Abstract
In eukaryotes, a pre-messenger RNA (pre-mRNA) must undergo several processing reactions before it is exported to the cytoplasm for translation. One of these reactions, endonucleolytic 3′ cleavage at the polyadenylation site, prepares the pre-mRNA for addition of the poly(A) tail and defines the 3′ untranslated region (UTR), which typically contains important gene expression regulatory sequences. While the protein factors responsible for the endonucleolytic cleavage have been largely identified, the means by which their action is limited to the 3′ end of the transcription unit and coordinated with other co-transcriptional events remains unclear. In this review, we summarize and review recent findings revealing that the mammalian 3′ cleavage factors undergo extensive post-translational modification. These modifications include: arginine methylation, lysine sumoylation, lysine acetylation, and the phosphorylation of serine, threonine and tyrosine residues. Every cleavage factor, though not every subunit, is affected. Human Fip1 and the 59 kD subunit of Cleavage Factor I emerge as the most frequently modified core cleavage factor subunits. We outline and compare the various proteomic methods that have uncovered these modifications, and review emerging hypotheses concerning their function. The roles of these covalent but reversible modifications in other systems suggest that 3′ end formation in mammals relies upon post-translational modification for proper function and regulation.
1. INTRODUCTION
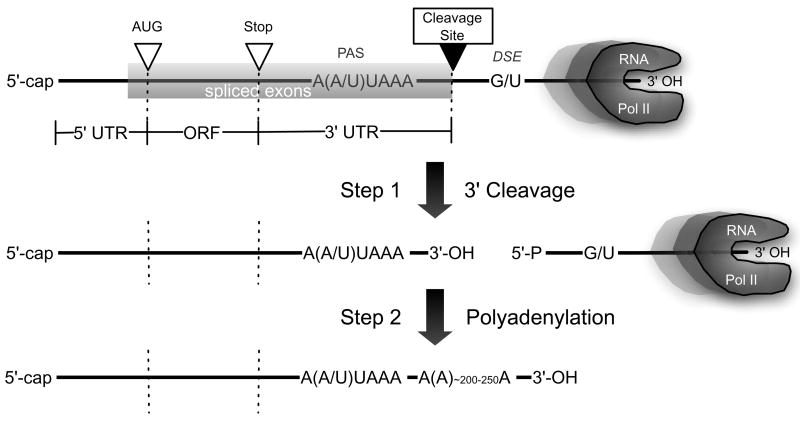
In eukaryotes, nearly all protein-encoding pre-messenger RNAs (pre-mRNAs) undergo a series of processing reactions before they are exported to the cytoplasm for translation. These reactions, now understood to occur for the most part co-transcriptionally, include the addition of the 5′-cap structure, removal of non-coding introns by the splicesome, 3′ cleavage and polyadenylation, and mRNA editing. The 3′ cleavage at the polyadenylation site (pA site) cuts the nascent mRNA into two fragments, producing the 3′ end of the templated portion of the transcript in preparation for the addition by poly(A) polymerase (PAP) of the untemplated poly(A) tail (Figure 1). The cleavage reaction therefore defines the 3′ untranslated region (3′ UTR), the regulatory element-rich segment of the transcript that lies between the translation stop codon and the poly(A) tail. Many human genes are subject to cleavage and polyadenylation at multiple pA sites (Edwalds-Gilbert, Veraldi, & Milcarek, 1997; Tian, Hu, Zhang, & Lutz, 2005), and so can possess variant 3′-UTRs and, in some cases, altered C-terminal protein sequences, though little is known about how regulatory choices are made among the different cleavage sites.
Figure 1.
Mammalian pre-mRNA 3′ end formation. Following transcription of the polyadenylation region, the capped and spliced pre-mRNA is cleaved at the pA site, between the PAS hexamer or variant, and the U-rich (or G/U-rich) DSE. The upstream fragment containing the coding region is then rapidly polyadenylated. (Not drawn according to scale.)
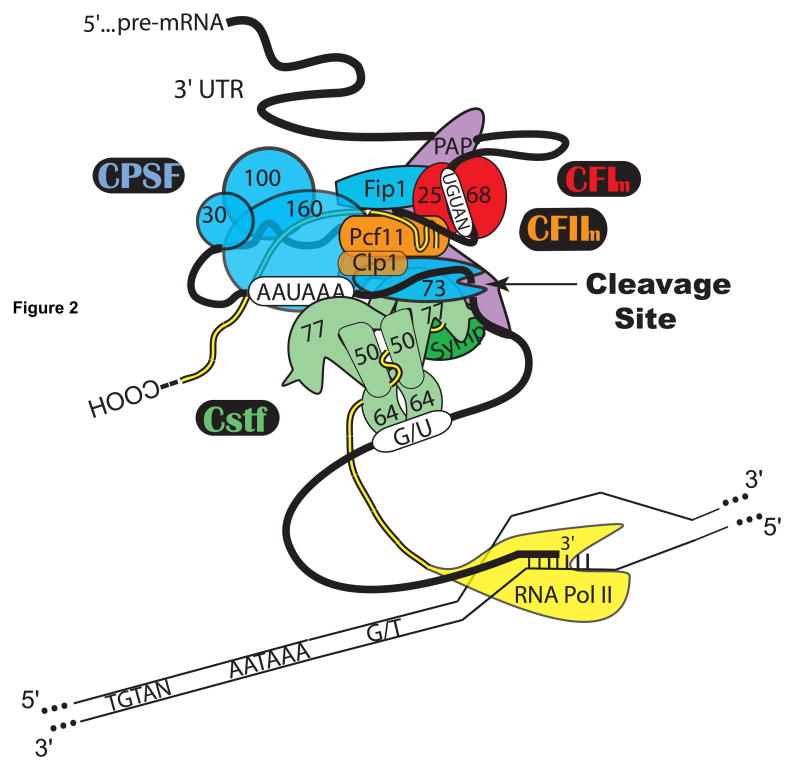
Many protein cleavage factors contribute to the recognition and endonucleolytic cleavage of the pA site in mammalian cells. For a recent review see (Mandel, Bai, & Tong, 2007). These factors include the Cleavage and Polyadenylation Specificity Factor (CPSF), which recognizes the highly conserved poly(A) signal (PAS) hexamer, A(A/U)UAAA, located 10–30 nucleotides (nt) upstream of the cleavage site, the Cleavage Stimulation Factor (CstF), which recognizes the less precisely conserved U-rich or G/U-rich downstream element (DSE) generally found within 30 nt downstream of the cleavage site, Cleavage Factors I (CFIm) and II (CFIIm), symplekin, PAP, and the RNA Polymerase II C-terminal Domain (CTD). These factors are shown as a hypothetical pre-cleavage complex in Figure 2.
Figure 2.
Protein factors involved in mammalian 3′ end cleavage. Depiction of the hypothetical pre-cleavage complex assembling co-transcriptionally on the cis-acting elements in a pre-mRNA 3′ UTR. Cleavage factors: CPSF (cleavage and polyadenylation specificity factor), CstF (cleavage stimulation factor), CFIm (mammalian cleavage factor I), CFIIm (mammalian cleavage factor II), PAP (poly(A) polymerase), Symp (symplekin), RNA Pol II (RNA Polymerase II, with CTD shown as trailing strand.)
Though most of the protein factors responsible for the endonucleolytic cleavage reaction have been identified, exactly how their action is limited to the 3′ end of the transcription unit and coordinated with other co-transcriptional events, including transcription termination, is poorly understood. Recent in vitro evidence indicates that selective post-translational modification of the 3′ cleavage factors may influence the control and coordination of 3′ cleavage during each transcription cycle. Post-translational modifications (PTMs) may be looked upon as the finishing touches put on certain proteins to aid in their function or regulation, while 3′ cleavage and polyadenylation constitutes another sort of finishing touch on the newly minted pre-mRNA. In this review, we focus on recent findings demonstrating that the mammalian 3′ cleavage factors are modified by a variety of covalent PTMs, many of which have been brought to light by large unbiased proteomic mass spectrometry (MS) screens. Alcohol residue phosphorylation, lysine acetylation, arginine methylation, and lysine sumoylation have all been found among the various 3′ cleavage factors. Every cleavage factor, though not every subunit, is affected. We also compile, summarize and discuss these modifications and the different contexts in which they have been identified. We begin with phosphorylation, the most widespread modification. We review the various phosphoproteomic approaches that have uncovered phosphate-modified cleavage factor residues, and then compile phosphorylation sites by cleavage factor. Also described are recent discoveries of cleavage factor acetylation, methylation and sumoylation. Where possible, we consider emerging hypotheses regarding the function of these modifications in mammalian pre-mRNA 3′ end formation.
2. PHOSPHORYLATION
Reversible phosphorylation is the most common post-translational protein modification in eukaryotes. It is thought to control some aspect of almost all cellular processes (Hubbard & Cohen, 1993), a scope consistent with the large number of mammalian genes that encode kinases and phosphatases (Cohen, 2002). Phosphorylation can be used by a cell to control a protein in many ways, from switching on or off a particular enzymatic activity, to modulating protein-protein interactions, intracellular localization or protein conformation. Phosphorylation cascades also amplify signal transduction effects. A phosphorylated amino acid distinguishes itself from the unmodified residue by being a large hydrophilic group with increased hydrogen-bonding, hydration and salt-bridge formation capability. At pH above ~7.5 approximately 90% of protein phosphates (pKa ~6.7 (Johnson & O’Reilly, 1996)) have a −2 charge, which cannot otherwise be obtained by any of the twenty proteinogenic amino acids. Perhaps because it is the one that has been most often looked for, phosphorylation is also the most common post-translational modification found among the 3′ cleavage factors to date. Phosphorylated serine, threonine and, to a lesser extent tyrosine, have been detected within the subunits of the 3′ cleavage factors.
2.1 Phosphoproteomic methods
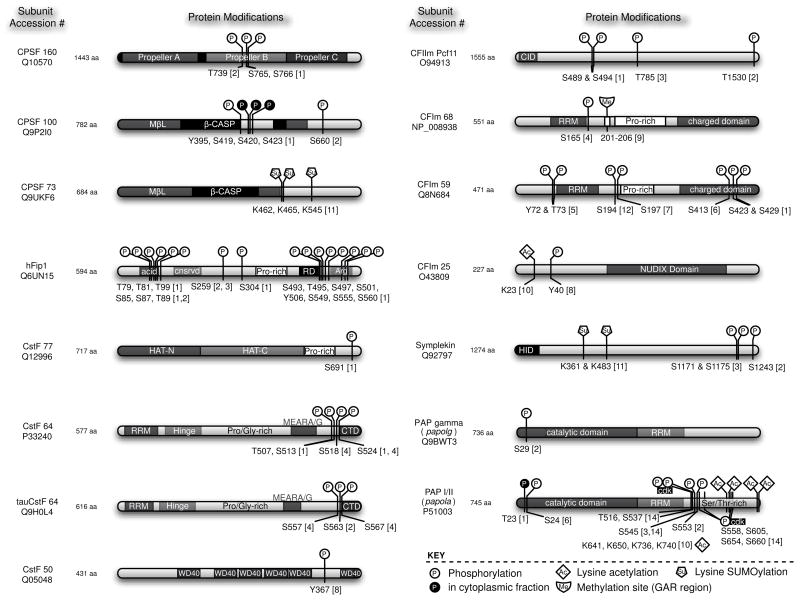
Most of the mammalian cleavage factor phosphorylation sites have been identified by large phosphoproteomic mass spectrometry (MS) screens. Such screens are to varying degrees unbiased, and therefore generally do not set out to identify modifications related to any particular biochemical pathway or process, let alone 3′ pre-mRNA processing. The interpretation of unbiased screen results can be frustrating because the modification sites are usually identified outside of a hypothetical context. Indeed, these methods are sometimes described as “hypothesis-free” (Olsen et al., 2006). In one sense, the results of hypothesis-free screens leap forward beyond current understanding of a given process. Nevertheless, these screens provide a database for future work to draw upon in the event that an identified phosphorylated residue is discovered to have particular importance, and they therefore help to push current understanding to catch up with their voluminous data. Before focusing on the details specific to each 3′ cleavage factor, we first describe and compare the different phosphoproteomic methods that have identified phosphorylation sites within the cleavage factors. The various phosphopeptide enrichment and mass spectrometric techniques are evolving rapidly, but no single method has emerged as optimal in all contexts. Therefore, the specific details of each method are important to consider when weighing their results because the techniques impose limitations on what can be found. In Figure 3, we depict the overall domain structure of each phosphorylated or otherwise post-translationally modified 3′ cleavage factor polypeptide. The subunit domains and the modified residue’s position are also indicated for quick reference, which should prove useful since most of these data are buried in large supplementary tables, in some cases in a form that cannot be searched by text matching.
Figure 3.
Atlas of post-translational modifications among the mammalian pre-mRNA 3′ cleavage factors. Only cleavage factor subunits known to be post-translationally modified are shown. Structural domains and documented modifications are indicated. All modifications were found in human proteins, except for the PAP cdk phosphorylation and acetylation sites. In those cases, the modifications were found in bovine PAP, but for consistency the homologous sites in the human protein are indicated here. Abbreviations: MβL, Metallo-β-lactamase domain; RD, Arg-Asp-rich domain; HAT, Half a TPR; RRM, RNA Recognition Motif; CTD, C-terminal domain CID, CTD-interacting domain; HID, HSF1 interacting domain. References: [1] Olsen et al., 2006; [2] Beausoleil et al., 2004; [3] Beausoleil et al., 2006; [4] Matsuoka et al., 2007; [5] Brill et al., 2004; [6] Yu et al., 2007; [7] Molina et al., 2007; [8] Rush et al., 2005; [9] Boisvert et al., 2003; [10] Shimazu et al., 2007; [11] Vethantham et al., 2007; [12] Cantin et al., 2006; [13] Amanchy et al., 2005; [14] Colgan et al., 1998.
One early phosphoproteomic study aimed to identify all HeLa cell nuclear phosphoproteins (Beausoleil et al., 2004). This approach used a strategy in which HeLa cell nuclear extract was digested with trypsin and then adjusted to pH 2.7, where most unphosphorylated peptides released during digestion will have a net charge of +2 (one positive charge from the C-terminal lysine or arginine, and one positive charge from the N-terminus). Those peptides bearing a single phosphate will have a net charge of +1 because in addition they have one phosphate negative charge, enabling enrichment of monophosphorylated peptides via early elution during strong cation exchange chromatography (SCX). The peptides captured during enrichment were then subjected to HPLC chromatography-tandem MS (LC-MS/MS). The “tandem” refers to the selection of one ion from the first spectrum for further fragmentation and analysis, producing a second spectrum. Some studies perform a third selection/fragmentation, known as LC-MS/MS/MS or LC-MS3. Enrichment of the phosphopeptides released by protease digestion is important because the digested lysate is complex, and the phosphorylated fraction of a given protein is usually small (Aebersold & Goodlett, 2001). Selected peptides showing a neutral loss of phosphate in the first MS scan were further fragmented and analyzed to yield the peptide sequence and site of modification. Using this method, Beausoleil et al. (Beausoleil et al., 2004) identified 2002 phosphorylation sites in 967 HeLa cell nuclear phosphoproteins, including sites in CPSF, CstF, CFIIm, Symplekin, PAP, and the tau variant of CstF-64 (Figure 3). A limitation of this technique to be kept in mind when evaluating its results is that most phosphopeptides with a net charge other than +1 are lost in the SCX step prior to LC-MS/MS. For example, singly phosphorylated peptides containing histidine, and multiply phosphorylated peptides without histidine are not collected for analysis. The latter problem is exemplified by the fact that only two peptides were detected from the highly phosphorylated RNA Polymerase II CTD (Pol II CTD). This early mammalian study, along with previous studies in yeast (S. B. Ficarro et al., 2002; Peng et al., 2003), plants (Nuhse, Stensballe, Jensen, & Peck, 2003) and mammalian reproductive cells (S. Ficarro et al., 2003), established the basic steps of the phosphoproteomic MS technique that have been used and built upon by many subsequent studies: tissue extraction, trypsin (or other protease) digestion, phosphopeptide enrichment and LC-MS/MS.
The phosphopeptide enrichment step is very important, and studies identifying phosphorylated 3′ cleavage factors have used a variety of methods, each having advantages and disadvantages. In place of, or in combination with, SCX (Beausoleil et al., 2004; Beausoleil, Villen, Gerber, Rush, & Gygi, 2006; Olsen et al., 2006), phosphopeptides have been selected and concentrated using immunoprecipitation with phosphoamino acid antibodies (Brill et al., 2004; Matsuoka et al., 2007; Rush et al., 2005), immobilized metal affinity chromatography (IMAC) (Brill et al., 2004; Cantin, Venable, Cociorva, & Yates, 2006), titanium dioxide (TiO2) matrix affinity (Molina, Horn, Tang, Mathivanan, & Pandey, 2007; Olsen et al., 2006; Yu et al., 2007) and isoelectric focusing (ISE) (Cantin et al., 2006). No method is without problems. For example, SCX does not capture all phosphopeptides, as discussed above. IMAC suffers from non-specific binding through peptide carboxylic acid residues, unless these are first chemically modified (Brill et al., 2004; S. B. Ficarro et al., 2002), an extra step that can lead to complicating side reactions (Yu et al., 2007). Like IMAC, TiO2 chromatography is subject to non-specific peptide binding in the complex milieu of cell lysates, though this can be partially alleviated using buffer additives (Larsen, Thingholm, Jensen, Roepstorff, & Jorgensen, 2005; Yu et al., 2007). Phosphoamino acid antibodies are generally influenced by the sequence context of the phoshophopeptides, and suffer from relatively low affinity due to the small antigen size. ISE depends on the pI of the peptide, which varies widely, resulting in many fractions only some of which are significantly enriched in phosphopeptides, and has to be combined with additional steps, such as IMAC (Cantin et al., 2006). Choosing a particular method involves compromise and, although it requires extra steps, the enrichment methods are sometimes combined. Thus, while not biased by hypotheses, phosphoproteomic methods are all subject to technical bias at the peptide selection stage, which should be taken into account when evaluating their results.
Researchers relying on phosphoproteomic data will naturally wonder about potential false negatives among the many serines, threonines and tyrosines that do not show up in the phosphopeptide lists, and from which, because of the technical limitations of each method, nothing should be concluded. Of greater concern is the possibility of false positives. The method described above (Beausoleil et al., 2004) that identified phosphorylation sites in CPSF, CstF, CFIIm, Symplekin, PAP, and the tau variant of CstF-64 was refined by the same research group in a study paying special attention to the minimization of false positives (Beausoleil et al., 2006). Here, phosphorylation sites were only found in CPSF, CFIIm and Symplekin, and in the latter two cases those were at different sites than were found in the first study. The source of discrepancy in the two studies should not be overly worrisome because there were significant experimental differences introduced in the second study, including the use of nocodazole-arrested HeLa cells. However, it is clear that researchers in this area are concerned with the balance between precision and sensitivity, i.e. minimizing false positives and false negatives, and are taking measures to improve confidence levels in their data (Beausoleil et al., 2006; Elias, Haas, Faherty, & Gygi, 2005).
Another frequent problem is uncertainty in the exact location of a phosphate within a tryptic phosphopeptide. Monophosphorylated peptides containing multiple serine, threonine and tyrosine residues are frequently detected, often leaving unclear the exact location of the phosphate. This problem can only be solved by careful analysis of the MS/MS fragmentation patterns. An automated approach has been devised to analyze the patterns by looking for and evaluating site-determining ions (Beausoleil et al., 2006), while other researchers even go so far as to perform MS/MS fragmentation on all possible synthetic phosphopeptides for comparison with the one derived from the biological source (Rush et al., 2005).
Stimulation or special treatment of cells prior to lysate preparation is another important variable that should be taken into consideration when evaluating the phosphorylated cleavage factor results compiled in this review. Pre-treatment agents include pervanadate, a tyrosine phosphatase inhibitor that leads to accumulation of phosphotyrosines (Amanchy, Kalume, Iwahori, Zhong, & Pandey, 2005; Rush et al., 2005), ionizing radiation (Matsuoka et al., 2007), nocodazole (Beausoleil et al., 2006), tumor necrosis factor-alpha (TNF-α) (Cantin et al., 2006) and epidermal growth factor (EGF) (Olsen et al., 2006). In order to compare treated with untreated cells, SILAC, stable isotope labeling with amino acids in cell culture, is frequently used (Ong et al., 2002). Treatment-specific effects on phosphorylation are detected by measurement of the isotope ratio in the mass spectrum of the specific phosphopeptides with and without treatment. Studies using pre-treatment of the cells have extra appeal because they use an experimental variable that can in some cases place the results in the context of a hypothesis. Where applicable, this aspect of the compiled results is described for the individual cleavage factors below.
2.2 In vitro methods
In spite of producing large tables of protein phosphorylation data, phosphoproteomic screens seldom provide functional information about what the phosphates actually do. In studying 3′ pre-mRNA processing, our own laboratory has investigated the role of cleavage factor phosphorylation in a different way. Using a functional in vitro reconstitution assay and dephosphorylating enzymes we found that treatment of HeLa cell nuclear extract with phosphatases strongly inhibited in vitro 3′ cleavage (Ryan, 2007). The susceptibility to phosphatase treatment was traced to a fraction containing CFIm and CFIIm activities (CFm). The same CFm fraction alone restored activity to phosphatase-treated HeLa nuclear extract. The simplest explanation for these results is that one of the CFIm or CFIIm subunits requires phosphorylation to function properly. Conceivably, phosphorylation within CFm could constitute a regulatory mechanism for turning on and off the enzymatic 3′ cleavage activity as needed during the transcription cycle. We are further fractionating CFm to identify the phosphorylated cleavage factor subunit, at which time the interpretation of our functional results may benefit from comparison with those of the phosphoproteomic approaches reviewed here, some of which have revealed phosphorylation sites in CFIm-68, -59, -25 and the CFIIm Pcf11 subunit, as described below. The intersection between phosphoproteomic studies and hypothesis-driven research such as ours highlights the complementary nature of the two experimental approaches.
2.3 Cleavage factor phosphorylation
The specific location of phosphorylated residues in the 3′ pre-mRNA cleavage factors are described here. Along with other PTMs, they are summarized in Figure 3. In cases where phosphorylation sites from potentially different tryptic peptides are listed, it is not known if they arose from the same molecule, that is, if they occur simultaneously in the factor. In contrast, modifications clustering in the same tryptic phosphopeptide occur simultaneously.
2.3.1. Cleavage Polyadenylation Specificity Factor (CPSF)
CPSF contains five subunits, four of which are commonly referred to by their apparent molecular mass: 160, 100, 73, 30, and Fip1 (Table 1). The DEAE-separated CPSF fraction from HeLa cell nuclear extract can be treated with a non-specific phosphatase without loss of the activity it contributes to the vitro 3′ cleavage reaction (Ryan, 2007). This observation implies that phosphorylation within CPSF is not important for its role in cleavage when assayed in vitro, where it is uncoupled from transcription and polyadenylation. The aforementioned phosphoproteomic study using HeLa cells (Beausoleil et al., 2004) detected phosphorylation of CPSF-160 (Thr739), CPSF-100 (Ser660) and Fip1 (Ser259), though only the Fip1 modification was found in a later study by the same group (Beausoleil et al., 2006). A study that used EGF stimulated cells found a different set of phosphorylation sites in the same three subunits: CPSF-160 (Ser765 and Ser766), Fip1 (Ser304), CPSF-100 (Tyr395), and, from the cytoplasmic fraction, CPSF-100 (Ser419, Ser420 and Ser423) (Olsen et al., 2006). Although this study used SILAC to look for EGF-dependent phosphorylation events, it also detected phosphorylation events not dependent on EGF. None of the cleavage factor phosphorylations showed definitive EGF dependence. This paper is one of the few studies that looked specifically at both the nuclear and cytoplasmic fractions. The cleavage factors are quite naturally expected to be found in the nuclear fraction, but the three CPSF-100 sites detected in the cytoplasmic fraction are not entirely unexpected as there is a cytoplasmic form of CPSF containing the 160, 100 and 30 subunits that assists in polyadenylation in that compartment (Mendez, Murthy, Ryan, Manley, & Richter, 2000). No phosphorylation sites have been reported in CPSF-30 or in the nuclease, CPSF-73.
In the EGF stimulation study, Olsen et al. (Olsen et al., 2006) found fourteen additional sites of EGF-independent phosphorylation in human Fip1 (Figure 3), though these were ascribed to an alternatively spliced Fip1 form apparently lacking three exons compared to the full length protein. This isoform is referred to in their supplementary table as hypothetical protein DKFZp586K0717, not Fip1. None of these sites are in peptides close to the missing exons, so it is just as likely that they arose from the full-length protein and are true Fip1 modifications. This would make Fip1 the most highly phosphorylated of the core 3′ cleavage factors (second if PAP is counted) and indeed multiple forms have been observed and suggested to be due to this PTM (Kaufmann, Martin, Friedlein, Langen, & Keller, 2004). There are notably two large clusters of phosphorylated residues, one mainly in the acidic region near the N-terminus, and the other in the extended arginine-rich region near the C-terminus. Interestingly, the yeast homolog also appears to be a phosphoprotein (He & Moore, 2005; Zielinski, Hellman, Kubinski, & Szyszka, 2006).
The first 339 amino acids of Fip1 have been found attached to the platelet-derived growth factor receptor alpha (PDGFRA) as the result of an aberrant gene deletion and fusion event (FIPL1-PDGFRA) whose product is a constitutively active tyrosine kinase (Cools et al., 2003). The resulting dysregulated kinase is responsible for F/P-positive chronic eosinophilic leukemia. One report detected phosphorylation of hFip1 at Ser85, Ser87 and Ser89 but ascribed these modifications to the FIPL1-PDGFRA fusion protein (Beausoleil et al., 2004). Since this study used HeLa cells they more likely arose from hFip1 itself. These three sites are identical to three found in the N-terminal acidic region by the aforementioned HeLa cell study that attributed them to hypothetical protein DKFZp586K0717 (Olsen et al., 2006).
2.3.2. Cleavage Stimulation Factor (CstF)
Unlike CPSF, CstF is not required for the polyadenylation step that immediately follows 3′ cleavage. In the pre-cleavage complex, CstF binds the G/U-rich DSE and, along with CPSF, to which it also binds, defines the approximately 50 nucleotides centering on the cleavage site. Similar to CPSF, in vitro evidence suggests that phosphorylation of this factor is not important to the uncoupled in vitro cleavage reaction (Ryan, 2007). Structural and biochemical studies indicate that CstF-77, the largest subunit, functions as an important protein-protein interaction scaffold within the pre-cleavage complex (Bai et al., 2007; Murthy & Manley, 1995). This subunit was found in one proteomic study to be phosphorylated at Ser691 (Olsen et al., 2006), near the C-terminus of the subunit.
The CstF-64 subunit contains the RNA binding domain that recognizes the DSE (Takagaki & Manley, 1997). This subunit has long been suspected of being phosphorylated (Takagaki, MacDonald, Shenk, & Manley, 1992; Wallace et al., 1999) and differently migrating forms have been detected by 2-D electrophoresis (Edwalds-Gilbert & Milcarek, 1995), but no definitive phosphorylation study has been done. Proteomic analyses have uncovered a cluster of four non-tyrosine phosphorylation sites in between the helical CstF-64 MEARA/G repeat domain (Richardson, McMahon, MacDonald, & Makhatadze, 1999) and the highly conserved C-terminal three-helix bundle domain (Figure 3) (Qu et al., 2007). Olsen et al. (Olsen et al., 2006) found phosphates on Thr507, Ser513 and Ser524, while Matsuoka et al., studying phosphorylation as a function of exposure to ionizing radiation (Matsuoka et al., 2007), found radiation-dependent phosphates on Ser518 and Ser524. These four modified sites are spaced with noticeable regularity: 507-pTGMQGApSIQGGpSQPGGFpS-524, such that two pairs of phosphates would be displayed for recognition by another protein on the same face of an alpha helix – Thr507 paired with Ser518, or Ser513 paired with Ser524 - were they to be simultaneously phosphorylated and to adopt alpha helices like the sequences that flank them.
A CstF-64 gene paralog called τCstf-64 was recently discovered (Dass et al., 2001; Wallace et al., 1999). Although its mRNA is present in most tissues (Huber, Monarez, Dass, & MacDonald, 2005), in mouse the protein is found almost exclusively in testis (Wallace et al., 1999). The τCstf-64 protein has a partially repetitive Gly/Gln-rich insertion of 30 amino acids in between the second and third CstF-64 phosphorylation sites described above, between Gly517 and Ser518. Proteomic analyses in HeLa and 293T cells have shown that τCstf-64 can be phosphorylated at the two serines corresponding to the third and fourth of the CstF-64 sites, namely, τCstf-64 Ser557 (Matsuoka et al., 2007), corresponding to CstF-64 Ser518, and τCstf-64 Ser563 (Beausoleil et al., 2004), corresponding to CstF-64 Ser524. τCstf-64 Ser567 has also been detected in phosphorylated form (Matsuoka et al., 2007). The modifications at Serines 557 and 567 are enriched by ionizing radiation treatment, possibly signaling a role in DNA repair (Matsuoka et al., 2007).
Phosphorylation of the smallest CstF subunit, CstF-50, at Tyr367 was detected in two anaplastic large cell lymphoma cell lines but not in Jurkat cells, which come from a leukemic T-cell line (Rush et al., 2005). The failure to find a phosphate on the same residue in Jurkat cells, or in other studies, may indicate that this tyrosine is abnormally phosphorylated in the two lymphoma cell lines, perhaps due to the oncogenic fusion tyrosine kinase that they express (Rush et al., 2005).
2.3.3. Mammalian Cleavage Factor I (CFIm)
CFIm is involved early in the assembly of the pre-cleavage complex, according to in vitro evidence (Ruegsegger, Blank, & Keller, 1998). HeLa CFIm activity can be replaced in vitro by a heterodimer composed of recombinant CFIm-68 expressed in P. pastoris and CFIm-25 expressed in E. coli (Ruegsegger et al., 1998). Two other heterodimers made from the 25 kD subunit paired with either a 59 kD subunit, expressed from a paralogous gene, or a 72 kD subunit, likely a splicing isoform, are believed to exist in human cells. All four subunits co-purified during extensive fractionation of CFIm activity from HeLa cells (Ruegsegger, Beyer, & Keller, 1996; Ruegsegger et al., 1998). The CFIm larger subunits have a domain organization similar to that found in the splicing SR proteins, including an N-terminal RNA recognition motif (RRM) and, near the C-terminus, several RS and alternating charge dipeptides which in SR proteins become phosphorylated (Ngo et al., 2005). CFIm has been suggested to be capable of a general regulatory function in 3′ processing (Brown & Gilmartin, 2003). As noted above, our laboratory has found that a fraction containing both CFIm and CFIIm (the DEAE CFm fraction) loses its ability to function in 3′ cleavage when treated with a phosphatase (Ryan, 2007). This finding points to the CFIm larger subunits as candidates to be the phosphoprotein(s) whose contribution to 3′ cleavage is lost upon phosphatase treatment. However, the 68 kD subunit has been found with only one phosphate, on Ser165, five residues beyond the end of the RNA binding RRM sequence (Matsuoka et al., 2007). The phosphopeptide implicating this site could not have come from the 59 kD paralog, but could have come from the less abundant 72 kD isoform.
Although the corresponding location in the 59 kD paralog has not been found to be phosphorylated, phosphorylation at seven other positions unique to CFIm-59 has been detected, making it the third most highly phosphorylated cleavage factor subunit after PAP and Fip1. Six different proteomic studies – more than for any other cleavage factor subunit – have uncovered phosphorylation sites in CFIm-59, though no single site was identified in more than one study. Using IMAC and anti-phosphotyrosine immunoprecipitation, Brill et al. found phosphorylations on Tyr72 and Thr73, just N-terminal to the RRM (Brill et al., 2004). Proceeding in the N to C direction, no phosphorylation sites have been reported in the RRM, but 33 amino acids beyond this region phosphorylation sites have been detected on Ser194 (Cantin et al., 2006) and Ser197 (Molina et al., 2007). No sites have thus far been reported within the proline-rich region, a likely protein-protein interaction site (Ingham et al., 2005; Sudol & Hunter, 2000), but one serine, Ser413, was found phosphorylated between this region and the charged domain, which includes the RS dipeptides (Yu et al., 2007). Within the C-terminal charged domain itself, two RS dipeptides serines, Ser423 and Ser429, have been found in phosphorylated form (Olsen et al., 2006). Using anti-phosphotyrosine antibody enrichment before protease treatment, another study identified CFIm-59 as a phosphoprotein but did not identify the modified residues (Amanchy et al., 2005). No functional roles have yet been ascribed to any of the CFIm-59 phosphorylation sites, but the sensitivity of CFm 3′ cleavage activity to dephosphorylation (Ryan, 2007), and the establishment of a CFIm-59-mediated link between splicing and 3′ cleavage (Millevoi et al., 2006) make this factor one of possible interest for future phospho-regulation studies.
CFIm-25 is the smallest 3′ cleavage factor subunit. It contains a Nudix hydrolase domain, which is predicted to bind a nucleotide diphosphate linked to some other functional group, X. Substrate examples for other Nudix proteins include ATP and NADH (Bessman, Frick, & O’Handley, 1996). In addition to binding the larger CFIm subunit, CFIm-25 binds PAP (H. Kim & Lee, 2001) and nuclear Poly(A) binding protein (PABPN1) (Dettwiler, Aringhieri, Cardinale, Keller, & Barabino, 2004), which itself has been found to be phosphorylated in three studies (Amanchy et al., 2005; Olsen et al., 2006; Rush et al., 2005). Tyr40 was found to be phosphorylated in two cancer cell lines expressing a mutant tyrosine kinase (Rush et al., 2005). This study is one of the few to use proteases other than trypsin. The lower frequency of cutting by alternative proteases produces fewer peptides in the optimal size range for LC-MS/MS analysis (Rush et al., 2005), which is why trypsin is normally used. Phosphopeptides containing Tyr40 were produced by digestion with both chymotrypsin and endoproteinase GluC. Using HeLa cells, Amanchy et al. found evidence for tyrosine phosphorylation on CFIm-25, but no modified residues were identified (Amanchy et al., 2005).
2.3.4. Mammalian Cleavage Factor II (CFIIm)
Mammalian cleavage factor II is the least understood of the 3′ cleavage factors. Unlike CFIm, its activity has not been reconstituted from recombinant proteins. Extensive fractionation of HeLa nuclear extract resulted in a group of fifteen identifiable proteins co-eluting with CFIIm activity (de Vries et al., 2000). Splicing, DNA repair and transcription factor proteins, among others, were found within this fraction, designated as CFIIAm. Inexplicably, all four CFIm subunits were found within CFIIAm, even though it required supplementation by a separately isolated CFIm fraction to reconstitute in vitro 3′ cleavage activity. No explanation has been proposed for the inactivity of the CFIm subunits in the CFIIAm fraction, though post-translational modification, or lack thereof, of the CFIm subunits is one possibility that remains to be explored. Another fraction, designated CFIIBm, was found to stimulate the in vitro 3′ cleavage activity, but not to be required for it. CFIIBm did not contain any of the known cleavage factors. Two of the CFIIAm fraction proteins, hClp1 and Pcf11, were identified as yeast 3′ processing orthologs. One of these, hClp1, was verified to interact with CFIm and CPSF, and to contribute to the 3′ cleavage reaction in vitro (de Vries et al., 2000). The drosophila ortholog of the other one, Pcf11, is involved in transcription termination (Zhang & Gilmour, 2006). Although no phosphorylation sites have been detected for hClp1, it was recently found to be a kinase itself, being able to 5′ phosphorylate oligoribonucleotides (Weitzer & Martinez, 2007). Phosphorylated residues have however been detected in Pcf11. In two separate studies, Beausoleil et al. found two different Pcf11 threonines, Thr785 (Beausoleil et al., 2006) and Thr1530 (Beausoleil et al., 2004), in phosphorylated form, while Olsen et al. found phosphorylation of Ser489 and Ser494 (Olsen et al., 2006). So little work has been done on the human Pcf11 that nothing can be said regarding any functional role for these phosphates.
2.3.5. Poly(A) Polymerase (PAP)
In addition to its adenylate-adding activity, poly(A) polymerase (PAP) is required for in vitro 3′ cleavage at most viral and pre-mRNA pA sites. The SV40L cleavage site is one notable and commonly used exception, demonstrating that the basal 3′ cleavage activity does not absolutely require PAP. PAP’s role in the cleavage reaction, in those cases where it is required, is not understood, though bacterially expressed PAP, presumably unphosphorylated, can replace HeLa PAP in this regard. The polymerase activity of the nuclear PAPI and PAPII forms, alternative splicing products from the PAPOLA gene, is inhibited during M-phase by complete phosphorylation of seven cyclin-dependent kinase (cdk) sites in the C-terminal Ser/Thr-rich domain (Colgan, Murthy, Prives, & Manley, 1996; Colgan, Murthy, Zhao, Prives, & Manley, 1998). While these reports showed that hyperphosphorylation can lead to the inhibition of PAP activity, an earlier paper, using an in vitro assay, found the opposite effect, that is, that CPSF-dependent PAP activity in HeLa nuclear extract was inhibited by phosphatase treatment (Chrislip, Hengst-Zhang, & Jacob, 1991). In proteomic screens, only two residues in PAP’s Ser/Thr-rich region have been found to be phosphorylated: Ser553 (Beausoleil et al., 2004), a cdk non-consensus site, and Ser545 (Beausoleil et al., 2006), which is in between two cdk sites (Colgan et al., 1998). Presumably, the cdk sites have not been found in these or other proteomic studies because very few cells are at any given time in M-phase. In the EGF stimulation study described above, Thr23 was found to be phosphorylated in an EGF-independent manner, but this modification was only detected in the cytoplasmic PAP (Olsen et al., 2006). Its neighbor, Ser24, was found phosphorylated in HeLa whole cell lysate (Yu et al., 2007).
A second nuclear poly(A) polymerase has been discovered (Perumal, Sinha, Henning, & Reddy, 2001; Topalian et al., 2001). Known alternatively as PAP gamma and neo-PAP, it is closely homologous to PAP but arises from a different gene. While its domain organization and biochemical activity are similar to that of PAP, it appears not to be regulated by phosphorylation in the same way as PAP (Topalian et al., 2001). In this enzyme, Ser29 has been found in phosphorylated form (Beausoleil et al., 2004).
2.3.6. Symplekin
Symplekin was not initially identified as a co-fractionating subunit of any of the core cleavage factors, i.e., CPSF, CstF, CFIm or CFIm. Rather, it was discovered in nuclear extract and in a crude CFm-like fraction by virtue of its ability to bind labeled CstF-77 during far western analysis (Takagaki & Manley, 2000). While it has an established role in cytoplasmic polyadenylation (Barnard, Ryan, Manley, & Richter, 2004), and is required for the nuclear processing of the cell-cycle dependent histone pre-mRNAs (Kolev & Steitz, 2005), which do not become polyadenylated, symplekin’s function in the 3′ cleavage of polyadenylated pre-mRNAs is not entirely clear, though it has been proposed to promote the assembly of the pre-cleavage complex through binding CstF-77 (Takagaki & Manley, 2000), a suggestion that may take on new significance now that it appears CstF is assembled co-transcriptionally (Glover-Cutter, Kim, Espinosa, & Bentley, 2008). Phosphorylation of the symplekin yeast homolog, Pta1, is known to inhibit polyadenylation (but not 3′ cleavage) possibly by altering the composition of the large yeast PCF complex, which contains yeast PAP and CPSF orthologs (He & Moore, 2005). Since the level of conservation between the two proteins is low, and phosphorylation does not appear to affect the cleavage reaction in yeast, it seems unlikely that phosphorylation of symplekin is involved in the regulation of 3′ cleavage in mammals. Nevertheless, one group has in two different studies identified three phosphoserines in HeLa-derived symplekin: Ser1171 and Ser1175 (Beausoleil et al., 2006) and Ser1243 (Beausoleil et al., 2004), but their functions have not been investigated.
2.3.7. RNA Polymerase II C-Terminal Domain (CTD)
RNA Pol II stimulates the in vitro cleavage reaction, and this activity has been traced to its unique and extensively phosphorylated C-terminal domain (CTD) (Hirose & Manley, 1998). More generally, this large repetitive domain serves as a coordinating protein-protein interaction platform, and phosphorylation contributes to this function (Hirose & Ohkuma, 2007). However, in vitro experiments using the hyper-, hypo- and non-phosphorylated forms of the CTD have shown that the domain’s phosphorylation status plays no role in the 3′ cleavage reaction (Hirose & Manley, 1998; Ryan, Murthy, Kaneko, & Manley, 2002).
3. LYSINE ACETYLATION
Whereas Ser/Thr/Tyr phosphorylation creates from a neutral side chain one having a −2 charge, lysine acetylation neutralizes the +1 charge found on the primary amino group of this amino acid at physiological pH. This modification has been most often studied in the context of the chromatin histone proteins, but other proteins undergo acetylation as well, and more continue to be found (Kouzarides, 2000). Proteomic screens for acetylation are less numerous than for phosphorylation. One proteomic mass spectrometry study has, however, identified a large number of acetylated nuclear and cytoplasmic proteins, but none of the 3′ cleavage factors was found (S. C. Kim et al., 2006).
Another acetylation proteomic study identified an acetylated lysine (K-Ac) at position 23 in CFIm-25, and furthermore provides an excellent example of how post-translational modification proteomics can be leveraged by hypothesis-driven research. Shimazu et al. first identified an acetylated peptide in CFIm-25 by using an anti-acetyl lysine antibody to select proteins from COS-7 cell lysate, followed by LC-MS/MS analysis of its tryptic digest (Shimazu et al., 2006). In a manner analogous to the use of pervanadate to enhance accumulation of phosphotyrosines, these researchers had first treated the COS-7 cells with class I, II and III histone deacetylase (HDAC) inhibitors to increase the accumulation of K-Ac (Shimazu, Horinouchi, & Yoshida, 2007). Using these inhibitors and a combination of immunoprecipitation and Western blotting, it was found that CFIm-68 recruits the CBP acetyltransferase to CFIm-25. These experiments led to the discovery that PAP can also be acetylated (at lysines 641, 650, 736 and 740) and that acetylation on both CFIm-25 and PAP causes a decrease in their affinity for one another, a mechanism that may regulate their association and recycling during processing. (Bovine PAP was used in these experiments, but all four lysines are conserved in human PAP; the numbering here and in Figure 3 refers to the human protein.) The same acetyl transferase, CBP, appears to acetylate both proteins, and the same group of HDACs removes the acetyl groups from both factors. Acetylation also influences PAP intracellular localization: the deacetylated form is exclusively nuclear, whereas acetylation increases partitioning to the cytoplasm. Overall, this post-translational modification has strong effects on the behavior of these two cleavage factors, though unlike PAP phosphorylation, no direct effect on the polymerization activity was observed, and the effect on the 3′ cleavage reaction has not yet been reported.
4. ARGININE METHYLATION
In contrast to phosphorylation and acetylation, arginine methylation does not alter the charge of the affected amino acid. It does however increase the size of the arginine side chain, reduce the number of its hydrogen bond donors by the number of methyl groups added, and make the residue more hydrophobic. Methyl groups are added to arginine residues by the protein arginine methyltransferases (PRMTs) in either a symmetric or asymmetric arrangement. For recent reviews of this large family of enzymes see references (Bedford & Richard, 2005; Pahlich, Zakaryan, & Gehring, 2006). PRMT1 is the predominant PRMT in human cells. It methylates a diverse group of proteins and has a preference for glycine-arginine rich (GAR) sequences, which are relatively common among RNA-binding proteins, for example the hnRNPs (Herrmann, Bossert, Schwander, Akgun, & Fackelmayer, 2004). Several of the other PRMTs also prefer GAR sequences. This modification can be removed by arginine demethylase activity, though only one such enzyme has been found (Chang, Chen, Zhao, & Bruick, 2007). Arginine methylation is thought to have a relatively subtle effect on protein function (Pahlich et al., 2006), being less prone to act as an on/off switch than phosphorylation (Johnson & O’Reilly, 1996) or acetylation (Verdone, Caserta, & Di mauro, 2005), but able nevertheless to affect protein-protein and protein-RNA binding (Bedford & Richard, 2005; Pahlich et al., 2006). Like the other post-translational modifications discussed here, it may occur on a very small percentage of the substrate protein present in a cell, making it difficult for MS based assays to detect all modified proteins (Huang et al., 2002).
Proteomic screens for methylated proteins are less common than for those of lysine acetylation and phosphorylation. Nevertheless, a few of studies have been reported, and one in particular found evidence for arginine methylation among the cleavage factors (Boisvert, Cote, Boulanger, & Richard, 2003). Boisvert et al. immunoprecipitated HeLa whole cell lysate with antibodies raised against various methylated GAR peptides and then used LC-MS/MS to analyze the trypsin digest of the precipitate. When an antibody recognizing a symmetrically dimethylated GAR epitope was used, peptides for CFIm-68, CFIm-25, CPSF-100 and poly(A) binding protein 1 (PABP1) were found. The precise methylation sites were not identified, but within this set of proteins CFIm-68 alone contains a sequence closely matching the GAR peptide antigen. (CFIm-59 lacks a GAR site.) It was concluded that CFIm-68 was likely methylated in its GAR region and recognized by the immunoprecipitation antibody. Moreover, it was likely responsible for the co-immunoprecipitation of the other proteins, which lack a close match to the antibody’s preferred binding site. Methylation within CFIm-68’s GAR sequence has recently been verified, and evidence has been found that PRMT5 is the methyl transferase responsible (G. Martin and W. Keller, personal communication).
This finding places the methylated section of CFIm-68 just before its long proline-rich sequence (Figure 3). Interestingly, methylation of another protein, SAM68, a Src-kinase adaptor protein, in a GAR sequence similarly flanking a proline-rich region, albeit a shorter one, was found to reduce the binding of the proline-rich region to an SH3 domain-containing binding partner, while interaction with a WW domain-containing binding partner was not affected (Bedford et al., 2000). This precedent raises the possibility that methylation within CFIm-68 might modulate interaction of its proline-rich domain with other proteins involved in 3′ processing.
5. LYSINE SUMOYLATION
The only other PTM that has been documented within the cleavage factors is the SUMO (small ubiquitin-related modifier) group (Vethantham, Rao, & Manley, 2007), recently reviewed in reference (Geiss-Friedlander & Melchior, 2007). The human genome encodes four distinct SUMOs, designated SUMO-1 to SUMO-4 (Melchior, 2000). Though small by protein standards at about 10 kD, the SUMO protein is relatively large compared to the other cleavage factor PTMs, and consequently alters a protein’s activity mainly by changing protein-protein interactions in either an inter- or intra-molecular fashion (Geiss-Friedlander & Melchior, 2007). SUMO’s C-terminal glycine becomes covalently ligated to a substrate protein’s lysine side-chain via an isopeptide bond formed in a multi-step process similar to ubiquitination. SUMO chains and single SUMO groups are found appended to human proteins.
Many proteomic SUMO mass spectrometry studies have been carried out on eukaryotic cell extracts. One of these studies found that symplekin is modified by SUMO-1 (Gocke, Yu, & Kang, 2005). In a subsequent study, using an inhibitor of desumoylating enzymes to allow for SUMO accumulation, Vethantham et al. found both HeLa symplekin and CPSF-73, the 3′ cleavage endonuclease, to be modified by SUMO-2/3, but not SUMO-1 (Vethantham et al., 2007). (SUMOs-2 and -3 are 97% identical, and share 50% sequence identity with SUMO-1 (Geiss-Friedlander & Melchior, 2007)). This study used site-directed mutagenesis to deduce the locations of the modified residues. Symplekin appeared to be sumoylated on Lys361 and Lys483, as numbered using the longer form of the protein (GenBank accession no. Q92797, used in Figure 3). The studies that found phosphorylation in symplekin used this form for numbering (Beausoleil et al., 2004; Beausoleil et al., 2006). (As reported by Vethantham et al., who number according to the shorter form, GenBank accession no. CAA71861, the sumoylated residues are Lys229 and Lys351). The probable sites of CPSF-73 sumoylation, Lys462, Lys465 and Lys545, all lie outside of that portion of the protein found in vitro to possess non-specific ribonuclease activity (Mandel et al., 2006). The functional effect of sumoylation on cell growth and 3′ processing was also investigated. RNAi knock-down of symplekin slowed cell growth in a manner that could be restored by wild-type symplekin, but not by a sumoylation-deficient mutant. RNAi knockdown of HeLa ubc9, the enzyme that catalyzes the formation of the isopeptide bond between SUMO and its substrates, yielded nuclear extracts with significantly reduced polyadenylation activity, but only slightly reduced 3′ cleavage activity. When pre-incubated with HeLa nuclear extracts at high concentration, SENP2, one of the SUMO-removing enzymes, but not a catalytically impaired SENP2 mutant, strongly reduced polyadenylation activity. 3′ cleavage activity was reduced as well, but to a much lesser extent than when treated similarly with phosphate-removing enzymes (Ryan, 2007). These results demonstrate a positive correlation between symplekin sumoylation and polyadenylation activity, and a to a lesser degree 3′ cleavage activity. Overall, this study is notable for having identified sites of post-translational modification on specific cleavage factors, and for having demonstrated the functional consequences of removing the modification (Vethantham et al., 2007).
6. REVERSIBLE POST-TRANSLATIONAL MODIFICATION: HOW MIGHT IT REGULATE 3′ CLEAVAGE?
In vitro RNA processing assays carried out in the absence of transcription will continue to be useful for understanding the function of PTMs, but most pre-mRNA 3′ processing is thought to take place while the nascent RNA is still attached to the elongating, though likely paused, RNA Pol II (Bentley, 2005; Glover-Cutter et al., 2008; Hirose & Manley, 2000; Rigo, Kazerouninia, Nag, & Martinson, 2005). Several of the 3′ cleavage factors are recruited to the elongating RNA Pol II complex long before they are needed for 3′ end formation. CPSF (Dantonel, Murthy, Manley, & Tora, 1997; Glover-Cutter et al., 2008; Venkataraman, Brown, & Gilmartin, 2005), CFIm-25 (Venkataraman et al., 2005) and CstF-64 (Glover-Cutter et al., 2008; Venkataraman et al., 2005) are loaded at or near the 5′ end and are present throughout model transcription units. In contrast, CstF-77 is present at maximal occupancy near and beyond the polyadenylation site (Glover-Cutter et al., 2008). Maximal occupancy for several of the 3′ cleavage factors tested thus far coincides with an RNA Pol II pause site about 1 kb downstream of the pA site, but above-background levels of all 3′ cleavage factors tested thus far, even CstF-77, are present throughout most of the transcription unit (Glover-Cutter et al., 2008; Venkataraman et al., 2005).
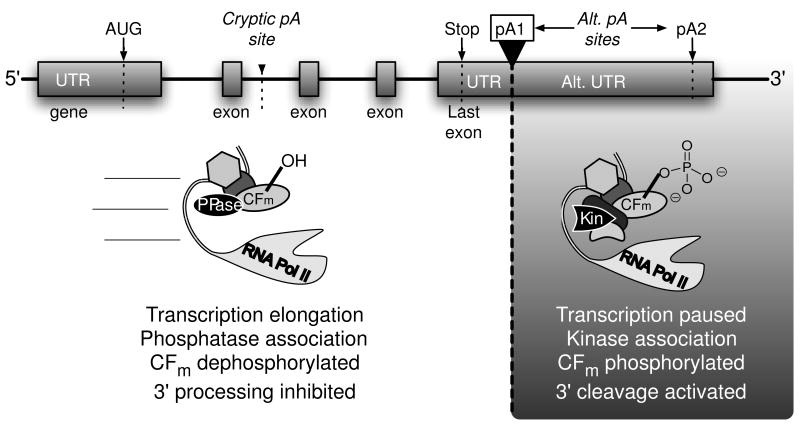
Cryptic pA sites resembling the PAS and DSE sequences are known to occur in transcription units, though during evolution there appears to have been selective pressure to minimize their accumulation on the coding strand (Glusman et al., 2006). Cryptic sites must obviously be bypassed to complete a transcription cycle. What, then, prevents assembly of the productive pre-cleavage complex at cryptic sites occurring early in a transcription unit? One of the determinants for triggering co-transcriptional 3′ cleavage may be missing from our understanding of the process. This determinant could be as simple as the recruitment of CFIIm downstream of the pA site, but unfortunately ChIP data for this factor is lacking. If CFIIm turns out to be freely available to the assembling complex via diffusion, or if it too is present at above-background levels throughout Pol II-transcribed genes, then some additional mechanism might exist to suppress the triggering of 3′ cleavage by prematurely encountered, inauthentic PAS/DSE-like sequences.
We suggest a model in which a reversible post-translational modification of one or more 3′ cleavage factors might act as a safety-lock to limit co-transcriptional 3′ cleavage activity to the 3′ end of a gene. The sharp but reversible loss of in vitro 3′ cleavage activity upon phosphatase treatment (Ryan, 2007) makes cleavage factor phosphorylation the best candidate thus far for this model. As depicted in Figure 4, we envision a protein phosphatase associating with the elongation complex to keep one or more co-transcriptionally recruited cleavage factors in the dephosphorylated state throughout most of the transcription unit, somehow preventing co-transcriptional assembly of the productive 3′ pre-cleavage complex. Some unknown signal in the 3′ UTR might then cause the phosphatase to yield to a kinase, which would phosphorylate the factor and allow pre-cleavage complex assembly to proceed. The post pA site pause might prevent the elongation complex from moving too far away before the cleavage factors can be transferred from the elongation complex to the pA site, and pre-cleavage complex assembly can be completed. Positioning the phosphorylatable cleavage factor in the RNA Pol II ternary complex, along with, perhaps sequentially, the postulated phosphatase/kinase pair, would allow for release of the safety-lock in a position-specific manner along the transcription unit, and make it independent of the majority of the cleavage factor fraction diffusing within the nucleus. Release of the safety-lock in between alternative cleavage sites might differentially expose the sites to the activated nuclease in the mature pre-cleavage complex, thereby influencing alternative polyadenylation.
Figure 4.
A speculative model for the regulation of 3′ cleavage by phosphorylation. In vitro data show that dephosphorylation within CFm (consisting of CFIm and CFIIm) prevents 3′ cleavage. Here, we propose that an unphosphorylated CFm factor subunit associating with the transcription elongation complex suppresses transcription-coupled processing at cryptic sites by preventing pre-cleavage complex formation. Once the 3′ UTR is reached, and the pA site transcribed, a protein phosphatase yields to a kinase, and the phosphorylated cleavage factor promotes pre-cleavage complex formation, possibly during the downstream pause. PPase, hypothetical protein phosphatase; Kin, hypothetical protein kinase.
7. SUMMARY AND OUTLOOK
We have reviewed here the methods used to discover PTMs within the human 3′ pre-mRNA cleavage factors, and catalogued the known sites of phosphorylation, acetylation, methylation and sumoylation. Post-translational modification adds to the energy cost of making a functional protein, so it is reasonable to propose that there is a functional purpose for creating the modifications in most cases, and we hypothesize that the reasons here may involve 3′ cleavage activity regulation. Preliminary in vitro evidence for 3′ cleavage control through phosphorylation and sumoylation is now documented, and more PTMs will undoubtedly be found to have an effect on 3′ end formation. Meanwhile, the group of known modifications provides a database to be drawn upon in the future study of mammalian 3′ end formation of polyadenylated transcripts.
Acknowledgments
This work was supported by the City College of New York Science Division. We also acknowledge NIH grant R21GM073944 to K.R. and the NIH Research Centers in Minority Institutions grant 5G12RR03060. D.L.V.B. is a Barry M. Goldwater Scholarship and Intel Science Talent Search award recipient and gratefully acknowledges their support. We thank G. Martin and W. Keller for sharing unpublished results, and B. Blagoev and B. Tian for helpful discussions.
Abbreviations
- CFIm
mammalian cleavage factor I
- CFIIm
mammalian cleavage factor II
- CFm
DEAE fraction containing CFIm and CFIIm
- CPSF
cleavage and polyadenylation specificity factor
- CstF
cleavage stimulation factor
- CTD
C-terminal domain of RNA Pol II
- DSE
downstream G/U- or U-rich element
- GAR
glycine and arginine-rich
- HDAC
histone deacetylase
- IMAC
immobilized metal affinity chromatography
- K-Ac
acetylated lysine
- MS
mass spectrometry
- pA site
polyadenylation site
- PAP
poly(A) polymerase
- PAS
polyadenylation signal hexamer
- PRMT
protein arginine methyl transferase
- PTM
post-translational modification
- cdk
cyclin dependent kinase
- RNA Pol II
RNA Polymerase II
- SCX
strong cation exchange chromatography
- SUMO
small ubiquitin-related modifier
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101(2):269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4(5):1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25(6):863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119(5):641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275(21):16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18(3):263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17(3):251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271(41):25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2(12):1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem. 2004;76(10):2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12(6):1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- Cantin GT, Venable JD, Cociorva D, Yates JR., 3rd Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J Proteome Res. 2006;5(1):127–134. doi: 10.1021/pr050270m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Chrislip KM, Hengst-Zhang JA, Jacob ST. Polyadenylation of SV40 late pre-mRNA is dependent on phosphorylation of an essential component associated with the 3′ end processing machinery. Gene Expr. 1991;1(3):197–206. [PMC free article] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115(Pt 2):241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384(6606):282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Zhao W, Prives C, Manley JL. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. Embo J. 1998;17(4):1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348(13):1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389(6649):399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J Biol Chem. 2001;276(11):8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. Embo J. 2000;19(21):5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279(34):35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol Cell Biol. 1995;15(11):6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25 (13):2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Ficarro S, Chertihin O, Westbrook VA, White F, Jayes F, Kalab P, et al. Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J Biol Chem. 2003;278(13):11579–11589. doi: 10.1074/jbc.M202325200. [DOI] [PubMed] [Google Scholar]
- Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15(1):71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Qin S, El-Gewely MR, Siegel AF, Roach JC, Hood L, et al. A third approach to gene prediction suggests thousands of additional human transcribed regions. PLoS Comput Biol. 2006;2(3):e18. doi: 10.1371/journal.pcbi.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J Biol Chem. 2005;280(6):5004–5012. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- He X, Moore C. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol Cell. 2005;19(5):619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Herrmann F, Bossert M, Schwander A, Akgun E, Fackelmayer FO. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J Biol Chem. 2004;279(47):48774–48779. doi: 10.1074/jbc.M407332200. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395(6697):93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14(12):1415–1429. [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141(5):601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- Huang HM, Tam MF, Tam TC, Chen DH, Hsieh M, Li C. Proteomic analysis of stable protein methylation in lymphoblastoid cells. J Biochem. 2002;132(5):813–818. doi: 10.1093/oxfordjournals.jbchem.a003291. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci. 1993;18(5):172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- Huber Z, Monarez RR, Dass B, MacDonald CC. The mRNA encoding tauCstF-64 is expressed ubiquitously in mouse tissues. Ann N Y Acad Sci. 2005;1061:163–172. doi: 10.1196/annals.1336.017. [DOI] [PubMed] [Google Scholar]
- Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25(16):7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN, O’Reilly M. Control by phosphorylation. Curr Opin Struct Biol. 1996;6(6):762–769. doi: 10.1016/s0959-440x(96)80005-4. [DOI] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. Embo J. 2004;23(3):616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee Y. Interaction of poly(A) polymerase with the 25-kDa subunit of cleavage factor I. Biochem Biophys Res Commun. 2001;289(2):513–518. doi: 10.1006/bbrc.2001.5992. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19(21):2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? Embo J. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2007 doi: 10.1007/s00018-007-7474-3. http://www.springerlink.com//p3r903543p4n4687/ [DOI] [PMC free article] [PubMed]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444(7121):953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6(5):1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. Embo J. 2006;25(20):4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9(21):2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, et al. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20 (1):77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2(11):1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: Cellular functions and methods of analysis. Biochim Biophys Acta. 2006;1764(12):1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Perumal K, Sinha K, Henning D, Reddy R. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J Biol Chem. 2001;276(24):21791–21796. doi: 10.1074/jbc.M101905200. [DOI] [PubMed] [Google Scholar]
- Qu X, Perez-Canadillas JM, Agrawal S, De Baecke J, Cheng H, Varani G, et al. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J Biol Chem. 2007;282(3):2101–2115. doi: 10.1074/jbc.M609981200. [DOI] [PubMed] [Google Scholar]
- Richardson JM, McMahon KW, MacDonald CC, Makhatadze GI. MEARA sequence repeat of human CstF-64 polyadenylation factor is helical in solution. A spectroscopic and calorimetric study. Biochemistry. 1999;38(39):12869–12875. doi: 10.1021/bi990724r. [DOI] [PubMed] [Google Scholar]
- Rigo F, Kazerouninia A, Nag A, Martinson HG. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell. 2005;20(5):733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1996;271(11):6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1(2):243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Ryan K. Pre-mRNA 3′ cleavage is reversibly inhibited in vitro by cleavage factor dephosphorylation. RNA Biol. 2007;4(1):26–33. doi: 10.4161/rna.4.1.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Murthy KG, Kaneko S, Manley JL. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol Cell Biol. 2002;22(6):1684–1692. doi: 10.1128/MCB.22.6.1684-1692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Horinouchi S, Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J Biol Chem. 2007;282(7):4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Komatsu Y, Nakayama KI, Fukazawa H, Horinouchi S, Yoshida M. Regulation of SV40 large T-antigen stability by reversible acetylation. Oncogene. 2006;25(56):7391–7400. doi: 10.1038/sj.onc.1209731. [DOI] [PubMed] [Google Scholar]
- Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103(7):1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, MacDonald CC, Shenk T, Manley JL. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci U S A. 1992;89(4):1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17(7):3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20(5):1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Kaneko S, Gonzales MI, Bond GL, Ward Y, Manley JL. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21(16):5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19(11):1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Caserta M, Di mauro E. Role of histone acetylation in the control of gene expression. Biochemistry and Cell Biology. 2005;83(3):344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol Cell Biol. 2007;27(24):8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, et al. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci U S A. 1999;96(12):6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447(7141):222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- Yu LR, Zhu Z, Chan KC, Issaq HJ, Dimitrov DS, Veenstra TD. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6(11):4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21(1):65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Zielinski R, Hellman U, Kubinski K, Szyszka R. Fip1--an essential component of the Saccharomyces cerevisiae polyadenylation machinery is phosophorylated by protein kinase CK2. Mol Cell Biochem. 2006;286(1–2):191–197. doi: 10.1007/s11010-005-9104-4. [DOI] [PubMed] [Google Scholar]