA4, respectively, in DH10B cells. pAC-tRNA encoded a single suppressor tRNA and pAC-tRNA4 encoded multiple copies of suppressor tRNA. Positive selection was carried out in GMML liquid medium with 100 μg/mL chloramphenicol and p-azido-L-phenylalanine. (B) pBR-SD-TyrRS4 encoding the mutant MjTyrRS was cotransformed with either pAC-tRNA or pAC-tRNA4 in DH10B cells. Positive selection was carried out in GMML liquid medium with 100 μg/mL chloramphenicol and p-azido-L-phenylalanine. (C) pBR-SD-TyrRS6 encoding the mutant MjTyrRS was cotransformed with either pAC-tRNA or pAC-tRNA4 in DH10B cells. Positive selection was carried out in GMML liquid medium with 100 μg/mL chloramphenicol and p-azido-L-phenylalanine. (D) In concordance with the positive selections of A–C, positive selection was carried out in GMML liquid medium with various concentrations of chloramphenicol in the presence of p-azido-L-phenylalanine in order to detect the survivability with chloramphenicol. (E) Negative screen was carried out in GMML liquid medium with various concentrations of chloramphenicol in the absence of p-azido-L-phenylalanine.
Expression and purification of the mutant uricase
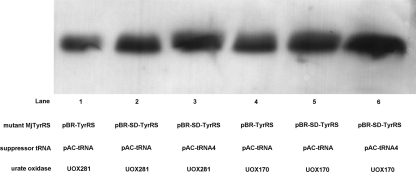
Based on the crystal structure of Aspergillus flavus uricase, whose sequence is 49% identical to that of Candida utilis, we hypothesized that Phe170 and Phe281 were two positions nonessential for enzyme activity (Hongoh et al. 2000). We thus chose these two positions to insert pAzpa into uricase and obtained uox-Phe170-Azp and uox-Phe281-Azp. The expression levels of uox-Phe170-Azp and uox-Phe281-Azp were analyzed by immunoblotting. Immunoblot analysis showed that the yield of uox-Phe170-Azp was significantly higher than that of uox-Phe281-Azp and the high yields of both uox-Phe170-Azp and uox-Phe281-Azp could be attributed to the efficient synthesis of the mutants MjTyrRS and suppressor tRNA. We also observed that the optimization of adding the SD sequence did not obviously increase the yields of the mutant uricase, though this optimization can be detected by the measurement of Cm concentrations. These two optimizations—placement of a SD sequence before the start codon for the plasmid-encoded mutant MjTyrRS and the incorporation of multiple copies of plasmid-encoded Methanococcus jannaschii-derived suppressor tRNA—increased the yields of the mutant uricase remarkably (Fig. 4). Moreover, in the presence of Cm (≥40 μg/mL) in the GMML, cells did not survive without pAzpa, and expression of uricase containing natural amino acids at 281 and 170 were prevented. When we purified and quantified the mutant proteins by Ni-NTA affinity chromatography, the yield of uox-Phe281-Azp was 1∼2 mg/L of culture, and the yield of uox-Phe170-Azp was 3∼6 mg/L.
Figure 4.
Western blot analysis of the expression levels of uox-Phe170-Azp and uox-Phe281-Azp under various conditions. An anti-His6 antibody was used to detect the expression levels of the uricase mutants. Cells from 200 μL of the cultures were pelleted and applied to gels for analysis.
Analysis of uricase activity
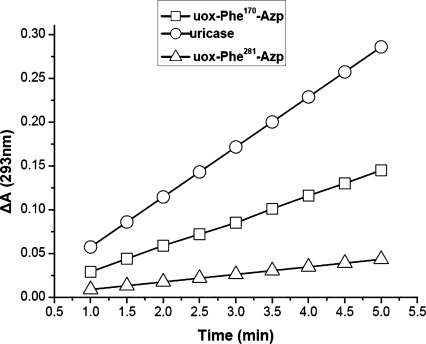
The assay mixture contained about 10 μL of protein (0.1 mg/mL) solution in borate buffer (pH 8.4, 0.1 M) and 59.84 μM uric acid in a final volume of 3 mL. One unit is defined as the amount of enzyme necessary to transform 1 μM of uric acid into allantoin in 1 min at 25°C (pH 8.4). As shown in Figure 5, based on the kinetics parameter of uric acid concentration decrease (uricase 0.0571; uox-Phe170-Azp 0.0288; uox-Phe281-Azp 0.0086), the activity of natural uricase was 16.33 U/mg, uox-Phe170-Azp was 8.26 U/mg and uox-Phe281-Azp was 2.49 U/mg. Compared with the natural uricase, the enzymatic activity of the mutant uricase containing pAzpa at site 170 amino acid was reduced to 50% and the enzymatic activity of the mutant uricase containing pAzpa at site 281 amino acid was reduced to 12%
Figure 5.
Kinetics of uric acid concentration decrease during conversion of uric acid to allantoin by uricase.
Discussion
Uricase is an endogenous enzyme found in most mammals, but not in humans. During primate evolution, the inactivation of the hominoid uricase gene was caused by independent nonsense or frameshift mutations and has taken a two-step deterioration process, first in the promoter and second in the coding region (Oda et al. 2002). The loss of uricase led to diseases such as gout, which is caused by the deposition of urate crystals. Uricase analog Rasburicase has been exploited as a therapeutic tool to treat gout (Coiffier et al. 2003; Li et al. 2006). But, it has a short half-life and requires a daily administration. However, studies have shown that PEGylation can prolong its half-life and reduce its immunogenicity (Sherman et al. 2008). In this study, we substituted pAzpa for Phe170 or Phe281 of uricase, and the resulting mutants will be used to perform site-specific PEGylation with alkynyl-PEG derivates without influencing its activity. This method of site-directed mutagensis not only generated uniform PEGylation production, but can also be utilized for research on conversion between the multimer and monomer (Zaremba et al. 2005; Aslan et al. 2007). This is important, as active uricase from Candida utilis often exists as a tetramer or even an octamer, and the enzyme activities of uricase are mainly related to its structure.
After optimization, the yield of uox-Phe170-Azp increased to 3∼6 mg/L, but the yield of uox-Phe281-Azp was only 1∼2 mg/L. This phenomenon was similar to that observed by other researchers on site-specific incorporation of unnatural amino acids (Chen et al. 2007; Ye et al. 2008). Remarkably, the yield of the mutant uricase correlated closely to the purity of pAzpa (pAzpa was synthetized by our lab). However, both mutations decreased uricase activity.
In summary, we have optimized and characterized a system for pAzpa mutagenesis in E. coli and have used it to introduce pAzpa at two sites in uricase. In addition, the mutant uricase retained 50% activity of natural uricase
Materials and Methods
Strains and plasmids
Plasmid pBR322 was obtained from TaKaRa. Plasmid pACYC184 and M. jannaschii total DNA were obtained from ATCC. DH10B was a gift of Shanghai Jiao Tong University. Plasmid pET28a-uox, encoding the Candida utilis uricase with a His6 tag at the COOH-terminal under the control of a T7 promoter and a T7 terminator was stored in our laboratory. All of the plasmids were verified by DNA sequencing.
Construction of tyrosyl tRNA/tRNA-synthetase pair
PCR fragments of the MjTyrRS mutants were generated from M. jannaschii total DNA using the following primers:
5P1: 5′-GGCAGATCTATGGACGAATTTGAAATGATA-3′
3P1: 5′-AAACCTATGGTAGCAGATTTTTC-3′
5P2: 5′-GATGAAAAATCTGCTACCATACG-3′
3P2: 5′-AAGCTACTTCCATAAACATA-3′
5P3: 5′-ATATGTTTATGGAAGTAGCTTCC-3′
3P3: 5′-CCTTGATAATGAAGAGGATTAACCTGC-3′
5P4: 5′-TTAATCCTCTTCATTATCAAGGCGTTG-3′
3P4: 5′-GCATCTAGATTATAATCTCTTTCTAATTG-3′.
The fragments of MjTyrRS mutants under the control of the E. coli GlnRS promoter and terminator were digested with restriction enzymes BamHI and SalI, and inserted into the predigested pBR322 to afford plasmid pBR-TyrRS. An amber codon was substituted for Asp112 in the CAT gene of pACYC184 to afford plasmid pAC-TAG (Wang and Schultz 2005). The gene encoding tyrosyl suppressor tRNA was chemically synthesized based on the Schultz research team reports (Wang and Schultz 2001). Suppressor tRNA under the control of the lpp promoter and rrnC terminator was then digested with restriction enzymes BclI and AvaI and inserted into the predigested pAC-TAG to afford plasmid pAC-tRNA.
Positive selection and negative screen
The positive selection and the negative screen were carried out in liquid medium. Competent DH10B cells were cotransformed with pBR-TyrRS and pAC-tRNA. Single colonies were inoculated into LB liquid medium supplemented with tetracycline (Tet, 40 μg/mL) and ampicillin (Amp, 100 μg/mL). Cultures were grown at 37°C with shaking until the OD600 was ∼1.0. Cells from 100 μL of these cultures were washed twice with GMML medium (1×M9/1 mM MgSO4/0.1 mM CaCl2/8.5 mM NaCl/5 μM FeSO4/1% glycerol/0.3 mM leucine) and resuspended in 20 mL of GMML liquid medium containing 1 mM pAzpa, 100 μg/mL Amp, 40 μg/mL Tet, and various concentrations of chloramphenicol (Cm), and GMML medium were supplemented with 100 μg/mL Amp, 40 μg/mL Tet, and various concentrations of Cm (Anderson et al. 2004). The cultures were incubated at 37°C with shaking for 72 h. Cell growth was monitored by OD600. Candidate clones were those that survived in GMML with pAzpa and a high concentration of Cm (e.g., 80 μg/mL or higher), but died in GMML without pAzpa and a low concentration of Cm (e.g., 20 μg/mL or lower) (Wang and Schultz 2005). Candidate clones were inoculated into LB liquid medium with tetracycline (Tet, 40 μg/mL) and ampicillin (Amp, 100 μg/mL), and grown to saturation at 37°C with shaking. Then, their plasmid DNA was isolated and sequenced.
Optimization of tyrosyl tRNA/tRNA-synthetase pair
An SD sequence was inserted before the gene of the mutant MjTyrRS in order to increase the amount of MjTyrRSs and enhance efficiency of the tyrosyl suppressor tRNA/aminoacyl-tRNA synthetase system. PCR fragments containing SD-MjTyrRSs were generated from the mutant MjTyrRSs using the following oligonucleotides:
5P1SD: 5′-AGATCTAGGAAAGGTCTATGGACGAATTTGAAATGATA-3′
3P4: 5′-GCATCTAGATTATAATCTCTTTCTAATTG-3′.
SD-MjTyrRSs genes under the control of the E. coli GlnRS promoter and terminator were excised with enzymes BamHI and SalI, and inserted into the predigested pBR322 to afford plasmid pBR-SD-TyrRS. Competent DH10B cells were cotransformed with pBR-SD-TyrRS and pAC-tRNA for positive selective and negative screen. Candidate clones were inoculated into LB liquid medium with tetracycline (Tet, 40 μg/mL) and ampicillin (Amp, 100 μg/mL) and grown to saturation in a 37°C shaker. Then, plasmid DNA was isolated.
The (lpp-tRNA-rrnC)4 sequence was designed for the high-efficient orthogonal system. A PCR fragment of (lpp-tRNA-rrnC)4 was generated from the suppressor tRNA using the following oligonucleotides:
tRNA1P5: 5′-GTATGATCAGGATAACCAGAAGCAAT-3′
tRNA1P3: 5′-CCCGAGTCGAAGCTTACGCTGCAGTAAAAAAAATCCT-3′
tRNA2P5: 5′-GTATCTGCAGGATAACCAGAAGCAAT-3′
tRNA2P3: 5′-GAAGCTTACGGTCGACTAAAAAAAATTCC-3′
tRNA3P5: 5′-GTCGACTCAGGATAACCAGAAGCAAT-3′
tRNA3P3: 5′-GAAGCTTACGCTCTAGAAAAAAAAATCCTTA-3′
tRNA4P5: 5′-TCTAGATCAGGATAACCAGAAGCAAT-3′
tRNA4P3: 5′-CGCAAGCTTAAAAAAAATCCTTA-3′.
The (lpp-tRNA-rrnC)4 fragment was excised with BclI and AvaI, and inserted into the predigested pAC-tRNA to afford plasmid pAC-tRNA4. Competent DH10B cells were cotransformed with pBR-SD-TyrRS and pAC-tRNA4 and performed positive selection and negative screen. Candidate clones were inoculated into LB liquid medium containing tetracycline (Tet, 40 μg/mL) and ampicillin (Amp, 100 μg/mL) and grown to saturation in a 37°C shaker. Then, plasmid DNA was isolated.
Expression and purification of uricase containing p-azido-L-phenylalanine
Plasmid pET28a-uox was used to encode uricase of Candida utilis. An amber codon was substituted for Phe170 or Phe281 in the uricase gene using the following oligonucleotides based on pET28a-uox:
uox5P1: 5′-ACGGGATCCATGTCAACAACGCTCTCATC-3′
uoxzs3P1: 5′-TGGAGAGGTGTACTACAACTCTTG-3′
uoxzs5P2: 5′-GACAACGAGTTGTAGTACCC-3′
uox3P2: 5′-ACTGTCGACTTACAACTTGGTCTTCTCCT-3′
uox170P5: 5′-AGTGTGACTAGACCACCTTG-3′
uox170P3: 5′-GGTCTAGTCACACTTGTTGT-3′.
The genes of uricase amber170 mutant and uricase amber281 mutant had a His6 tag at their COOH-terminal and their expressions were under the control of the T7 promoter and T7 terminator. These genes were excised with BclI and inserted into the predigested pAC-tRNA4 to afford plasmids pAC-tRNA4-uox281 and pAC-tRNA4-uox170.
E. coli DH10B cells were cotransformed with pAC-tRNA4-uox170/pAC-tRNA4-uox281 and pBR-SD-TyrRS. Single colonies were inoculated into LB with Amp (100 μg/mL) and Tet (40 μg/mL) and grown at 37°C to saturation. The cells were harvested and washed twice with GMML. The pellets were resuspended in GMML medium and inoculated into 1 L of GMML medium with 1 mM pAzpa, 100 μg/mL Amp, 40 μg/mL Tet, and 40 μg/mL Cm, and the cells were grown at 37°C to an OD600 of 0.5. Isopropyl-β-D-thiogalactopyranoside (1 mM) was added to induce the expression of the proteins. After 24 h, cells from 200 μL of the cultures were pelleted and loaded onto gels for SDS-PAGE and Western Blot analysis. An anti-His6 antibody was used to detect the expression levels of the mutant proteins. The mutant proteins were purified by Ni2+ affinity chromatography and were then stored in 0.1 M (pH 8.4) borate-sodium biborate buffer. A Coomassie brilliant blue assay kit was purchased from Jiancheng Bioengineering Institute and used to determine the protein content.
Determination of uricase activity
Uricase activity was assayed by detecting the consumption of uric acid. In detection of uricase activity, at 25°C, uric acid was dissolved in 0.1 M (pH 8.4) borate-sodium biborate buffer to a final concentration of 59.48 μM. After the enzyme was added, absorbance at 293 nm was monitored for 5 min every 30 s.
Acknowledgments
We acknowledge the Chinese National Natural Science Foundation (30772679, 30672559) and Hi-Tech Research and Development Program of China-863 Program (2007AA02Z101) for their support.
Footnotes
Reprint requests to: Wenbing Yao, 24 Tongjia Xiang, Nanjing 210009, People's Republic of China; e-mail: wbyao@cpu.edu.cn; fax: 86-025-83302827.
Abbreviations: pAzpa, p-azido-L-phenylalanine; uox-Phe281-Azp, urate oxidase with p-azido-L-phenylalanine at the site of 281; uox-Phe170-Azp, urate oxidase with p-azido-L-phenylalanine at the site of 170; MjTyrRS, Methanococcus jannaschii aminoacyl-tRNA synthetase.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.034587.108.
References
- Anderson, J.C., Wu, N., Santoro, S.W., Lakshman, V., King, D.S., Schultz, P.G. An expanded genetic code with a functional quadruplet codon. Proc. Natl. Acad. Sci. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan, F.M., Yu, Y., Vajda, S., Mohr, S.C., Cantor, C.R. Engineering a novel, stable dimeric streptavidin with lower isoelectric point. J. Biotechnol. 2007;128:213–225. doi: 10.1016/j.jbiotec.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Bardin, T. Current management of gout in patients unresponsive or allergic to allopurinol. Joint Bone Spine. 2004;71:481–485. doi: 10.1016/j.jbspin.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Cammalleri, L., Malaguarnera, M. Rasburicase represents a new tool for hyperuricemia in tumor lysis syndrome and in gout. Int. J. Med. Sci. 2007;4:83–93. doi: 10.7150/ijms.4.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Schultz, P.G., Brock, A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae . J. Mol. Biol. 2007;371:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Coiffier, B., Mounier, N., Bologna, S., Fermé, C., Tilly, H., Sonet, A., Christian, B., Casasnovas, O., Jourdan, E., Belhadj, K., et al. Efficacy and safety of Rasburicase (recombinant urate oxidase) for the prevention and treatment of hyperuricemia during induction chemotherapy of aggressive non-Hodgkin's lymphoma: Results of the GRAAL1 (Groupe d'Etude des Lymphomes de l'Adulte Trial on Rasburicase Activity in Adult Lymphoma) study. J. Clin. Oncol. 2003;21:4402–4406. doi: 10.1200/JCO.2003.04.115. [DOI] [PubMed] [Google Scholar]
- Deiters, A., Cropp, T.A., Summerer, D., Mukherji, M., Schultz, P.G. Site-specific PEGylation of proteins containing unnatural amino acids. Bioorg. Med. Chem. Lett. 2004;14:5743–5745. doi: 10.1016/j.bmcl.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Dougherty, D.A. Unnatural amino acids as probes of protein structure and function. Curr. Opin. Chem. Biol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- Gustafsson, C., Govindarajan, S., Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Hongoh, Y., Sasaki, T., Ishikawa, H. Cloning, sequence analysis and expression in Escherichia coli of the gene encoding a uricase from the yeast-like symbiont of the brown planthopper, Nilaparvata lugens . Insect Biochem. Mol. Biol. 2000;30:173–182. doi: 10.1016/s0965-1748(99)00116-2. [DOI] [PubMed] [Google Scholar]
- Kim, K.J., Kim, H.E., Lee, K.H., Han, W., Yi, M.J., Jeong, J., Oh, B.H. Two-promoter vector is highly efficient for overproduction of protein complexes. Protein Sci. 2004;13:1698–1703. doi: 10.1110/ps.04644504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Chen, Z., Hou, L., Fan, H., Weng, S., Xu, C., Ren, J., Li, B., Chen, W. High-level expression, purification, and characterization of non-tagged Aspergillus flavus urate oxidase in Escherichia coli . Protein Expr. Purif. 2006;49:55–59. doi: 10.1016/j.pep.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Oda, M., Satta, Y., Takenaka, O., Takahata, N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol. Biol. Evol. 2002;19:640–653. doi: 10.1093/oxfordjournals.molbev.a004123. [DOI] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester, H.A., Dougherty, D. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: Minimizing misacylation. RNA. 2007a;13:1703–1714. doi: 10.1261/rna.666807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester, H.A., Dougherty, D. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: Evaluating suppression efficiency. RNA. 2007b;13:1715–1722. doi: 10.1261/rna.667607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, M.R., Saifer, M.G., Perez-Ruiz, F. PEG-uricase in the management of treatment-resistant gout and hyperuricemia. Adv. Drug Deliv. Rev. 2008;60:59–68. doi: 10.1016/j.addr.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Turner, J.M., Graziano, J., Spraggon, G., Schultz, P.G. Structural plasticity of an aminoacyl-tRNA synthetase active site. Proc. Natl. Acad. Sci. 2006;103:6483–6488. doi: 10.1073/pnas.0601756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Schultz, P.G. A general approach for the generation of orthogonal tRNAs. Chem. Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- Wang, L., Schultz, P.G. Expanding the genetic code. Angew Chem. Int Ed Engl. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- Ye, S., Köhrer, C., Huber, T., Kazmi, M., Sachdev, P., Yan, E.C., Bhagat, A., Rajbhandary, U.L., Sakmar, T.P. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- Zaremba, M., Sasnauskas, G., Urbanke, C., Siksnys, V. Conversion of the tetrameric restriction endonuclease Bse634I into a dimer: Oligomeric structure-stability-function correlations. J. Mol. Biol. 2005;348:459–478. doi: 10.1016/j.jmb.2005.02.037. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Wang, L., Schultz, P.G., Wilson, I.A. Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine. Protein Sci. 2005;14:1340–1349. doi: 10.1110/ps.041239305. [DOI] [PMC free article] [PubMed] [Google Scholar]