Abstract
Import of tRNA into the mitochondrial matrix of Trypanosoma brucei was reconstituted in vitro. Efficient import required the hydrolysis of externally added ATP and was shown to be a carrier-mediated process depending on proteinaceous receptors on the surface of mitochondria. A partly synthetic tRNATyr as well as a physiological tRNALys were imported along the same pathway. Contrary to import of all matrix-localized proteins, tRNA import does not require a membrane potential. Furthermore, addition of an excess of import-competent tRNA had no effect on import of a mitochondrial matrix protein. In summary, these results show that tRNAs and proteins in T. brucei are imported by fundamentally different mechanisms.
INTRODUCTION
Mitochondria contain their own genome and are able to transcribe and translate the genes encoded on their DNA. There are, however, only a limited number of proteins (12 in yeast) encoded and synthesized in that compartment, indicating that the majority of organellar proteins are imported from the cytosol (Attardi and Schatz, 1988). In contrast to proteins, mitochondria generally encode all the structural RNAs (e.g., rRNA and tRNAs) that are needed for organellar translation. However, there is an increasing number of evolutionary divergent organisms whose mitochondrial genome does not contain a full set of tRNA genes and which, therefore, have to import nuclear encoded tRNAs (Schneider, 1994). In most of these cases only a small amount of a given tRNA is targeted to mitochondria, whereas the remaining nonimported fraction is being used in cytosolic translation.
Saccharomyces cerevisiae is the system in which most is known about tRNA import. It is a highly specific process, since only one of two cytosolic tRNALys isoacceptors is targeted to mitochondria (Martin et al., 1979). Using an in vitro import system, it was shown that import of the tRNA required the presence of the precursor of mitochondrial lysyl-tRNA synthetase as well as the membrane potential. In addition, import of the tRNALys was abolished in mutants deficient in the protein-import pathway (Tarassov et al., 1995; Tarassov and Martin, 1996). In summary, these data strongly suggest that the tRNA is cotransported as a complex with the mitochondrial precursor of lysyl-tRNA synthetase across the protein-import channel (Tarassov and Martin, 1996).
Less is known about tRNA import in plants as no in vitro import system is yet available. Plants import a subset of their mitochondrial tRNAs (Dietrich et al., 1992). In vivo data have shown that in potato a single-point mutation within the acceptor stem of the imported tRNAAla concomitantly abolishes import as well as charging, providing indirect evidence that in plants, aminoacyl-tRNA synthetases might also be involved in import (Dietrich et al., 1996).
Recently, much work on mitochondrial tRNA import has been focused on trypanosomatids such as Trypanosoma brucei and Leishmania. Indeed, these parasitic protozoa are excellent systems in which to study the process. In contrast to yeast and plants, the whole set of mitochondrial tRNAs needs to be imported as no tRNA genes have been found in the mitochondrial genome. The majority of tRNAs from trypanosomatids have a dual location in the cytosol and the mitochondrion, resulting in almost identical sets of tRNAs in the two compartments (Simpson et al., 1989; Hancock and Hajduk, 1990; Mottram et al., 1991). Few compartment-specific tRNAs, however, do exist; one of which, a cytosol-specific tRNAGln, has been cloned in Leishmania tarentolae (Lye et al., 1993). It has been shown in an in vivo study that replacing the D-loop of this cytosolic tRNAGln by one of the imported tRNAIle leads to import of the resulting mutant tRNA, indicating that the D-loop region contributes to the import signal (Lima and Simpson, 1996). A similar conclusion was reached in an in vitro analysis using sequences derived from the tRNATyr of Leishmania tropica (Mahapatra et al., 1998). When cytosolic and mitochondrial isotypes of tRNAs in T. brucei were compared, high molecular forms of mitochondrial tRNAs mainly caused by 5′-extensions were detected. The exact sequence of these extensions is unknown, but they can be processed in vitro by RNase P from Escherichia coli, suggesting that tRNAs carrying 5′-extensions are the physiological import substrates (Hancock et al., 1992). However, in an in vivo study it was shown that tRNAs can be imported into mitochondria of T. brucei even if they are expressed in heterologous genomic contexts. Nevertheless, in this case 5′-extended forms have also been detected, demonstrating that variable 5′-sequences, even of nontrypanosomal origin, may be able to direct import (Hauser and Schneider, 1995).
Not much is known about the actual mechanism of tRNA import in trypanosomatids. Indirect data suggest that, at least for the imported tRNAGln in L. tarentolae, the corresponding mitochondrial glutaminyl-tRNA synthetase is not involved in import (Nabholz et al., 1997). Furthermore, in a pioneering study, ATP-driven cytosol-independent import of tRNA-like substrates into isolated mitochondria of L. tropica has been shown (Mahapatra et al., 1994; Mahapatra and Adhya, 1996). However, in that study no comparative analysis of tRNA and protein import was performed. Here we have developed in vitro import systems derived from T. brucei for both groups of macromolecules to asses the relation between tRNA and protein import.
MATERIALS AND METHODS
Cells
Procyclic T. brucei, stock 427, were grown at 27°C in SDM-79 medium supplemented with 5% fetal bovine serum. Cells were harvested at late-log phase corresponding to 2.5 × 107 to 3.5 × 107 cells/ml. Cells were washed once in cold 20 mM sodium phosphate buffer (pH 7.9) containing 150 mM NaCl and 20 mM glucose.
Isolation of Mitochondria
Mitochondria were isolated as described by Hauser et al. (1996) except that a low-speed spin (300 × g) was routinely performed before loading of the Nycodenz gradients. This step removed all the remaining intact cells, resulting in a highly purified mitochondrial fraction. Routinely, cells from 5 l of cultures were being used and yielded 50–100 mg of mitochondria. Mitochondria were resuspended at very high concentrations (30–50 mg/ml) in SoTE (0.6 M sorbitol, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 4 mM DTT) containing 10 mg/ml fatty acid-free BSA and frozen in aliquots in liquid N2. The samples were kept at −70°C and retained tRNA and protein import capacity for at least 2 months.
In Vitro Import Assays for tRNA and Proteins
tRNA import was performed in 20 mM Tris-HCl, pH 7.2, 15 mM KH2PO4, 0.6 M sorbitol, 5 mM succinate, 0.1 mM of each of the biological amino acids, 20 mM MgSO4, 5 mM NADH2, 5 mg/ml fatty acid-free BSA. A standard import reaction was performed in 50 μl volume and contained 200 μg of mitochondria and 0.2 pmol of in vitro transcribed substrate tRNA. In most cases the 5′-syntRNATyr (Figure 1A) was used as substrate. Where indicated, 4 mM of ATP (pH 7.0) was added. Import was done for 60 min at 25°C. Nonimported tRNAs were digested for 10 min at 25°C by the addition of 5 μg of RNase A (Pharmacia LKB, Uppsala, Sweden) and 100 U of RNase T1 (Boehringer Mannheim, Mannheim, Germany). A small amount of nonimported tRNA was protected from RNase attack. Therefore, the sample was further treated for 10 min on ice with 5 μg of proteinase K. Finally, mitochondria were reisolated by centrifugation for 3 min at 5200 × g, and RNA was extracted from the resulting pellets (Chomczyinski and Sacchi, 1987) and analyzed on 8 M urea/10% polyacrylamide gels. Protein import was done as described by Hauser et al. (1996). Due to the optimization of the purification method for mitochondria, much higher import efficiencies were reached than previously published.
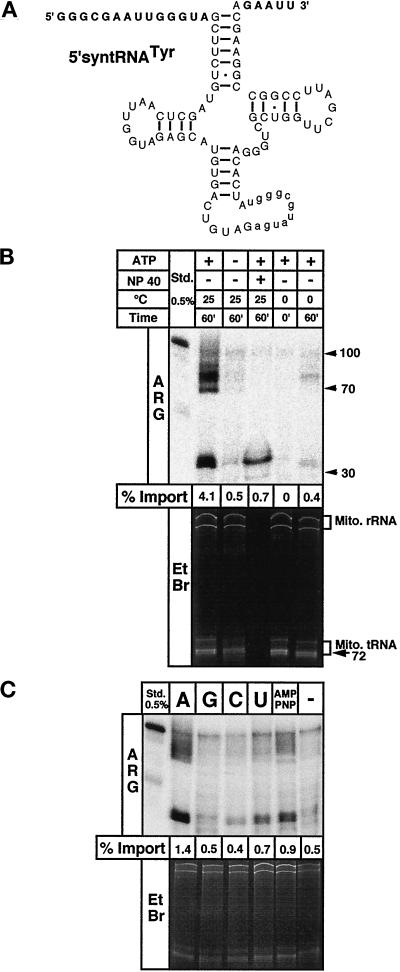
Figure 1.
tRNA import is time, temperature, and ATP dependent. (A) Substrate for standard in vitro import assays consisting of the T. brucei tRNATyr derivative (5′-syntRNATyr) containing synthetic 5′- and 3′-flanking regions (shown in bold). The substrate also contains an 11-nucleotide intron (shown in lowercase), which does not interfere with import as was shown in T. brucei (Schneider et al., 1994) and Leishmania in vivo (Sbicego et al., 1998). (B) In vitro import assays using the 5′-syntRNATyr as a substrate. ATP, reactions were performed in the presence (+) or absence (−) of 4 mM ATP. NP 40, sample was treated with 0.5% of Nonidet P 40 before RNase digestion. The length in nucleotides of single-stranded DNA fragmentsused as molecular weight markers and the positions of mitochondrial rRNAs (Mito. RNA) and tRNAs (Mito. tRNA) are indicated on the right. (C) Import assays were performed in the presence (+ATP) or absence of 1 mM ATP (−ATP) or 1 mM of the indicated nucleotides or nucleotide analogues. RNAs extracted from reisolated mitochondria of all samples were analyzed on 8 M urea/10% polyacrylamide gels. The relevant regions of the ethidium bromide-stained (EtBr) gels and of the corresponding autoradiograms (ARG) are shown. For the stained gels, the panels correspond to the top part of the gel showing the mitochondrial rRNAs and the tRNA region, whereas for the autoradiograms, only the lower portions of the gels are shown. The two panels, therefore, overlap only partially but cover all observable signals. The percentage of the substrate that was imported (% Import) is indicated at the bottom of each autoradiogram. To determine import efficiencies the signals of the two sets of RNase-resistant fragments (see Figure 2) were combined and quantified on a phosphorimager, and 0.5% of the added substrate was run on the gels and used as a standard (Std.).
Preparation of Substrate tRNAs
The substrate tRNAs for in vitro import were prepared by in vitro transcription using T7 polymerase and α32P-GTP using standard procedures. For chemical amounts of substrates, which were used in the competition experiments in Figure 3, trace amounts of α32P-GTP were added to allow the determination of the exact chemical concentrations. Double-stranded DNA templates for T7 transcription were obtained by PCR using a 5′-oligonucleotide having a T7 promoter sequence and the corresponding 3′-oligonucleotides.
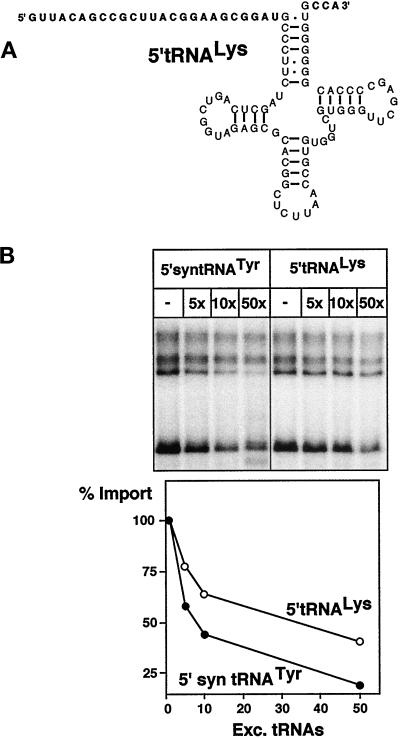
Figure 3.
A putative physiological substrate competes with import of the standard substrate. (A) Predicted secondary structure of a putative physiological import substrate, termed 5′-tRNALys, consisting of the tRNALys carrying a 5′-extension of 25 nucleotides (bold), corresponding to the 5′-flanking sequence found in the genome. It does not contain an intron and has a mature CCA 3′-end (bold). (B) Import of 0.2 pmol of radiolabeled 5′-syntRNATyr was challenged by adding an excess of unlabeled tRNAs. The molar excess of the competitor tRNA is indicated at the top of the panels. In vitro transcribed 5′-syntRNATyr as well as the 5′-tRNALys were used as competitors. Quantification of the competition experiments is shown in the graph.
Submitochondrial Fractionation by Digitonin Extraction
For the digitonin extraction an upscaled standard import reaction in a total volume of 1 ml using 4 mg of mitochondria and 4 pmol of labeled 5′-syntRNATyr was performed. The reaction was digested with 100 μg of RNase A. The RNase T1 and the proteinase K steps were omitted. The reaction was divided into nine aliquots of 100 μl each. Mitochondria were reisolated and resuspended in 30 μl of SoTE containing 120 U of RNasin (Promega, Madison, WI). This amount of RNasin was sufficient to completely inhibit the residual RNase A activity (data not shown). Finally, after the addition of 30 μl of SoTE containing twice the final concentration of digitonin (Fluka Chemical, Buchs, Switzerland), the samples were mixed, incubated for 5 min on ice, and separated into pellets and supernatants; 15% of each fraction was used for immunoblot analysis with antisera directed against marker proteins. Adenylate kinase activity for each supernatant was determined using fractions obtained from a duplicate experiment starting with 12 mg of mitochondria (Schmidt et al., 1984). No radioactive substrate was added in this case.
Submitochondrial Fractionation by Hypotonic Treatment and Sonication
Import reactions using 700 μg of mitochondria and 0.7 pmol of labeled 5′-syntRNATyr were performed in the absence and the presence of 2 μM valinomycin and 50 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP). After incubation the reactions were treated with RNase A as described but not with RNase T1 and proteinase K and mitochondria were reisolated. The pellets were resuspended in 100 μl of 0.1× SoTE containing 320 U of RNasin. After 10 min at 0°C, 29 μl of 2.4 M sorbitol were added, and the samples were centrifuged. The resulting supernatant corresponded to the intermembrane space fraction. The pellet was resuspended in 129 μl of 0.1× SoTE containing 320 U of RNasin, extensively sonicated and centrifuged. The obtained supernatant fraction corresponds to the mitochondrial matrix. RNA was isolated from the intermembrane space and the matrix fractions of both samples and analyzed for the presence of imported 5′-syntRNATyr and endogenous tRNAs. Adenylate kinase activity was determined using fractions obtained in a duplicate experiment in which no radioactive substrate was added (Schmidt et al., 1984).
Trypsin Pretreatment of Mitochondria
The appropriate amount of mitochondria (200 μg/assay) was resuspended in import buffer (20 μl/assay) containing 10% glycerol instead of BSA and treated with the corresponding concentration (10, 30, and 100 μg/ml) of sequencing grade-modified porcine trypsin (Promega) for 20 min at 25°C. After incubation the protease was stopped by the addition of trypsin inhibitor to a final concentration of 10 mg/ml, and 75 μl of import buffer containing either in vitro transcribed labeled substrate tRNA or in vitro translated precursor protein were added, resulting in a volume of 100 μl. Import was then performed according to the standard protocol as described above.
RESULTS
In Vitro Import System
In order to investigate the mechanism of mitochondrial tRNA import in T. brucei, we have reconstituted the process in vitro using isolated organelles and radiolabeled substrate tRNAs. Most published isolation procedures for trypanosomal mitochondria involve an initial hypotonic lysis of the cells resulting in biochemically pure organelles which, however, do not exhibit a membrane potential (Braly et al., 1974; Harris et al., 1990). Mitochondria used in this study were prepared by isotonic lysis of the cells using nitrogen cavitation followed by separation on Nycodenz gradients (Hauser et al., 1996). This procedure yields pure mitochondria having an intact outer and inner membrane that are devoid of cytosolic contamination (see MATERIALS AND METHODS). They exhibit a membrane potential, are competent for protein import (Hauser et al., 1996), and are able to synthesize proteins (Nabholz et al., 1999).
The standard substrate tRNA used in the in vitro import assays was prepared by in vitro transcription using T7 polymerase. It consists of a trypanosomal tRNATyr derivative carrying an artificial 5′-flanking sequence of 14 and a 3′-trailer of 5 nucleotides (Figure 1A) and is essentially identical to the one utilized in the L. tropica import system (Mahapatra and Adhya, 1996). The substrate tRNA also contains an 11-nucleotide intron which, however, does not interfere with import as was shown in T. brucei (Schneider et al., 1994a) and L. tarentolae in vivo (Sbicego et al., 1998). Import assays were performed in the presence or the absence of ATP. After incubation of the reactions, RNases were used to remove the nonimported tRNAs, and mitochondria were reisolated by centrifugation. Finally, RNA was extracted from the pellets and resolved on a denaturing polyacrylamide gel. In order to ensure that equal amounts of RNA were loaded, the gel was first stained with ethidium bromide to visualize endogenous mitochondrial rRNA and tRNAs (Figure 1B, lower panel). No cytosolic RNAs were found, indicating that the mitochondrial preparations were of high purity. In addition, the inaccessibility of the endogenous RNAs to the added RNases demonstrates that the mitochondria were still intact after the import assays. Import of the radiolabeled substrate tRNA was then analyzed by autoradiography. The upper panel of Figure 1B shows that the tRNA was imported into isolated mitochondria, as evidenced by the appearance of two sets of RNase-resistant fragments that were recovered providing ATP was added to the reaction. Only a residual signal is observed when the mitochondrial membranes were destroyed by NP-40 before the RNase digestion. RNase-resistant fragments do not appear when the incubation step is omitted or performed at 0°C. In vitro import does not require cytosolic factors. Import efficiencies up to 4% of added substrate were reached, which is comparable to the physiological situation in which an estimated 4–6% of a given tRNA are found in mitochondria. Efficient tRNA import requires the hydrolysis of ATP: no other nucleotides including the nonhydrolyzable 5′-adenylimidodiphosphate (AMP-PNP) are able to substitute for ATP (Figure 1C). The residual nuclease-resistant fragments observed in the reactions containing nucleotides other than ATP are most likely due to conversion of these nucleotides to ATP or to ATP contaminations of the chemicals (Horst et al., 1996). Even in the absence of added ATP or other nucleotides (Figure 1, B and C) a residual amount of protected fragments is observed. These might be caused by ATP leaking out of mitochondria or indicates that a small level of import occurs independently of added ATP.
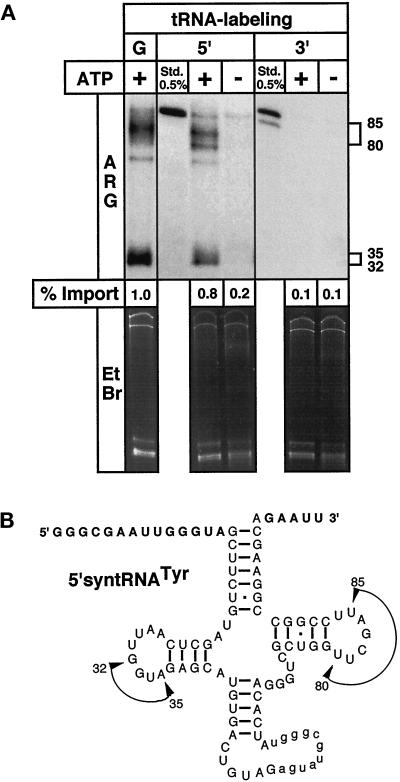
Unexpectedly, two sets of tRNA fragments rather than the intact substrate tRNA were recovered after import. To characterize these fragments in more detail, import was performed with the same substrate, which was either uniformly labeled as in the previous experiments or labeled at the 5′- or the 3′-end only. As shown in Figure 2A, the RNase-resistant fragments of the uniformly and the 5′-end-labeled substrates looked very similar. The protected bands therefore represent sequences with an intact 5′-end. The size was 70–90 nucleotides (the main products being 80–85 nucleotides) for the upper set of bands, corresponding to fragments ranging from the 5′-extension to the T-loop region. The lower two bands are 32 and 35 nucleotides in length and are due to fragments carrying the 5′-extension and sequences up to the D-loop region (Figure 2B). No protected fragments were observed when the 3′-end labeled tRNA was used, implying the activity of a 3′-exonuclease that completely degrades the 3′-fragments of the tRNA. The distinct 5′-fragments, however, are protected from complete degradation by the added RNases and only appear in the presence of ATP, which strongly suggests that the fragmentation occurs inside mitochondria. This is supported by the fact that the added RNases (RNase A and RNase T1) show neither ATP-dependent nor 3′-exonuclease activity. The fragmentation is, however, specific for the imported radiolabeled substrate, since the endogenous rRNAs and tRNAs are not degraded (Figure 2A, lower panel). This observation might be explained by the fact that the tRNA used in the assay was transcribed in vitro and therefore is devoid of nucleotide modifications. In vivo, however, the majority of the nucleotide modifications are known to occur in the nucleus; therefore, the in vivo import substrate would be completely modified (Schneider et al., 1994b). Nucleotide modifications are known to protect RNAs from nuclease attack. tRNA is the most extensively modified RNA in nature, with many of the conserved nucleotide modifications clustered in the D- and the T-loop. The modifications in the D- and T-loop provide, therefore, a plausible explanation why these regions, in a nonmodified tRNA such as the in vitro import substrate, are the preferred sites of attack by the mitochondrial RNase activity. Furthermore, high RNase activity is expected in trypanosomes, as many primary transcripts of mitochondrial mRNAs are extensively processed by RNA editing, a process in which various nucleases have been implicated (Cruz-Reyes and Sollner-Webb, 1996; Piller et al., 1997; Alfonzo et al., 1998).
Figure 2.
Imported tRNA is fragmented by mitochondrial 3′-exonuclease activity. (A) 5′-syntRNATyr was labeled uniformly by in vitro transcription using α32P-GTP (G), at the 5′-end by polynucleotide kinase and γ32P-ATP (5′), or at the 3′-end by splint labeling using α32P-ATP (3′) (Hausner et al., 1990; (Schneider et al., 1994). All substrates were tested in standard import reactions. (B) The size of the two sets of fragments have been calculated, and the regions of their estimated 3′-ends are indicated by arrows in the sequence of the 5′-syntRNATyr.
tRNA Import Is a Carrier-mediated Process
Import of the labeled standard substrate could be competed with an excess of the homologous substrate, indicating that mitochondrial tRNA import is a saturatable carrier-mediated process (Figure 3B). All the experiments described above have been performed using the standard substrate containing 5′- and 3′-flanking sequences of synthetic origin. So far, only indirect evidence concerning the nature of the in vivo import substrate exists. However, the fact the 5′-extended tRNAs have been detected in mitochondria makes it likely that these molecules are the import substrates (Hancock et al., 1992). Furthermore, experiments using transgenic trypanosomes have suggested that, while 5′-extended tRNAs were detected in these experiments, there is no requirement for a specific sequence of the 5′-extension (Hauser and Schneider, 1995). This is in agreement with the fact that a synthetic 5′-extension is functional on the standard import substrate. In order to show that import of a putative physiological substrate follows the same pathway than the semisynthetic standard substrate, we used the trypanosomal tRNALys. This tRNA carries a 5′-extension of 25 nucleotides, which corresponds to the 5′-flanking sequence found in the genome (Mottram et al., 1991). It does not contain an intron and has a CCA at its 3′-end (Figure 3A). Figure 3B shows that this putative physiological tRNA is able to compete for import of the labeled standard substrate, suggesting that they both use the same pathway. Competition of import with an excess of the tRNALys is slightly less efficient than with the corresponding homologous substrate. This might be due to the different lengths of their 5′-flanking sequences (25 nucleotides for the tRNALys and 14 for the tRNATyr), or it might reflect the requirement for cytosolic factors for correct targeting.
tRNAs Are Imported into the Matrix of Mitochondria
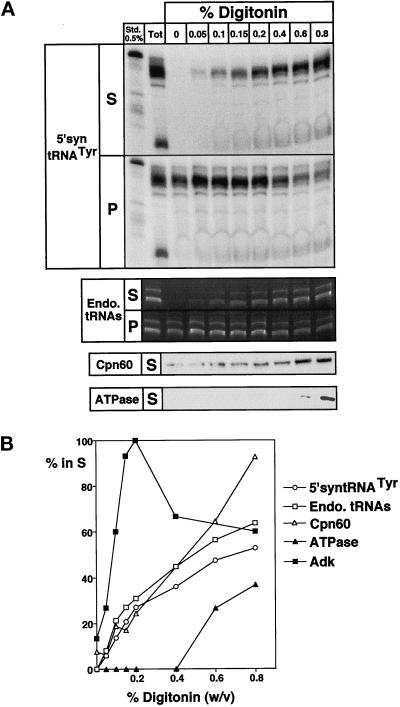
In order to be physiologically relevant, the in vitro tRNA import system should result in the transport of the tRNA into the matrix, the mitochondrial subcompartment where translation occurs. The submitochondrial localization of the imported tRNA fragments was established by digitonin extraction (Hartl et al., 1986). Samples of mitochondria containing imported tRNAs were treated with increasing concentrations of the detergent, separated into pellets and supernatants, which were then analyzed for the presence of labeled and endogenous tRNAs, as well as for different marker proteins. Figure 4 shows that the labeled tRNA fragments were released into the supernatant at very similar digitonin concentrations as the endogenous tRNAs and the matrix marker chaperonin 60. Adenylate kinase, a classic marker of the intermembrane space (Figure 4B) (Schmidt et al., 1984), was released at much lower concentrations, whereas a component of the F1Fo-ATPase was only partially released even at high concentrations of the detergent, as expected for an inner membrane protein. The obtained results establish that mitochondria with intact outer and inner membranes were used in the experiments and that the imported tRNA fragments are found in the same intramitochondrial fraction as the endogenous tRNAs.
Figure 4.
In vitro imported tRNAs are localized in the matrix. (A) A preparative RNase-treated import reaction was split into aliquots that were incubated with increasing concentrations of digitonin. The samples were separated into supernatants (S) and pellets (P) and subjected to RNA isolation. The upper panel shows the recovery of the labeled, imported tRNA fragments (5′-syntRNATyr). In the panel below, the endogenous (Endo.) tRNAs are visualized on the corresponding ethidium bromide-stained gel. In the lowest two panels, immunoblot analysis of the supernatant fractions using antisera directed against chaperonin 60 (Cpn60), a matrix marker, and a component of the FoF1-ATPase (an inner membrane marker) are shown. (B) The signals shown in panel A were quantified using a phosphorimager and densitometry. For all molecules the percentage released to the supernatant is indicated. In addition to the markers shown in panel A, the activity of adenylate kinase (Adk), a marker of the intermembrane space, measured in the supernatant fractions, is shown (Schmidt et al., 1984). Maximal released activity for Adk was set at 100%.
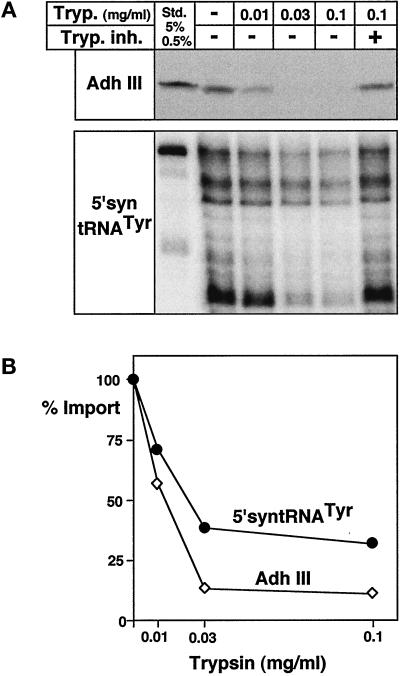
tRNA and Protein Import Depends on Proteinaceous Receptors
Mitochondrial protein import has been studied in great detail in many different systems (Schatz, 1996; Neupert, 1997; Glaser et al., 1998; Mori and Terada, 1998). In these studies the existence of receptors for precursor proteins on the surface of mitochondria has been established (Haucke and Lithgow, 1997). The competition experiments shown in Figure 3 showed that tRNA import is a carrier-mediated process in trypanosomes. The involvement of protein receptors on the surface of mitochondria is therefore expected for tRNA import as well. In order to test this prediction, mitochondria were treated with various concentrations of trypsin before import. After inactivation of the protease, the sample was split in half and analyzed for tRNA and as a control for protein import. Protein import was monitored by the appearance of proteinase K-resistant radiolabeled protein as described previously (Hauser et al., 1996). In vitro translated mitochondrial precursor of yeast alcohol dehydrogenase III (AdhIII), a mitochondrial matrix protein, was used as a substrate. However, it was previously shown that a precursor protein carrying a trypanosomal presequence behaves the same (Hauser et al., 1996). Figure 5 shows that trypsin pretreatment of mitochondria abolishes protein import as expected. Essentially the same result was obtained for tRNA import. A mock control was performed where trypsin at the highest concentration used in the experiment was mixed with trypsin inhibitor before it was added to mitochondria. This control was crucial as most commercial trypsin preparations tested contained RNases, and only protein sequencing-grade trypsin was devoid of such interfering activity. In summary, we can conclude from these experiments that tRNA as well as protein import need proteinaceous receptors on the surface of mitochondria. In addition, the observed inhibition of tRNA import by trypsin pretreatment of mitochondria precludes that the observed fragmentation of the imported tRNA is caused by the added RNases, since these proteins are only added after the inactivation of trypsin. These experiments therefore establish that the imported tRNA is cut into two sets of fragments by an as-yet-unidentified mitochondrial RNase activity.
Figure 5.
Membrane receptors are required for protein and tRNA import. Isolated mitochondria of T. brucei were pretreated with the indicated concentrations of trypsin (Tryp.), split in half, and analyzed for in vitro protein and tRNA import. For the mock control, trypsin was inactivated by trypsin inhibitor (Tryp. inh.) before the addition to mitochondria. (A) Upper panel, import of in vitro translated mitochondrial precursor of yeast alcohol dehydrogenase III (AdhIII) was analyzed by proteinase K protection assays and fluorography (Hauser et al., 1996). Lower panel, in vitro tRNA import assays using the labeled 5′-syntRNATyr as a substrate. Added AdhIII (5%) and 0.5% of added 5′syntRNATyr were used as standards. (B) Quantification of the results in panel A. Import in the mock control was set at 100%.
Distinct Mechanisms for tRNA and Protein Import
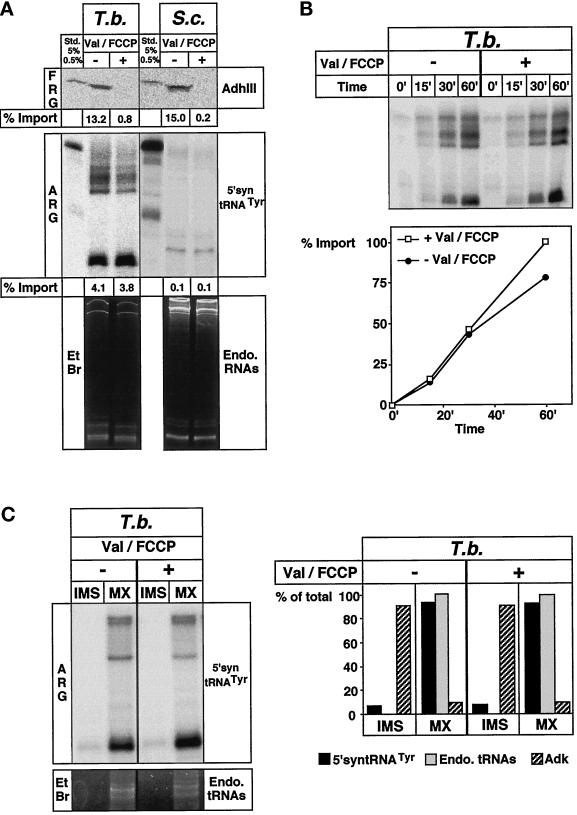
The mechanism of mitochondrial tRNA import has been studied most extensively in S. cerevisiae, and it was shown that the tRNA is coimported with a mitochondrial precursor protein across the protein import channel (Tarassov and Martin, 1996). To investigate whether this also applies for T. brucei, we compared in vitro import of proteins and tRNAs in both T. brucei and S. cerevisiae mitochondria. The top panel of Figure 6A shows that import of a yeast mitochondrial matrix protein (AdhIII) occurs in both organisms with comparable efficiencies. As expected, dissipation of the membrane potential by the ionophore valinomycin and the protonophore FCCP abolishes protein import in both species. Using the same mitochondrial preparations and conditions as used for protein import, tRNA import assays were performed in both organisms. As predicted, no import was observed in yeast under these conditions. tRNA import in yeast has previously been analyzed in great detail and was shown to be specific for a single tRNALys isoacceptor, to depend on the precursor of mitochondrial lysyl-tRNA synthetase, and to need the membrane potential (Tarassov et al., 1994). In T. brucei, in contrast to yeast, the tRNA was imported both in the presence and in the absence of a membrane potential. The time course experiment in Figure 6B shows that not only the standard import reaction performed for 60 min, but the whole kinetic of tRNA import, remains unaffected by the electrochemical potential. In order to define the submitochondrial localization of the tRNA fragments imported in the presence or the absence of the membrane potential, mitochondria were separated into an intermembrane space and a matrix fraction using hypotonic swelling and sonication. Analysis of the obtained fractions gave the same result as the digitonin extraction (Figure 4). Both the labeled tRNA fragments as well as the endogenous tRNAs were localized in the matrix, irrespectively of whether import has been performed in the presence or the absence of a membrane potential (Figure 6C). The hypotonic procedure to selectively disrupt the outer membrane was monitored by measuring the activity of adenylate kinase that is localized to the intermembrane space (Schmidt et al., 1984). Two conclusions may be derived from these results. First, tRNA import can occur in the presence of the membrane potential. This is not trivial, as in the living cell mitochondria certainly exhibit a membrane potential that renders the inside of the inner membrane acidic. Membrane translocation of tRNAs that are negatively charged would therefore be expected to be inhibited due to charge repulsion. Second, it is a well-established fact that all mitochondrial matrix proteins require a membrane potential to translocate the inner membrane (Wachter et al., 1994; Bauer et al., 1996). The absence of such a requirement for tRNA import in trypanosomes indicates that different mechanisms must exist for import of tRNAs and proteins.
Figure 6.
Distinct mechanisms for mitochondrial import of tRNAs and proteins in T. brucei. (A) Upper panel, in vitro translated mitochondrial precursor of AdhIII was imported into mitochondria of T. brucei (T. b.) or yeast (S. c.) in the absence (−) or the presence (+) of the uncouplers valinomycin and FCCP (Val/FCCP). Import was analyzed by fluorography (FRG). Import efficiencies of AdhIII are indicated. Lower panel, import of 5′syntRNATyr was analyzed under identical conditions as AdhIII import. Both the autoradiogramm (ARG) and the ethidium bromide-stained gel (EtBr) are shown. Import efficiencies of 5′-syntRNATyr are indicated. Standards as in Figure 5. (B) Time course of 5′-syntRNATyr import performed in the absence or the presence of Val/FCCP. Import at 60 min was set at 100%. (C) Mitochondria reisolated at the 60-min time point from import reactions performed in the presence and absence of uncouplers were separated into an intermembrane space (IMS) and a matrix (MX) fraction. All fractions were analyzed for the presence of radioactive substrate tRNAs (ARG), for the endogenous tRNAs (EtBr), and for adenylate kinase activity (Adk). Quantification of the results is shown on the graph below the panel. The total signal observed in matrix and the intermembrane space fraction was set at 100%.
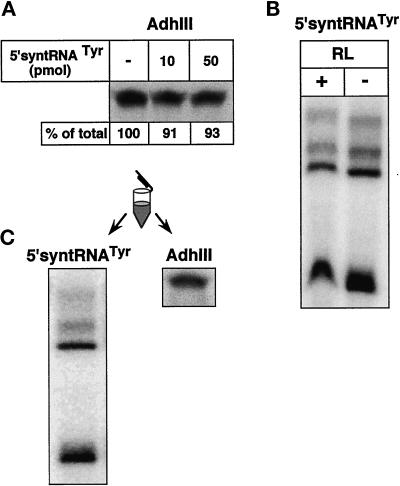
Additional evidence for the existence of two distinct mechanisms is provided by competition experiments. No competition for import of the mitochondrial matrix protein AdhIII is observed by the addition of an excess of the standard tRNA import substrate (Figure 7A), even though as little as 10 pmol of the tRNA is sufficient to efficiently compete for import of the corresponding labeled tRNA. Control experiments showed that the same amount of reticulocyte lysate that was added to the protein import assay does not inhibit tRNA import (Figure 7B). Finally, in Figure 7C, the result of a combined import reaction of protein and tRNA performed in the same tube is shown. Half of the reaction was analyzed for tRNA import, and the other half for protein import. Both macromolecules were shown to be imported, indicating that the selected conditions are compatible for tRNA as well as protein import.
Figure 7.
An excess of import-competent tRNA does not compete with import of a matrix protein. (A) Import of AdhIII precursor was performed after preincubation (20 min) of the mitochondria without (−) or with an excess (10, 50 pmol) of 5′-syntRNATyr (10 pmol of the tRNA corresponds to a 50-fold molar excess in Figure 3). Relative import efficiencies are indicated at the bottom of each lane. (B) Import of AdhIII precursor was performed in the absence and the presence of the same amount of reticulocyte lysate (RL) as was used in panel A. (C) Radioactive AdhIII precursor and radioactive 5′-syntRNATyr were incubated with mitochondria in the same tube. One-half of the reaction was analyzed for tRNA import, the other half for protein import.
DISCUSSION
Reconstitution of Mitochondrial tRNA Import
We have reconstituted an in vitro system for mitochondrial tRNA import containing isolated mitochondria of T. brucei and radiolabeled substrate tRNA. The mitochondria used in the assay had intact outer and inner membranes and were fully functional, which is illustrated by the presence of a membrane potential and competence for protein import (Hauser et al., 1996) as well as for mitochondrial translation (Nabholz et al., 1999). Incubation of these mitochondria with the substrate led to protection of radiolabeled tRNA fragments from added RNases. This protection is dependent on intact membranes, the hydrolysis of ATP, time, and elevated temperature. Recovery of the substrate tRNA fragments requires proteins on the surface of mitochondria. The appearance of protected tRNA fragments is due to a process that could be saturated by the addition of a chemical excess of unlabeled substrate tRNA. Using two independent fractionation procedures (digitonin extraction and hypotonic disruption of the outer membrane), the protected substrate fragments were found in the soluble matrix, the innermost subcompartment of mitochondria. The conclusion reached from these experiments is that translocation of the tRNA across the two mitochondrial membranes has been reconstituted in vitro. Surprisingly, import did not require cytosolic factors and led to fragmentation of the imported in vitro transcribed substrate (discussed in RESULTS). A similar in vitro import system has been reported for L. tropica (Mahapatra et al., 1994; Mahapatra and Adhya, 1996). However, the mitochondria used in that study have not been analyzed for an intact outer membrane and the presence of a membrane potential. Furthermore, the submitochondrial location of the imported tRNA has not been analyzed. The fact that a hypotonic procedure has been used to isolate the organelles in that study makes it likely that the membrane potential is not maintained and that the outer membrane is disrupted. Therefore, the L. tropica system may not, in these respects, reflect the physiological situation. A 12-kDa protein appears to play a role in import in the L. tropica import system. However, the intracellular localization of the protein is unclear: it has been found in isolated mitochondria but may also be present in other cellular fractions (Adhya et al., 1996). At present we have no evidence for the occurrence of a similar protein in the mitochondrial preparations used in the T. brucei import assays.
Mitochondrial tRNA and Protein Import
Mitochondrial protein import requires two complex import machineries, one in the outer and one in the inner mitochondrial membrane (Pfanner et al., 1997). The task of these machineries to transport proteins across the two membranes is, at least in principle, comparable to import of tRNAs, in that proteins and tRNAs are both macromolecules. Furthermore, it has been shown in vitro that a double-stranded DNA oligonucleotide that was covalently linked to either a mitochondrial precursor protein (Vestweber and Schatz, 1989) or to a presequence alone (Seibel et al., 1995) can be transported across the protein import pore. It was reasonable, therefore, to suggest that also in vivo imported tRNAs may bind to mitochondrial precursor proteins and use the protein import pathway. Indeed, physiological coimport of a tRNA complexed with a precursor protein has been shown in S. cerevisiae. As expected, in this case the requirements for import of the tRNA are identical to the requirements of protein import (Tarassov et al., 1994; Tarassov and Martin, 1996).
Surprisingly, we find in our comparative analysis of tRNA and protein import that the situation is different in T. brucei. Here the features for tRNA import are clearly distinct from the ones for protein import. Both processes need added ATP and proteinaceous receptors on the surface. The membrane potential, however, is needed for protein, but not for tRNA, import. This finding suggests that the protein import machinery of the mitochondrial inner membrane cannot be involved in tRNA import as it is only functional in the presence of a membrane potential (Wachter et al., 1994; Bauer et al., 1996). Further evidence that the two processes are not linked is provided by the observations that no cytosolic factors are needed for tRNA import in vitro. It should be emphasized, though, that whereas the actual membrane translocation of tRNA does not need cytosolic factors, soluble proteins may still play a role in import in vivo. These factors, analogous to cytosolic chaperones in protein import, may play a role in unfolding of the secondary or tertiary structure of the tRNA. Depending on the tRNA substrate used in the reconstituted system, the requirement for cytosolic factors might be bypassed. This could result in a different substrate specificity of the in vitro system when compared with the in vivo situation and might explain why the 5′-tRNALys, a putative natural import substrate, is a less efficient competitor of a standard import reaction than the semisynthetic 5′-syntRNATyr (Figure 3). Finally, it has been shown in a combined import assay that import of a matrix protein cannot be competed by import-competent tRNA even when added in large excess. This argues against the involvement of the outer membrane protein import machinery in tRNA import. In summary, these results clearly show that in trypanosomes, unlike in yeast, tRNAs and proteins are imported into mitochondria by fundamentally different mechanisms, suggesting the existence of a separate import machinery devoted to tRNAs only. It therefore appears that, during evolution, nature invented two different solutions to the problem of how hydrophilic RNAs are transported across the mitochondrial membranes. In yeast, where only a small amount of one specific tRNA is imported, the best solution for the cell may be to make use of the already existing protein import machinery. Trypanosomes are faced with quite a different situation: they must import the whole set of organellar tRNAs (30 species), in quantities sufficient for mitochondrial translation (Hancock and Hajduk, 1990). Coimport with protein may therefore not be an option; instead, a more efficient pathway solely devoted to import tRNAs has evolved. In many plants the situation may be very similar to trypanosomes: a large number of tRNAs must be imported in order to support mitochondrial translation (Dietrich et al., 1992). It will interesting to see, therefore, which of the two principal models, coimport or direct import, applies to plants.
Mitochondrial tRNA Import and Nuclear tRNA Export
Before nuclear-encoded tRNAs are imported into mitochondria they first must be exported from the nucleus. The two processes, nuclear tRNA export and mitochondrial tRNA import, however, are fundamentally different, since the nature of the nuclear and the mitochondrial membranes is not comparable. The nuclear membrane contains large pores that are permeable for molecules up to 20 kDa and across which proteins and RNAs are actively transported (Forbes, 1992; Mattaj and Englmeier, 1998). In mitochondria, the inner membrane represents a much tighter barrier. The oxidative phosphorylation in mitochondria is coupled to the membrane potential across the inner membrane, illustrating that not even protons can freely pass across this membrane. Considering these facts it is not surprising that completely different mechanisms are being used to transport the tRNAs across the nuclear and the mitochondrial membranes. Nuclear export of tRNAs has been analyzed in great detail and was shown to require proteins that are cotransported with the tRNA (Wolin and Matera, 1999). The situation is different for mitochondrial tRNA import in T. brucei: no soluble protein factors are needed on the cis side of the membrane, and no cotransport of the tRNA with proteins occurs.
The mitochondrial membranes are clearly the tightest barriers any RNA molecule faces within the cell. There appears to be two principal mechanisms by which tRNAs can be translocated across these membranes: coimport with mitochondrially destined proteins, as has been shown in yeast, and direct import, as presented in this study for T. brucei. tRNA import in T. brucei appears to be the only case in which RNA is transported across a membrane without any evidence for simultaneous translocation of associated proteins. Further work will now be concentrated on the identification of membrane factors involved in this unique process.
ACKNOWLEDGMENTS
We thank G. Schatz and W. Oppliger for antiserum against Cpn60, the AdhIII expression plasmid, and for purified yeast mitochondria; S. Adhya for the 5′-syntRNATyr expression plasmid; D. Speijer for the antiserum against T. brucei FoF1-ATPase; and A. Puoti, R. Pach, M. Wymann, M. Zetka, and S. Nabholz for helpful discussions and comments on the manuscript. This study was supported by grant 31–46628.96 from the Swiss National Foundation and by a fellowship of the Prof. Dr. Max Cloëtta Foundation (to A. S.).
REFERENCES
- Adhya S, Ghosh T, Das A, Bera SK, Mahapatra S. Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J Biol Chem. 1996;272:21396–21402. doi: 10.1074/jbc.272.34.21396. [DOI] [PubMed] [Google Scholar]
- Alfonzo JD, Thiemann OH, Simpson L. Purification and characterization of MAR1. A mitochondrial associated ribonuclease from Leishmania tarentolae. J Biol Chem. 1998;273:30003–30011. doi: 10.1074/jbc.273.45.30003. [DOI] [PubMed] [Google Scholar]
- Attardi G, Schatz G. The biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bauer MF, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- Braly P, Simpson L, Kretzer F. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J Protozool. 1974;21:782–790. doi: 10.1111/j.1550-7408.1974.tb03752.x. [DOI] [PubMed] [Google Scholar]
- Chomczyinski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Sollner-Webb B. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc Natl Acad Sci USA. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich A, Marechal-Drouard L, Carneiro V, Cosset A, Small I. A single base change prevents import of cytosolic analyl-tRNA into mitochondria in transgenic plants. Plant J. 1996;10:913–918. doi: 10.1046/j.1365-313x.1996.10050913.x. [DOI] [PubMed] [Google Scholar]
- Dietrich A, Weil JH, Maréchal-Drouard L. Nuclear-encoded tRNAs in plant mitochondria. Annu Rev Cell Biol. 1992;8:115–131. doi: 10.1146/annurev.cb.08.110192.000555. [DOI] [PubMed] [Google Scholar]
- Forbes DJ. Structure and function of the nuclear pore complex. Annu Rev Cell Biol. 1992;8:495–527. doi: 10.1146/annurev.cb.08.110192.002431. [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjoling S, Tanudji M, Whelan J. Mitochondrial protein import in plants. Signals, sorting, targeting, processing and regulation. Plant Mol Biol. 1998;38:311–338. doi: 10.1023/a:1006020208140. [DOI] [PubMed] [Google Scholar]
- Hancock K, Hajduk SL. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem. 1990;265:19208–19215. [PubMed] [Google Scholar]
- Hancock K, LeBlanc AJ, Donze D, Hajduk SL. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J Biol Chem. 1992;267:23963–23971. [PubMed] [Google Scholar]
- Harris ME, Moore DR, Hajduk SL. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- Hartl F-U, Schmidt B, Wachter E, Weiss H, Neupert W. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986;47:939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- Haucke V, Lithgow T. The first steps of protein import into mitochondria. J Bioenerg Biomembr. 1997;29:11–17. doi: 10.1023/a:1022451520203. [DOI] [PubMed] [Google Scholar]
- Hauser R, Pypaert M, Häusler T, Horn EK, Schneider A. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Sci. 1996;109:517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- Hauser R, Schneider A. tRNAs are imported into mitochondria of Trypanosoma brucei independent of their genomic context and of their genetic origin. EMBO J. 1995;14:4212–4220. doi: 10.1002/j.1460-2075.1995.tb00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner TP, Giglio LM, Weiner AM. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- Horst M, Oppliger W, Feifel B, Schatz G, Glick BS. The mitochondrial protein import motor: dissociation of mitochondrial hsp70 from its membrane anchor requires ATP binding rather than ATP hydrolysis. Protein Sci. 1996;5:759–767. doi: 10.1002/pro.5560050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima BD, Simpson L. Sequence-dependent in vivo import of tRNAs into the mitochondrion of Leishmania tarentolae. RNA. 1996;2:429–440. [PMC free article] [PubMed] [Google Scholar]
- Lye L-F, Chen D-HT, Suyama Y. Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol Biochem Parasitol. 1993;58:233–246. doi: 10.1016/0166-6851(93)90045-y. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Adhya S. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J Biol Chem. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Ghosh S, Bera SK, Ghosh T, Das A, Adhya S. The D arm of tyrosine tRNA is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 1998;26:2037–2041. doi: 10.1093/nar/26.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RP, Schneller J-M, Stahl AJC, Dirheimer G. Import of nuclear desoxyRNA coded lysine-accepting transfer RNA (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Mori M, Terada K. Mitochondrial protein import in animals. Biochim Biophys Acta. 1998;1403:12–27. doi: 10.1016/s0167-4889(98)00021-4. [DOI] [PubMed] [Google Scholar]
- Mottram JC, Bell SD, Nelson RG, Barry JD. tRNAs of Trypanosoma brucei: unusual gene organization and mitochondrial importation. J Biol Chem. 1991;266:18313–18317. [PubMed] [Google Scholar]
- Nabholz CE, Hauser R, Schneider A. Leishmania tarentolae contains distinct cytosolic and mitochondrial glutaminyl-tRNA synthetase activities. Proc Natl Acad Sci USA. 1997;94:7903–7908. doi: 10.1073/pnas.94.15.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz, C.E., Speijer, D., and Schneider, A. (1999). Chloramphenicol-sensitive mitochondrial translation in Trypanosoma brucei. Parasitol. Res. (in press). [DOI] [PubMed]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Honlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- Piller KJ, Rusche LN, Cruz-Reyes J, Sollner-Webb B. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA. 1997;3:279–290. [PMC free article] [PubMed] [Google Scholar]
- Sbicego S, Nabholz C, Hauser R, Blum B, Schneider A. In vivo import of unspliced tyrosyl-tRNA containing synthetic introns of variable length into mitochondria of Leishmania tarentolae. Nucleic Acids Res. 1998;26:5251–5255. doi: 10.1093/nar/26.23.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–3166. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Schmidt B, Wachter E, Sebald W, Neupert W. Processing peptidase of Neurospora mitochondria. Two-step cleavage of imported ATPase subunit 9. Eur J Biochem. 1984;144:581–588. doi: 10.1111/j.1432-1033.1984.tb08505.x. [DOI] [PubMed] [Google Scholar]
- Schneider A. Import of RNA into mitochondria. Trends Cell Biol. 1994;4:282–286. doi: 10.1016/0962-8924(94)90218-6. [DOI] [PubMed] [Google Scholar]
- Schneider A, Martin JA, Agabian N. A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol Cell Biol. 1994a;14:2317–2322. doi: 10.1128/mcb.14.4.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, McNally KP, Agabian N. Nuclear-encoded mitochondrial tRNAs of Trypanosoma brucei have a modified cytidine in the anticodon loop. Nucleic Acids Res. 1994b;22:3699–3705. doi: 10.1093/nar/22.18.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel P, Trappe J, Villani G, Klopstock T, Papa S, Reichmann H. Transfection of mitochondria: strategy toward a gene therapy of mitochondrial DNA diseases. Nucleic Acids Res. 1995;23:10–17. doi: 10.1093/nar/23.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AM, Suyama Y, Dewes H, Campbell DA, Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Entelis N, Martin RP. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J Mol Biol. 1994;245:315–323. doi: 10.1006/jmbi.1994.0026. [DOI] [PubMed] [Google Scholar]
- Tarassov I, Entelis N, Martin RP. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Martin R. Mechanisms of tRNA import into yeast mitochondria: an overview. Biochimie. 1996;78:502–510. doi: 10.1016/0300-9084(96)84756-0. [DOI] [PubMed] [Google Scholar]
- Vestweber D, Schatz G. DNA-protein conjugates can enter mitochondria via the import pathway. Nature. 1989;338:170–172. doi: 10.1038/338170a0. [DOI] [PubMed] [Google Scholar]
- Wachter C, Schatz G, Glick BS. Protein import into mitochondria: requirement for external ATP is precursor specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol Biol Cell. 1994;5:465–474. doi: 10.1091/mbc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Matera AG. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]