Abstract
p53 binds to some members of the S100 family (S100B, S100A4, S100A2, and S100A1). We previously showed that both S100B and S100A4 bind to the p53 tetramerization domain, and consequently control its oligomerization state, but only S100B binds to the C-terminal negative regulatory domain (NRD). Here, we investigate other binding partners for p53 within the S100 family (S100A6 and S100A11), and show that binding to the p53 tetramerization domain seems to be a general feature of the S100 family, while binding to the NRD is a characteristic of a subset of the family.
Keywords: p53, S100 proteins, tetramerization, negative regulatory domain, protein–protein interactions
The p53 tumor suppressor plays a key role in preventing tumorigenic transformation through the induction of cell cycle arrest or apoptosis in response to certain stress signals (Levine 1997; Vousden 2000). p53 works essentially as a homotetrameric transcription factor although some apoptotic activities seem to be transcriptionally independent. Owing to its importance in control of the cell cycle, p53 is the subject of a complex regulation, and is known to interact with many and diverse proteins. These interactions are important for the direct regulation of its activity as a transcription factor but also for the control of protein stability, subcellular localization, and tetramerization.
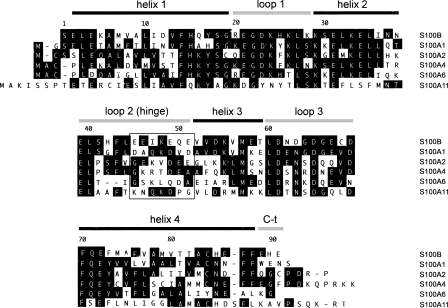
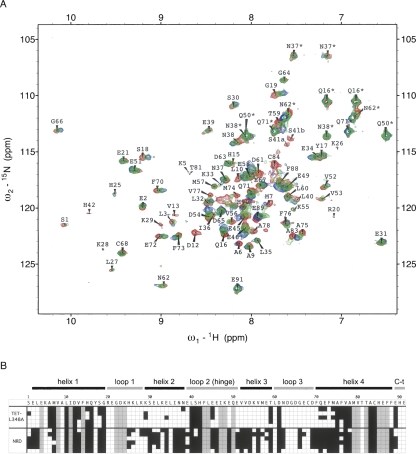
The S100 family is a highly conserved group of proteins that contains EF-hand calcium-binding domains and consists of more than twenty members that can form either homodimeric or heterodimeric complexes with each other (Fig. 1). In most cases, calcium binding induces a conformational change in the S100 dimer that facilitates binding to target proteins. The majority of S100 proteins are encoded in the epidermal differentiation complex (EDC) located on human chromosome 1 (locus 1q21), a region that is frequently rearranged in human cancer (Engelkamp et al. 1993). The exact function of S100 proteins is not fully understood, and sometimes seems contradictory, but they have been implicated in the regulation of a variety of processes, including cell proliferation, differentiation, and intracellular signaling (Schafer and Heizmann 1996; Donato 1999, 2001).
Figure 1.
Sequence comparison of S100 proteins. Alignment of the human S100 sequences studied. Sequences were aligned using Megalin (Laser Gene) software. Residues matching the consensus sequence are shaded. The secondary structure of calcium-bound S100B is indicated above the sequence. The net charge in the area delimited by a square in the loop 2 (hinge) region is negative in the proteins that bind to NRD peptide (S100B, S100A1, and S100A2) but zero or positive for those proteins that do not bind (or bind very weakly) to NRD peptide (S100A4, S100A6, and S100A11). This area contains negatively charged residues in S100B (E46, E49, E51), whose HSQC correlations were perturbed upon binding of NRD but not upon binding of TET-L348A (see Fig. 5B).
Some members of the S100 family are known to interact with p53. The most extensively studied are: S100B (Baudier et al. 1992; Rustandi et al. 1998, 2000; Delphin et al. 1999; Fernandez-Fernandez et al. 2005; Wilder et al. 2006) and S100A4 (Chen et al. 2001; Grigorian et al. 2001; Fernandez-Fernandez et al. 2005). Additionally, S100A1 binds to a peptide derived from the C-terminal negative regulatory domain (NRD) of p53 (Garbuglia et al. 1999). The interaction with S100A2 has been identified, with p53 NRD also being involved (Mueller et al. 2005). The interaction with S100 proteins affects the transcriptional activity of p53, but there is some debate about how transcription is affected (Scotto et al. 1998, 1999; Lin et al. 2001; Mueller et al. 2005).
Comparing the interactions of S100B or S100A4 with p53 (Fernandez-Fernandez et al. 2005) revealed that both bind to the p53 tetramerization domain (TET) and have a common role in regulating p53 tetramerization, but S100B also binds to the NRD. Here, we investigate whether binding to p53 is a general feature of the S100 family by characterizing the interactions with p53 TET and NRD derived peptides. In all cases, S100 proteins bind to p53 TET and consequently regulate its oligomerization state. But p53 NRD binds to only a subset of the S100 proteins. NMR results obtained with 15N-labeled S100B bound to NRD or a TET peptide suggest that the regions of S100B involved in the interaction with these peptides could be different. Nevertheless, a certain degree of competition was detected between the binding of NRD and TET peptides to S100B. Our results identify two categories of S100 proteins: those exemplified by S100A4 that exclusively bind to the p53 TET domain; and those exemplified by S100B that additionally bind to the NRD.
Results
The binding to p53 TET domain seems to be a general property of S100 proteins and is affected by p53 oligomerization
Human S100A1, S100A2, S100A6, and S100A11 were overexpressed and purified. The binding of S100 proteins to the p53 TET domain was monitored using fluorescein-labeled (fluo-) TET peptide (residues 325–355 of human p53) and the monomeric mutant TET-L344P in fluorescence anisotropy experiments at physiological ionic strength (Fernandez-Fernandez et al. 2005). S100 proteins were titrated into solutions of either 25 nM (fluo-) or 250 nM (25 nM fluo- + 225 nM nonlabeled) TET peptides to investigate whether tetramerization influences the binding affinity in a similar manner to its effect on S100B and S100A4 (Fernandez-Fernandez et al. 2005). We have quantified the Kd for the dissociation of TET peptide tetramers in the conditions used in these experiments (M.R. Fernandez-Fernandez and A.R. Fersht, unpublished results). Although we were unable to discern whether the complex dissociates into dimers or directly into monomers, quantitatively the value for the dissociation is ∼100 nM (Kd = [D]2/[T] or 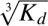 for the Kd = [M]4/[T]). Consequently, at 25 nM and 250 nM TET peptide, the proportion of tetramer present in solution is small and high, respectively.
for the Kd = [M]4/[T]). Consequently, at 25 nM and 250 nM TET peptide, the proportion of tetramer present in solution is small and high, respectively.
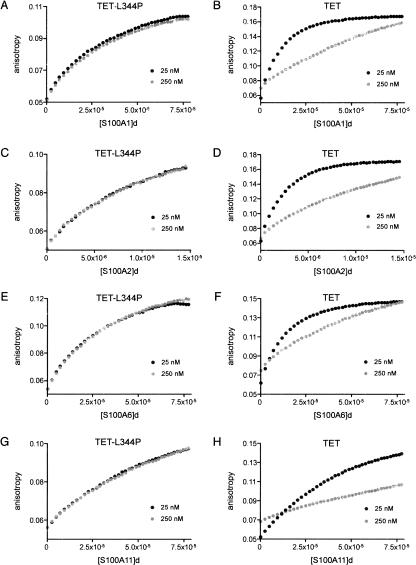
S100A1, S100A2, S100A6, and S100A11 bind to p53 TET peptide (Fig. 2B,D,F,H) and to TET-L344P (Fig. 2A,C,E,G). As expected, the binding of S100 proteins to the TET-L344P mutant is not affected by peptide concentration, as the peptide is monomeric. But for TET, the higher the concentration of peptide, the weaker the binding to S100 protein. Thus, as found for S100B and S100A4 (Fernandez-Fernandez et al. 2005), the proteins bind to the TET domain only when it is in its lower oligomerization states (i.e., monomers and dimers).
Figure 2.
Binding of S100 proteins to p53 TET and TET-L344P peptides. Binding of S100A1 (A,B), S100A2 (C,D), S100A6 (E,F), and S100A11 (G,H) to fluorescein-labeled TET-L344P (A,C,E,G) and TET (B,D,F,H) peptides by using fluorescence anisotropy is shown. The data shown are the mean of three independent measurements. Peptides were studied at two different concentrations, 25 nM and 250 nM. The 250 nM sample was prepared as a 9:1 mixture of nonlabeled:labeled peptide to normalize the fluorescence intensity to the 25 nM sample and to avoid energy transfer effects. The S100 protein dimer concentration is expressed in molar units (M). The S100 stock was 400 μM dimer, except in experiments with S100A2 (75 μM), which shows tight affinity for TET peptide.
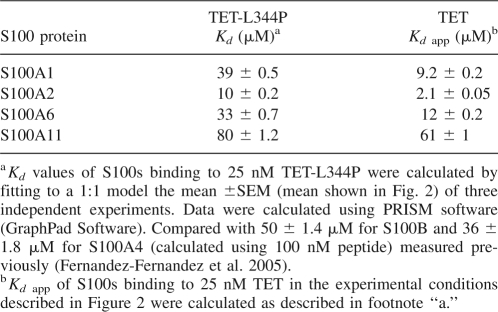
For a quantitative comparison, we measured the binding affinities of S100 proteins with TET using the TET-L344P mutant peptide to avoid the complications of different oligomerization states. Data from S100s binding to 25 nM TET-L344P (Fig. 2) were fitted to a 1:1 binding model (one molecule of peptide binding to a S100 dimer) to derive the Kd values shown in Table 1. S100A2 binds the tightest of all the S100 proteins analyzed.
Table 1.
S100 proteins binding to p53 TET and TET-L344P peptides by fluorescence anisotropy
Apparent Kd values were also obtained for S100s binding to 25 nM TET, in the experimental conditions shown in Figure 2 (Table 1). For each individual S100 protein, the binding to TET is considerably tighter than to TET-L344P despite a proportion of TET peptide being tetrameric at that concentration.
The data revealed new binding partners for p53 within the S100 family (S100A6 and S100A11). Moreover, the binding to the p53 TET domain seems to be a general property of the S100 protein family, including S100A1 and S100A2 for which the binding had been previously described to occur through the p53 NRD (Garbuglia et al. 1999; Mueller et al. 2005).
Binding of S100 proteins to the NRD of p53
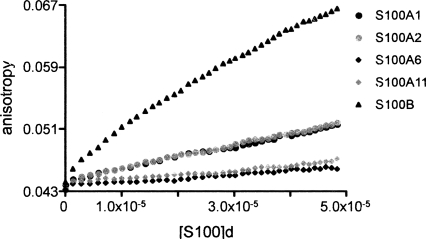
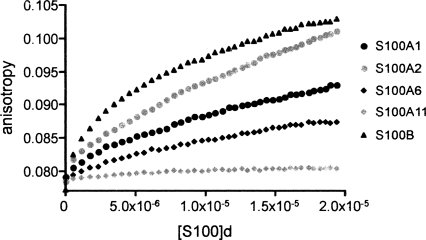
The binding of S100 proteins to p53 NRD (p53 residues 367–393) was also studied by fluorescence anisotropy, titrating S100 proteins into fluo-NRD peptide, using S100B as a control. Figure 3 shows the mean of five independent titrations for each S100 protein. S100A1 and S100A2 clearly show binding to the NRD peptide although the affinity is weaker than that of S100B. However, S100A6 and S100A11 show very weak or no binding to the peptide.
Figure 3.
Binding of S100 proteins to p53 NRD peptide (residues 367–393). Comparison of the binding of S100A1, S100A2, S100A6, S100A11, and S100B to 0.5 μM fluorescein-labeled NRD peptide using fluorescence anisotropy. Concentration of dimeric protein is in molar units (M).
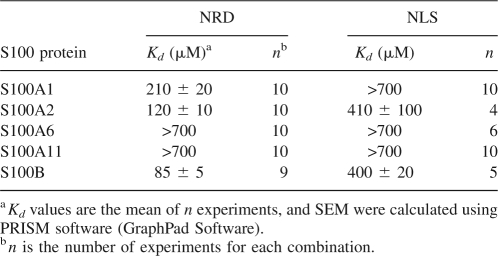
Analytical ultracentrifugation (AUC) was used to quantify and compare the Kd values of the weak complexes between the S100 proteins and fluo-NRD peptide. We also investigated the binding of S100 proteins to a peptide representing the nuclear localization signal of p53 (NLS) (residues 305–322) that binds weakly to S100B and does not bind to S100A4 (Fernandez-Fernandez et al. 2005). The AUC results (Table 2) confirm that S100B has the highest binding affinity for the NRD peptide, with S100A2 and S100A1 showing only slightly lower affinity. Both S100A6 and S100A11 do not bind (or bind very weakly) to NRD. The only protein binding to NLS with a comparable affinity to that of S100B was S100A2. For all other S100s there is no binding (or very weak binding) to NLS.
Table 2.
S100 proteins binding to p53 derived peptides NRD and NLS studied by AUC
Binding of S100 proteins to the C-terminal region of p53
There are different binding sites for S100B within the p53 C-terminal region (p53CT, residues 293–393), resulting in S100B binding to this region with a very high affinity compared to S100A4 (Fernandez-Fernandez et al. 2005). The other S100 proteins were titrated into fluo- p53CT, to compare their binding affinities with that of S100B. The results in Figure 4 are the mean of five independent titrations and show that the synergistic effect of the multiple binding sites applies to these proteins as well. Consequently, S100A2 binds p53CT almost as tightly as S100B, and S100A11 has no detectable binding under these experimental conditions.
Figure 4.
Binding of S100 proteins to p53CT (residues 293–393 of p53). Comparison of the binding of S100 proteins to 0.1 μM fluorescein-labeled p53CT using fluorescence anisotropy. Concentration of dimeric protein is in molar units (M).
Analysis of TET and NRD peptides binding to S100B by NMR spectroscopy
Despite the high sequence conservation among S100 proteins, our results show that there are differences in the way they bind to p53. The binding of S100B to a peptide similar to the NRD has already been characterized by NMR spectroscopy (Rustandi et al. 1998, 2000) and residues involved in the interaction have been mapped (Rustandi et al. 2000).
The two-dimensional {1H,15N}HSQC spectrum for 15N-labeled S100B in a physiological ionic strength buffer has a similar overall pattern to S100B in alternative solvent conditions (Fig. 5A). However, the differences were sufficient to preclude an unambiguous assignment of all correlations according to the published resonance assignments (Rustandi et al. 1998). As it was relevant to examine the binding in the buffer that is equivalent to the binding studies shown above, we confirmed the assignments using a 13C- and 15N-double-labeled sample. A subset of signals in the HSQC at physiological ionic strength (depicted with gray squares in Fig. 5B) was broadened, and because of the low intensity and poor resolution of these signals their connectivity and assignment could not be assigned with confidence.
Figure 5.
Perturbation of 15N-S100B chemical shifts upon binding to NRD and TET-L348A. (A) 500 MHz 2D fast-HSQC of 80 μM 15N-S100B alone (red), or in the presence of 100 μM TET-L348A (green) or 100 μM unlabeled NRD (blue). Assignments with an asterisk denote peaks for side-chain amides. (B) The amino acid sequence of S100B is indicated with a one-letter code, with elements of secondary structure shown above. 15N-S100B (80 μM) was mixed with either 80–100 μM TET-L348A or 100–300 μM NRD. Shading in the squares indicates residues for which the N-H correlation for S100B is perturbed by >0.04 ppm (black), unperturbed (white), or unavailable assignments (gray). Each line is an independent experiment with each peptide (five for TET-L348A and six for NRD).
The HSQC spectrum of 15N-S100B was unaffected by the presence of the TET peptide (data not shown). This is consistent with the earlier observation that S100 proteins can bind to the TET domain only when it adopts small oligomeric forms (monomers/dimers). The peptide is predominantly tetrameric at the concentrations of TET used in these experiments (35–100 μM). Therefore, the binding of p53 fragments was studied using the TET-L348A peptide, a dimeric mutant that associates into tetramers at high concentration (Mateu and Fersht 1998; Fernandez-Fernandez et al. 2005), and the NRD peptide (Fig. 5).
Figure 5B depicts the HSQC correlations of S100B that are significantly perturbed on binding (combined 1H and 15N weighted chemical shift perturbation > 0.04 ppm); either peptide induces a general perturbation of chemical shifts throughout the protein (Fig. 6), as reported for the binding of a shorter version of the NRD peptide (Rustandi et al. 1998, 2000). Binding to NRD induces more extensive changes than binding to TET-L348A (Fig. 6), both in terms of the number of resonances that are significantly affected and the magnitude of the perturbation. Qualitatively, the largest differences between the NRD and TET-L348A binding perturbations occur in a stretch of residues in loop 2 and helix 3. In particular, HSQC correlations for E46, E49, V52, V53, D54, and V56 are perturbed by at least 0.08 ppm on NRD binding but not on TET-L348A binding (perturbations below the threshold < 0.04 ppm).
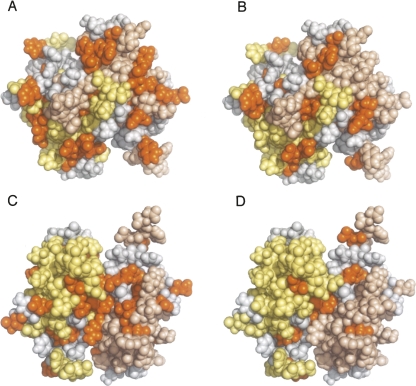
Figure 6.
Mapping of chemical shift changes on calcium-bound human S100B upon binding to TET-L348A and NRD. Monomers within S100B dimer are shown in pale yellow and wheat color. A and B are equivalent views showing the dimer interface, and C and D are a 180° rotation around the X-axis of A and B, respectively. Residues for which perturbations of chemical shifts are >0.04 ppm in all the experiments shown in Figure 5B upon binding of NRD (A,C) or TET-L348A (B,D) are shown in orange. Residues that we were not able to unambiguously reassign are shown in gray.
Loop 2 and helix 3 of S100B are involved in the interaction with the shorter version of NRD and, in particular, V52, V56, and 19 other residues are present in the NRD binding site (Rustandi et al. 2000). We could not identify the signal of any residue that changes upon binding of TET-L348A but not NRD. However, some residues (e.g., those from E89) change upon binding in all TET-L348A experiments but not in all of the NRD experiments (Fig. 5B). There could be more signals that change exclusively upon binding of NRD or TET-L348A among those that are unassigned. These results suggest that the binding sites for NRD and TET peptides could be different, but as there are widespread general perturbations (Fig. 6) resulting from reorganization of the dimer interface (Rustandi et al. 2000), chemical shift mapping alone did not identify the binding site for TET peptides.
Competition experiments of TET and NRD peptides binding to S100B
To investigate whether NRD and TET can bind simultaneously to form a ternary complex with S100B, we measured fluorescence anisotropy for samples containing 2 μM fluo-NRD, 15 μM S100B, and TET-L344P at increasing concentrations (0, 25, 75, and 125 μM) (Fig. 7). Replacing TET with the monomeric mutant TET-L344P eliminates the effect of TET oligomerization on binding to S100B. TET-L344P displaced S100B from binding to fluo-NRD in a concentration-dependent manner, although the displacement is not complete, and no ternary complex was observed. Therefore, NRD and TET-L344P bind competitively to S100B.
Figure 7.
Fluorescence anisotropy measurements of S100B in the presence of fluo-NRD and TET-L344P peptides. Data are the mean of duplicate measurements. Fluo-NRD and S100B concentrations were kept constant at 2 μM and 15 μM, respectively. The bottom value of the Y-axis 0.04845 is the mean anisotropy of two independent samples containing exclusively 2 μM NRD as a reference.
Discussion
Binding of S100 proteins to the TET domain of p53 seems to be a general property of the family; binding was previously shown for S100B and S100A4 (Fernandez-Fernandez et al. 2005) and is shown here for S100A1, S100A2, S100A6, and S100A11. Preliminary data (not shown) indicate that S100A9 also binds to the p53 TET domain but we could not obtain sufficient quantities of pure protein to make a comparison with the other proteins. In each case, binding affinities of the S100 protein are dependent upon the oligomerization state of the TET domain. S100 proteins bind only to monomers or dimers of p53, and thus regulate its oligomerization by displacing the equilibrium from the tetrameric form. This effect could account for previous observations that did not identify TET as a binding site for S100A2 using pulldown experiments (Mueller et al. 2005). These require high concentrations of protein and consequently, constructs containing TET but not NRD were not binding to S100A2 as the TET binding site was occluded in the tetramer. In contrast, binding to the NRD of p53 is not common to all S100 proteins. S100B, S100A1, and S100A2 have a comparable binding affinity for NRD, but S100A4 (Fernandez-Fernandez et al. 2005), S100A6, and S100A11 show no binding (or very weak binding).
S100 proteins can bind to their protein targets using different recognition sites as described for S100A10 (Kassam et al. 1998; Rety et al. 1999; Poon et al. 2004). Weber and co-workers described the NMR structure of S100B in complex with a shorter NRD peptide, and identified that loop 2 (hinge region) and helix 3 (Rustandi et al. 2000) are at the binding interface. We investigated the location of binding sites for TET and NRD on S100B by NMR. Binding to either peptide induces a general perturbation of resonances in S100B, due to minor reorganization of the EF-hand dimer interface (Rustandi et al. 1998, 2000), which hinders the identification of a binding surface by chemical shift mapping alone. However, resonances from the NRD binding site (Rustandi et al. 2000) are not perturbed upon binding of the TET domain. A correlation between the selectivity and structure can be obtained from a sequence comparison for the S100 family in equivalent positions to S100B loop 2 and helix 3 (Fig. 1). The C-terminal half of the loop 2 region (residues 45–51) has a net negative charge in S100 proteins that bind strongly to NRD, but has a neutral or net positive charge in the proteins that do not. Further investigation would be required to confirm the significance of this charge distribution to binding specificity. Our results do not distinguish whether NRD and TET-L344P peptides compete for partly overlapping binding sites or if they bind to distinct sites with a conformational change on S100B that prevents binding of the other peptide.
Proteins from the S100 family can be organized into two categories according to the manner in which they bind to p53. S100A1 and S100A2 resemble S100B, while S100A6 and S100A11 resemble S100A4. It would be of interest to further explore whether the binding characteristics correlate with the different ways in which S100 proteins modulate p53 function.
Materials and Methods
Plasmid construction
The coding sequence for S100A1, S100A2, S100A6, S100A9, and S100A11 was amplified from a human cDNA library (Clontech). Direct and reverse oligos were designed according to GenBank sequences NM_006271, NM_005978, NM_014624, NM_002965, and NM_005620. PCR products were cloned into the pGEMT vector system (Promega) and then expression plasmids were constructed as previously described for S100B (Fernandez-Fernandez et al. 2005).
Protein expression and purification
Escherichia coli C41 (Miroux and Walker 1996) cells were transformed with the expression plasmids, and cultures were grown and processed for protein production. S100 proteins were purified as described for S100B, with the exception of S100A9 for which a source 30Q anion exchange column was used instead of hydrophobic interaction chromatography. p53CT was purified and labeled with fluorescein as previously described (Fernandez-Fernandez et al. 2005).
15N-labeled S100B was prepared in minimal media with 15NH4Cl as the sole nitrogen source and then purified as described for the unlabeled protein. For reassignment purposes, 13C /15N double-labeled S100B was prepared in a similar way but using 13C-glucose as the sole carbon source.
Fluorescence anisotropy
Anisotropy was measured at 25°C as described previously (Fernandez-Fernandez et al. 2005). One milliliter of fluorescein-labeled (fluo-) peptide (NRD, TET, or TET-L344P) or labeled-protein (p53CT) was placed in a cuvette. Two hundred forty microliters of S100 protein (at different concentrations depending on the experiment) was dispensed in additions of 6 μL, titrated at 1 min intervals; the solution was stirred for 1 min, and the anisotropy measured. Competition experiments were performed in a volume of 100 μL with no titration, and sample mixtures were prepared in duplicate.
Analytical ultracentrifugation (AUC)
AUC was performed and analyzed as described previously (Fernandez-Fernandez et al. 2005), at 20°C in a volume of 110 μL. In each run the fluo- peptide concentration was 6 μM and the concentration of S100 protein ranged from 50 to 200 μM.
NMR experiments of S100B binding to NRD and TET peptides
Uniformly 15N-labeled S100B was used in the {1H,15N}HSQC experiments at a concentration of 80 μM in a physiological ionic strength buffer (25 mM Tris pH 7.2, 5 mM CaCl2, 114 mM NaCl, 1 mM DTT, 0.05 mM EDTA, 2.5% D2O). The experiments were recorded on Bruker DRX spectrometers equipped with cryogenic inverse triple resonance probeheads, operating at 500 or 600 MHz 1H frequency. Data were acquired at a sample temperature of 25°C in the presence or absence of fluo-NRD or TET-L348A (nonlabeled) peptides. HSQC time domain data (2048 in t 2, 128 complex points in t 1, 64 transients per row) were processed using NMRPipe software (Delaglio et al. 1995), using Lorentz-to-Gauss (8 Hz line narrowing, 14 Hz Gaussian broadening) and squared cosine bell window functions in t 2 and t 1, respectively, and peak positions were fitted manually using Sparky 3 (T.D. Goddard and D.G. Kneller, University of California, San Francisco). Per residue chemical shift perturbations were calculated as a combined 1H and 15N chemical shift change index, |Δδ1H| + |Δδ15N|/5, of peaks in the HSQC spectra as described by (Hajduk et al. 1997). Reassignment from a 13C and 15N S100B double-labeled sample was performed with standard triple resonance techniques for backbone resonance assignment (HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HN(CA)CO, and HNCO), using unmodified pulse programs from the Bruker pulse program library.
Acknowledgments
M.R.F.F. is a Career Development Fellow of Medical Research Council-UK. We thank Karoly von Gloss for excellent technical assistance and Dr. Fiona Townsley for helpful discussion.
Footnotes
Reprint requests to: Alan R. Fersht, Centre for Protein Engineering, Medical Research Council, Department of Chemistry, Cambridge University, Lensfield Road, Cambridge CB2 1EW, UK; e-mail: arf25@cam.ac.uk; fax: 44-1223-402140.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035527.108.
References
- Baudier, J., Delphin, C., Grunwald, D., Khochbin, S., Lawrence, J.J. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc. Natl. Acad. Sci. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Fernig, D.G., Rudland, P.S., Sparks, A., Wilkinson, M.C., Barraclough, R. Binding to intracellular targets of the metastasis-inducing protein, S100A4 (p9Ka) Biochem. Biophys. Res. Commun. 2001;286:1212–1217. doi: 10.1006/bbrc.2001.5517. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Delphin, C., Ronjat, M., Deloulme, J.C., Garin, G., Debussche, L., Higashimoto, Y., Sakaguchi, K., Baudier, J. Calcium-dependent interaction of S100B with the C-terminal domain of the tumor suppressor p53. J. Biol. Chem. 1999;274:10539–10544. doi: 10.1074/jbc.274.15.10539. [DOI] [PubMed] [Google Scholar]
- Donato, R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim. Biophys. Acta. 1999;1450:191–231. doi: 10.1016/s0167-4889(99)00058-0. [DOI] [PubMed] [Google Scholar]
- Donato, R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Engelkamp, D., Schafer, B.W., Mattei, M.G., Erne, P., Heizmann, C.W. Six S100 genes are clustered on human chromosome 1q21: Identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc. Natl. Acad. Sci. 1993;90:6547–6551. doi: 10.1073/pnas.90.14.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez, M.R., Veprintsev, D.B., Fersht, A.R. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl. Acad. Sci. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuglia, M., Verzini, M., Rustandi, R.R., Osterloh, D., Weber, D.J., Gerke, V., Donato, R. Role of the C-terminal extension in the interaction of S100A1 with GFAP, tubulin, the S100A1- and S100B-inhibitory peptide, TRTK-12, and a peptide derived from p53, and the S100A1 inhibitory effect on GFAP polymerization. Biochem. Biophys. Res. Commun. 1999;254:36–41. doi: 10.1006/bbrc.1998.9881. [DOI] [PubMed] [Google Scholar]
- Grigorian, M., Andresen, S., Tulchinsky, E., Kriajevska, M., Carlberg, C., Kruse, C., Cohn, M., Ambartsumian, N., Christensen, A., Selivanova, G., et al. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: Functional consequences of their interaction. J. Biol. Chem. 2001;276:22699–22708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- Hajduk, P.J., Dinges, J., Miknis, G.F., Merlock, M., Middleton, T., Kempf, D.J., Egan, D.A., Walter, K.A., Robins, T.S., Shuker, S.B., et al. NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J. Med. Chem. 1997;40:3144–3150. doi: 10.1021/jm9703404. [DOI] [PubMed] [Google Scholar]
- Kassam, G., Le, B.H., Choi, K.S., Kang, H.M., Fitzpatrick, S.L., Louie, P., Waisman, D.M. The p11 subunit of the annexin II tetramer plays a key role in the stimulation of t-PA-dependent plasminogen activation. Biochemistry. 1998;37:16958–16966. doi: 10.1021/bi981713l. [DOI] [PubMed] [Google Scholar]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lin, J., Blake, M., Tang, C., Zimmer, D., Rustandi, R.R., Weber, D.J., Carrier, F. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J. Biol. Chem. 2001;276:35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- Mateu, M.G., Fersht, A.R. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J. 1998;17:2748–2758. doi: 10.1093/emboj/17.10.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux, B., Walker, J.E. Overproduction of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Mueller, A., Schafer, B.W., Ferrari, S., Weibel, M., Makek, M., Hochli, M., Heizmann, C.W. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 2005;280:29186–29193. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- Poon, W.Y., Malik-Hall, M., Wood, J.N., Okuse, K. Identification of binding domains in the sodium channel NaV1.8 intracellular N-terminal region and annexin II light chain p11. FEBS Lett. 2004;558:114–118. doi: 10.1016/S0014-5793(03)01512-6. [DOI] [PubMed] [Google Scholar]
- Rety, S., Sopkova, J., Renouard, M., Osterloh, D., Gerke, V., Tabaries, S., Russo-Marie, F., Lewit-Bentley, A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat. Struct. Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- Rustandi, R.R., Drohat, A.C., Baldisseri, D.M., Wilder, P.T., Weber, D.J. The Ca2+-dependent interaction of S100B(ββ) with a peptide derived from p53. Biochemistry. 1998;37:1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- Rustandi, R.R., Baldisseri, D.M., Weber, D.J. Structure of the negative regulatory domain of p53 bound to S100B(ββ) Nat. Struct. Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- Schafer, B.W., Heizmann, C.W. The S100 family of EF-hand calcium-binding proteins: Functions and pathology. Trends Biochem. Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- Scotto, C., Deloulme, J.C., Rousseau, D., Chambaz, E., Baudier, J. Calcium and S100B regulation of p53-dependent cell growth arrest and apoptosis. Mol. Cell. Biol. 1998;18:4272–4281. doi: 10.1128/mcb.18.7.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto, C., Delphin, C., Deloulme, J.C., Baudier, J. Concerted regulation of wild-type p53 nuclear accumulation and activation by S100B and calcium-dependent protein kinase C. Mol. Cell. Biol. 1999;19:7168–7180. doi: 10.1128/mcb.19.10.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden, K.H. p53: Death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Wilder, P.T., Lin, J., Bair, C.L., Charpentier, T.H., Yang, D., Liriano, M., Varney, K.M., Lee, A., Oppenheim, A.B., Adhya, S., et al. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim. Biophys. Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]