Abstract
The mushroom-producing fungus Schizophyllum commune has thousands of mating types defined, in part, by numerous lipopeptide pheromones and their G protein-linked receptors. Compatible combinations of pheromones and receptors encoded by different mating types regulate a pathway of sexual development leading to mushroom formation and meiosis. A complex set of pheromone–receptor interactions maximizes the likelihood of outbreeding; for example, a single pheromone can activate more than one receptor and a single receptor can be activated by more than one pheromone. The current study demonstrates that the sex pheromones and receptors of Schizophyllum, when expressed in Saccharomyces cerevisiae, can substitute for endogenous pheromone and receptor and induce the yeast pheromone response pathway through the yeast G protein. Secretion of active Schizophyllum pheromone requires some, but not all, of the biosynthetic machinery used by the yeast lipopeptide pheromone a-factor. The specificity of interaction among pheromone–receptor pairs in Schizophyllum was reproduced in yeast, thus providing a powerful system for exploring molecular aspects of pheromone–receptor interactions for a class of seven-transmembrane-domain receptors common to a wide range of organisms.
INTRODUCTION
Schizophyllum commune, a filamentous wood-rotting fungus, belongs to a class of mushroom-producing fungi known as homobasidiomycetes. Such fungi typically have many different mating types in nature – Schizophyllum is known to have thousands of “sexes” (Raper, 1966). Mate recognition and sexual development leading to formation of fruiting bodies (mushrooms) and meiosis require the action of two unlinked genetic complexes, called A and B. Each complex is composed of two linked, but genetically separable, loci: Aα and Aβ for the A complex, and Bα and Bβ for the B complex. Each locus exists in multiple versions or specificities within the worldwide population. The Aα locus has 9 different specificities, Aβ has 32, and Bα and Bβ have 9 specificities each (Raper et al., 1960; Koltin et al., 1967; Stamberg and Koltin, 1972). The minimal requirement for a fertile pairing among these numerous, haploid mating types is a difference in specificity at either Aα or Aβ and a difference in specificity at either Bα or Bβ (Raper, 1966).
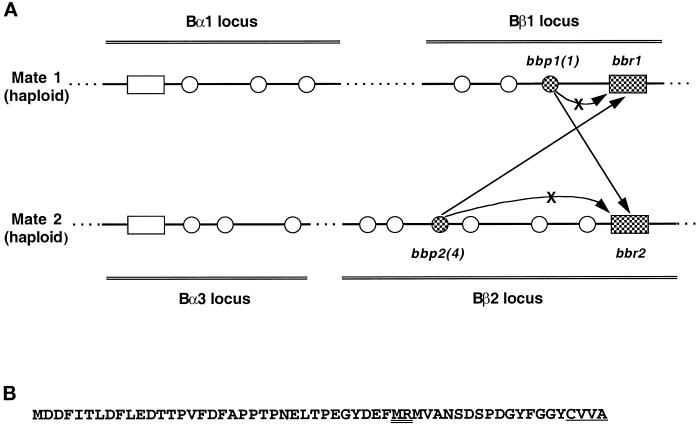
The genes contained within the B mating-type loci regulate a process of reciprocal fertilization in which nuclei of one mate migrate into and throughout the hyphal cells of the other (Raper, 1966; Koltin and Flexer, 1969; Wessels and Marchant, 1974; reviewed by Raudaskoski, 1998). Characterization of several specificities of the two B mating-type loci indicates that each locus encodes one seven-transmembrane-domain receptor and several putative lipopeptide pheromones (Wendland et al., 1995; Vaillancourt et al., 1997; Fowler et al., 1998). The Bβ1 locus, for example, contains three unique pheromone genes, called bbp1(1), bbp1(2), and bbp1(3), and one unique pheromone receptor gene called bbr1. The protein products of these genes are symbolized Bbp1(1), Bbp1(2), Bbp1(3), and Bbr1 accordingly. Functional analyses of cloned genes revealed the fundamentals of self/non-self-recognition (Figure 1A). No wild-type pheromone–receptor pair encoded within a haploid individual, such as Bbr1 and Bbp1(1) shown in Figure 1A, can activate the B-regulated pathway of development in “self,” where self is defined to include the haploid individual and any other individual with identical B mating-type loci. A single pheromone encoded within a specific Bβ locus can activate a subset of the receptors encoded by all other Bβ specificities. For example, pheromone Bbp1(1) activates the Bβ2 receptor (Figure 1A) as well as four additional Bβ receptors (Vaillancourt et al., 1997). Collectively, the pheromones of any one Bβ specificity can trigger the receptors of all eight non-self Bβ specificities, and similarly, the pheromones of any one Bα specificity can trigger the receptors of all eight non-self Bα specificities (Wendland et al., 1995; Vaillancourt et al., 1997; Fowler et al., 1998). However, no pheromone encoded by any specificity of the Bα locus can activate receptors encoded within any specificity of the Bβ locus and vice versa. This complex set of pheromone–receptor interactions governing mating maximizes the likelihood of outbreeding while minimizing inbreeding.
Figure 1.
(A) Interactions of two pheromones and two receptors encoded in the Schizophyllum mating-type loci Bβ1 and Bβ2. The linked Bα and Bβ loci contain open reading frames for putative lipopeptide pheromones (circles) and seven-transmembrane domain receptors (rectangles). The distances between and within the loci are not shown to scale. Bβ genes examined in this study, obtained from a Bα1-Bβ1 strain (Mate 1) and a Bα3-Bβ2 strain (Mate 2), are shown as checkered symbols and designated with the gene name (Vaillancourt et al., 1997; Fowler et al., 1998). Bbp1(1) can activate Bbr2 but not Bbr1, and Bbp2(4) can activate Bbr1 but not Bbr2, as indicated by arrows. Additional interactions among gene products encoded in these loci are not indicated. (B) Predicted amino acid sequence of the Bbp2(4) pheromone precursor. The CaaX box recognition motif for protein prenylation is single underlined, and a postulated N-terminal cleavage site for maturation is double underlined (Casselton and Olesnicky, 1998). The predicted amino acid sequence of Bbp1(1) was previously published (Vaillancourt et al., 1997).
The numerous variants of pheromones and receptors that naturally exist in Schizophyllum make this organism an attractive system for investigations of the molecular and structural basis for specificity of pheromone–receptor interactions. However, the complexity of this system confounds analysis of its components and the role they play in signal transduction. We therefore attempted to reconstitute Schizophyllum pheromone–receptor interactions in a more genetically tractable system, Saccharomyces cerevisiae. Development of a yeast system would allow the examination of individual pheromone–receptor pairs in isolation and facilitate genetic analysis of the specificity determinants of pheromone– receptor interactions.
S. cerevisiae has two mating types, MATa and MATα, and mating involves two pheromone–receptor pairs (reviewed by Sprague and Thorner, 1992; Kurjan, 1993). MATa cells express a-factor pheromone and the α-factor receptor Ste2p; MATα cells express α-factor pheromone and the a-factor receptor Ste3p. The two receptors are characterized by seven-transmembrane domains that span the plasma membrane. When bound by pheromone secreted from cells of the opposite mating type, each receptor couples with the same heterotrimeric G protein to initiate a signal-transduction pathway, known as the pheromone-response pathway. Defined effects of pheromone response include transcriptional activation of a large set of genes, cell-cycle arrest, cell fusion, and nuclear fusion (for review, see Sprague and Thorner, 1992; Kurjan, 1993). The mature α-factor pheromone is a simple 13-amino acid peptide. The active form of the a-factor pheromone is a mixture of two farnesylated peptides of 12 amino acids that differ in sequence in one position; these lipopeptides are processed from two similar precursors of 36 and 38 amino acids encoded by the genes MFA1 and MFA2 (Michaelis and Herskowitz, 1988).
The presumptive Schizophyllum pheromone precursors appear to be comparable to a-factor in that they are small, ranging in size from 40 to 75 amino acids, and end in a C-terminal signal for farnesylation. This signal is a CaaX motif, where a cysteine residue is followed by two aliphatic residues and ends with any of five specific amino acids (Schafer and Rine, 1992). Processing of the N termini of these pheromone precursors may occur, but has yet to be shown (Casselton and Olesnicky, 1998). A comparison of predicted amino acid sequences from nine Schizophyllum pheromone-precursor genes that have been cloned and tested for function reveals considerable variation except for the CaaX motif. All five Schizophyllum pheromone-receptor genes analyzed so far are predicted to encode proteins with seven-transmembrane domains. Amino acid sequence comparisons show that these receptors are significantly similar to the pheromone receptors of S. cerevisiae (Wendland et al., 1995; Vaillancourt et al., 1997; Fowler, Mitton, and Raper, unpublished).
Previous studies demonstrated that some mammalian G protein-coupled receptors expressed in S. cerevisiae showed membrane localization and allowed antagonist and/or agonist binding (King et al., 1990; Price et al., 1995). In one case, a rat somatostatin receptor treated with somatostatin could couple with the yeast G protein to activate the yeast pheromone-response pathway. Here we demonstrate that Schizophyllum receptors can be expressed in yeast and can couple with the yeast G protein. In addition, this study presents evidence that S. cerevisiae can process and secrete functional pheromones encoded by putative pheromone genes of Schizophyllum, thus confirming that these genes encode bona fide sex pheromones. Combinations of pheromones and receptors that are naturally compatible in Schizophyllum activate the yeast pheromone-response pathway, while incompatible combinations do not. This system will make the numerous genetic tools applicable to S. cerevisiae available for the exploration of interactions among the numerous pheromones and pheromone receptors of Schizophyllum.
MATERIALS AND METHODS
Plasmid Isolation
Escherichia coli strains TG1, HB101, and DH5α were used for plasmid production. E. coli transformations were done by electroporation using the Gene Pulser (Bio-Rad, Hercules, CA), and plasmids were isolated with the Qiaprep kit (Qiagen, Valencia, CA).
Yeast Cultures, Transformations, and Gene Disruptions
S. cerevisiae strains (Table 1) were grown at 30°C on YEPD, synthetic drop-out (SD) media lacking uracil, or SD media lacking both uracil and tryptophan (Treco and Lundblad, 1997). Plasmids were introduced into yeast using the PLAG (polyethylene glycol-lithium acetate-glycerol) method (Chen et al., 1992).
Table 1.
S. cerevisiae strains used in this study
| Isogenic to: | Strain | Genotypea | Reference/source |
|---|---|---|---|
| W303-1A | W303-1A | MATa | Kurjan and Dietzel, 1987 |
| W303-1B | MATα | Kurjan and Dietzel, 1987 | |
| RAK2 | MATαste4::HIS3 | R. Akada | |
| RAK32 | MATaste6::HIS3 | R. Akada | |
| Tn44-1B | MATαsst2::LEU2 | Kurjan and Dietzel, 1987 | |
| SDK45 | MATαste3::ADE2 | This study | |
| SDK47 | MATαste3::ADE2 sst2::LEU2 | This study | |
| SDK48 | MATαste3::ADE2 ste4::HIS3 | This study | |
| SM1058 | SM1058 | MATa | Michaelis and Herskowitz, 1988 |
| SM2331 | MATamfa1-Δ1 mfa2-Δ1 | Chen et al., 1997b | |
| SM1188 | MATaΔste14-3::TRP1 | Hrycyna et al., 1991 | |
| SM1865 | MATaΔram1::URA3 | S. Michaelis | |
| LHK1 | MATaΔram1::LEU2 | This study | |
| SM2744 | MATaΔaxl1::LEU2 Δste23::LEU2 | S. Michaelis | |
| SM3614 | MATa Δste24::LEU2 Δrce1::TRP1 | Tam et al., 1998 | |
| None | SM1872 | MATaram2-1 | S. Michaelis |
Strains isogenic to W303-1A contain the markers ade2-1 his3-11,15 leu2-3, 112 trp-1 ura3-1 can1-100; strains isogenic to SM1058 contain the markers trp1 leu2 ura3 his4 can1; and SM1872 contains the markers leu2 ura3 his3 ade8 lys2 can1 cyh4 in addition to the markers shown.
pSK-STE3 was constructed by subcloning the MfeI–SacI fragment from pSL1 (Hagen et al., 1986) into EcoRI-SacI–digested pBluescriptSK+ (Stratagene, La Jolla, CA). The 2.3-kilobase (kb) ADE2 BglII fragment was subcloned into the BamHI site of pBluescriptSK+, and the SpeI–PstI ADE2 fragment from this plasmid was subcloned into pSK-STE3 to make pSK-ste3::ADE2. The KpnI–SacI ste3::ADE2 fragment from pSK-ste3::ADE2 was used to make STE3 gene replacements by lithium acetate transformation (Chen et al., 1992) in strains Tn44–1B and RAK2 (Table 1). The ram1::URA3 disruption in SM1865 was changed to a ram1::LEU2 disruption by cleaving pUL9 (Cross, 1997) with XbaI to obtain a ura3::LEU2 fragment and transforming SM1865.
Schizophyllum Mating, RNA Extraction, and cDNA Synthesis
The following method for growth and mating of Schizophyllum was adapted from Vaillancourt et al. (1997). S. commune strains 4–40 (Aα4-Aβ6/Bα1-Bβ1) and 4–8 (Aα4-Aβ6/Bα3-Bβ2) grown on CYM-agar plates (Raper and Hoffman, 1974) were cut away from the agar and separately macerated in CYM liquid media in a Waring blendor to provide inocula for 100-ml liquid cultures. These liquid cultures were grown 24 h at 30°C with shaking at 200 rpm and then macerated again. After this process was repeated, ∼1 ml of the final macerates was spread on separate 5-cm squares of semipermeable cellophane membrane (Dupont, Wilmington, DE) placed on CYM agar plates and grown at 30°C for 48 h. The membranes on which strain 4–40 was growing were lifted and placed hyphae-side down onto the strain 4–8 cultures. Genes within the B loci are known to be up-regulated after contact between individuals with B loci of different specificities (Vaillancourt et al., 1997). After 8 h of contact, the hyphal mats were stripped from the membranes and flash frozen in liquid nitrogen. Total RNA was isolated by a hot phenol:SDS method as described previously (DeVries et al., 1988). Total RNA (10 μg) was treated with RNAse-free DNAse I (Life Technologies, Gaithersburg, MD) and then converted to cDNA with oligo(dT)12–18 primers using the Superscript Preamplification System (Life Technologies) according to the manufacturer’s protocol.
Construction of Pheromone- and Receptor-Expression Plasmids
DNA fragments containing Schizophyllum pheromone gene- and receptor gene-coding sequences were amplified by PCR, as described below, using oligonucleotides (Genosys, Woodlands, TX) that incorporate EcoRI and BamHI recognition sites at the 5′- and 3′-ends of the open reading frames, respectively. The EcoRI–BamHI fragments were subcloned into pPGK (Kang et al., 1990) to allow expression under the control of the high-expression promoter of the yeast phosphoglycerate kinase gene (PGK).
pPGK-bbr1: oligonucleotides 980420–3 and 980420–4 (Table 2) were used as primers for PCR amplification of the coding region of bbr1 using the pool of cDNAs obtained from the 4–40 × 4–8 mating as template. A standard 50-μl reaction using Taq DNA polymerase (Life Technologies) and a 480 thermocycler (Perkin Elmer-Cetus, Norwalk, CT) was performed.
Table 2.
Oligonucleotides
| Gene | Oligonucleotide | Position | Sequencea |
|---|---|---|---|
| bbr1 | 980420-3 | Upstream | TTCGAATTCATGCACCCCGAGTTTGCCCCCGTAG |
| 980420-4 | Downstream | TCCGGATCCCTACCTGCGCCCACCGGGGAACAC | |
| bbr2 | 980730-1 | Upstream | TTCGAATTCATGTACTCCAACGACCCGACG |
| 980730-2 | Downstream | TCCGGATCCTTAAAGGACGACGCGGTGTG | |
| bbp1(1) | 980730-5 | Upstream | TTCGAATTCATGGACGCCTTCACCGCC |
| 980730-6 | Downstream | TCCGGATCCCTACGCAACCACGCACCAC | |
| bbp2(4) | 980227-2 | Upstream | CGGAATTCATGGACGACTTCATCACC |
| 980227-3 | Downstream | CGGGATCCGGCTACGCCGGGTCACGCC |
The oligonucleotide sequences are shown in the 5′ to 3′ direction. Single underlines indicate EcoRI sites in the upstream primers or BamHI sites in the downstream primers. Double underlines indicate an ATG initiation codon in upstream primers or a termination codon in the reverse strand of downstream primers; 980730-2 binds downstream of the termination codon.
pPGK-bbr2: Full-length cDNA clones of bbr2 (GenBank Accession AF148501) from a mating of strains 4–40 and 4–39 (Aα4-Aβ6/Bα3-Bβ2) were kindly provided by Dr. Marjatta Raudaskoski. The coding region of bbr2 was amplified by PCR from a cDNA template using oligonucleotide primers 980730–1 and 980730–2 (Table 2).
pPGK-bbp2(4) and pPGK-bbp1(1): PCR amplification products containing the bbp1(1) (Vaillancourt et al., 1997) and bbp2(4) (Fowler, Mitton, and Raper, unpublished; GenBank Accession AF148500) coding sequences were generated from genomic clones using oligonucleotide pairs 980730–5 and 980730–6 for bbp1 and 980227–2 and 980227–3 for bbp2(4) (Table 2). No intron interrupts the bbr1 coding region (Raudaskoski et al., 1998) or the bbp2(4) coding region (our unpublished data).
DNA sequences were confirmed using the dideoxynucleotide termination sequencing method and the fluorescent label system of the ABI Prism kit (Perkin Elmer-Cetus). Sequencing reactions were run on an ABI 373 DNA Sequencer by the Vermont Cancer Center DNA Analysis Facility.
Yeast FUS1-lacZ Induction Assays
Cell culture supernatants were obtained from MATa strains containing pPGK-bbp1(1), pPGK-bbp2(4), or the control vector pPGK in the following manner. Fresh cultures grown to an OD600 of ∼1.0, in SD(ura−) medium to maintain plasmids, were diluted to OD600 = 0.1 in SD(ura−) medium and grown for an additional 4 h at 30°C in glass culture tubes. The cultures were centrifuged to obtain the cell culture supernatants. In one set of experiments, secretion of Bbp2(4) by a MATα strain (W303–1B) was tested.
MATα cells containing pPGK-bbr1, pPGK-bbr2, or the control vector pPGK (URA3) as well as the pheromone-inducible reporter construct FUS1::lacZ (pTCFL1, TRP1; Trueheart and Fink, 1989; Chen and Kurjan, 1997) were grown in SD(ura−, trp−) as described for the MATa cells, but after centrifugation the cells were resuspended in 0.5 vol of fresh selective media. MATα cells (1 ml) in fresh medium were mixed with an equal volume of MATa cell supernatant (or fresh SD(ura−) medium for the control) and incubated at 30°C for 2 h in glass culture tubes in a roller drum. The cells were harvested and permeabilized, and β-galactosidase units were calculated as described previously (Reynolds et al., 1997) using o-nitrophenyl β-d-galactopyranoside as the colorimetric substrate. The β-galactosidase data are presented as representative results from experiments repeated two to five times. The activities shown are averages from at least three measurements taken from assays of at least two independent transformants of each strain analyzed. Error bars indicate 1 SD.
Halo Assays of S. cerevisiae Cell Cycle Arrest
MATa strains containing pPGK or pPGK-bbp2(4) were grown in patches on SD(ura−) master plates 24–48 h. MATα cells (0.3 ml of cells at OD600 = 0.3) were spread on either YEPD plates for the MATα STE3 sst2 strain or SD(ura−) plates for the MATα ste3 sst2 strains containing pPGK or pPGK-bbr1 constructs. The master plates containing the MATa patches were replica plated to the appropriate MATα lawn plates and grown 24–48 h. The plates were analyzed for induction of cell-cycle arrest by secreted pheromone resulting in inhibition of growth of the surrounding lawn (halo formation).
Mating Assays
Overlapping perpendicular streaks of MATa strains (his4) and MATα strains (his3) were grown overnight on SD(ura−) medium to maintain pPGK-based plasmids and allow diploids to form. These plates were then replica plated onto SD(his−) medium, which provides a selection for growth of the nutritionally complemented diploid cells produced by mating.
RESULTS
A Schizophyllum Receptor–Pheromone Pair Is Functional in S. cerevisiae
In this study, we asked whether Schizophyllum receptors and pheromones could substitute for their S. cerevisiae counterparts to activate the pheromone-response pathway in S. cerevisiae. The gene encoding the Bβ1 receptor Bbr1, called bbr1, and the gene encoding Bβ2 pheromone Bbp2(4), called bbp2(4) (Figure 1 and Table 3), were placed under the control of a constitutive promoter derived from the yeast phosphoglycerate kinase (PGK) gene (Kang et al., 1990) to create the plasmids pPGK-bbr1 and pPGK-bbp2(4), respectively. Any possible competition between Bbr1 and the a-factor receptor, Ste3p (Hagen et al., 1986), was avoided by transforming pPGK-bbr1 into a MATα ste3 mutant. Schizophyllum pheromones, including Bbp2(4), are predicted from DNA sequences to be small peptides that are modified with a farnesyl group (Wendland et al., 1995; Vaillancourt et al., 1997; Fowler, Mitton, and Raper, unpublished), the same lipid moiety that is attached to the yeast a-factor (Anderegg et al., 1988; Marcus et al., 1991). Expression of Bbp2(4) from pPGK-bbp2(4) was attempted in a MATa strain because these cells modify, process, and secrete the lipopeptide a-factor and thus were thought to provide the best possibility of producing mature Bbp2(4).
Table 3.
Genes/gene products relevant to this study
| Gene(s) | Protein product(s) | Reference |
|---|---|---|
| S. cerevisiae | ||
| STE4 | Gβ subunit | Whiteway et al., 1989 |
| STE6 | a-factor transporter | McGrath and Varshavsky, 1989; Kuchler et al., 1989 |
| SST2 | Regulator of G protein signaling | Dietzel and Kurjan, 1987 |
| STE3 | a-factor receptor | Hagen et al., 1986 |
| MFA1, MFA2 | a-factor peptide precursors | Michaelis and Herskowitz, 1988 |
| RAM1, RAM2 | α- and β-subunits of farnesyl transferase | He et al., 1991 |
| STE14 | Carboxyl methylase | Sapperstein et al., 1994 |
| STE24 | N- and C-terminal protease of a-factor precursors | Boyartchuk et al., 1997; Tam et al., 1998; Fujimura-Kamada et al., 1997 |
| RCE1 | C-terminal protease of a-factor precursors | Boyartchuk et al., 1997 |
| STE23, AXL1 | N-terminal proteases of a-factor precursors | Adames et al., 1995; Chen et al., 1997b |
| S. commune | ||
| bbr1 | Pheromone receptor from specificity 1 of the Bβ locus | Vaillancourt et al., 1997 |
| bbr2 | Pheromone receptor from specificity 2 of the Bβ locus | This study |
| bbp1(1) | Pheromone 1 from specificity 1 of the Bβ locus | Vaillancourt et al., 1997 |
| bbp2(4) | Pheromone 4 from specificity 2 of the Bβ locus | This study |
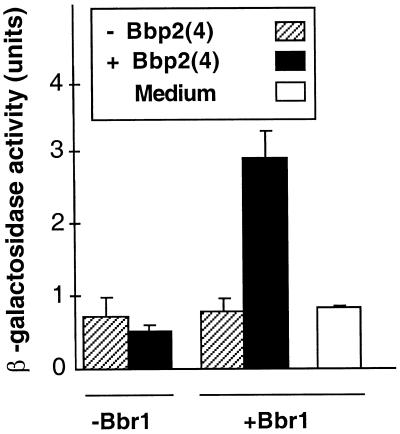
The first assay for activation of the pheromone-response pathway by the Bbp2(4)–Bbr1 pheromone–receptor pair utilized the pheromone-inducible FUS1-lacZ reporter gene (Trueheart and Fink, 1989; Chen and Kurjan, 1997). The procedure involved treating cells, containing the receptor and reporter gene constructs, with supernatants from cultures of cells containing the pheromone gene construct. In four paired combinations of MATa culture supernatants with MATα cells, cell culture supernatants of the MATa strains containing pPGK-bbp2(4) or the control plasmid pPGK were mixed with MATα ste3 strains containing pPGK-bbr1 or the control plasmid. The combination of supernatant from the MATa strain containing pPGK-bbp2(4) and MATα cells containing pPGK-bbr1 (and the FUS1-lacZ reporter) showed three- to fourfold higher β-galactosidase levels in comparison with the negative controls (Figure 2). The elevated β-galactosidase activity was ∼40% of the activity seen in concurrent a-factor/Ste3p controls, where the genes are expressed from their native chromosomal positions (our unpublished data). The increased β-galactosidase level above background in this test of Schizophyllum gene products required expression of bbp2(4) by the MATa cells and expression of bbr1 by the MATα cells. These results indicate that an active Schizophyllum pheromone Bbp2(4) is secreted by S. cerevisiae and that the Schizophyllum receptor Bbr1 is also functionally expressed. Furthermore, Bbr1 is inactive until stimulated by Bbp2(4), at which time the receptor transduces a signal that elicits a response from the FUS1-lacZ reporter gene, indicating that the receptor is capable of coupling with the S. cerevisiae pheromone-response pathway (see below).
Figure 2.
Activation of a yeast pheromone-inducible gene by a Schizophyllum pheromone–receptor pair. Cell culture supernatants were collected from MATa strains (W303–1A) containing pPGK-bbp2(4), which encodes the Schizophyllum pheromone Bbp2(4) (black), or the control vector pPGK (stripes). Response to culture supernatants or a fresh medium control (white) was tested using a MATα ste3 strain (SDK45) containing the pheromone-inducible FUS1-lacZ reporter plasmid and either pPGK-bbr1, which encodes Schizophyllum receptor Bbr1, or pPGK. β-Galactosidase activity was assayed to assess pheromone response. Error bars here and in subsequent figures represent 1 SD.
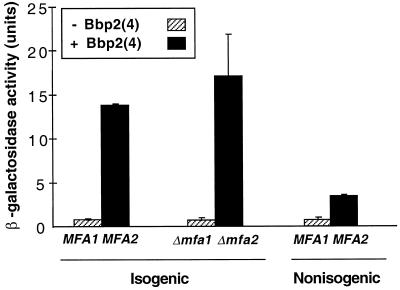
In the previously described experiment, the MATa strain used to secrete Bbp2(4) also secreted a-factor, but the a-factor could not induce signaling in the MATα ste3 strain due to the absence of the a-factor receptor, Ste3p. It was possible, however, that the simultaneous expression of the a-factor and Bbp2(4) precursors might influence Bbp2(4) maturation or secretion or that secreted a-factor could interfere with the interaction between Bbp2(4) and the Bbr1 receptor. We therefore compared Bbp2(4) activity in culture supernatants from an a-factor-deficient strain (SM2331, MATa mfa1 mfa2; Michaelis and Herskowitz, 1988) with that from an isogenic wild-type strain (Figure 3, isogenic strains; SM1058, MATa MFA1 MFA2). Activation of the FUS1-lacZ reporter gene through Bbr1 was comparable for these culture supernatants, indicating that a-factor production does not interfere with Bbp2(4) production or affect interaction of Bbp2(4) with its receptor.
Figure 3.
Production of Bbp2(4) and response to this pheromone are not influenced by the presence of a-factor, but Bbp2(4) production is affected by yeast strain differences. The MATα ste3 strain (SDK45) tested for pheromone response contained the pheromone-inducible FUS1-lacZ reporter plasmid and pPGK-bbr1. To test for any effect on Bbp2(4) signaling by the presence of a-factor, culture supernatants from isogenic MATa MFA1 MFA2 (SM1058) and MATa mfa1 mfa2 (SM2331) strains containing either pPGK-bbp2(4) (black) or pPGK (stripes) were compared. Comparison to a nonisogenic MATa MFA1 MFA2 strain (W303–1A) showed a strain-related difference in Bbp2(4) activity. β-Galactosidase assays were done as in Figure 2.
The two isogenic strains (SM2331, SM1058) tested above for any interference between a-factor and Bbp2(4) are not isogenic with the strain (W303–1A) used in the initial assay. Culture supernatants containing Bbp2(4) obtained from the MATa MFA1 MFA2 strain SM1058 induced four- to sixfold higher β-galactosidase levels than did culture supernatants from the nonisogenic MATa MFA1 MFA2 strain W303–1A (Figure 3). This result suggests that there are strain differences in production of the secreted, active Schizophyllum pheromone by S. cerevisiae.
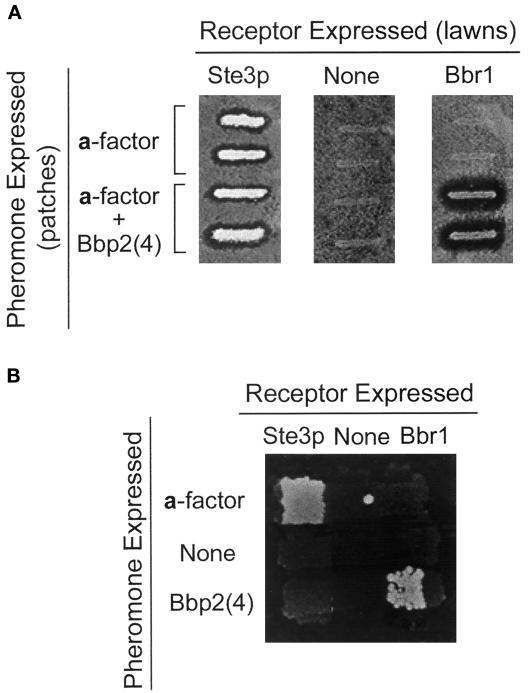
An important response to pheromone in S. cerevisiae is arrest in the G1 phase of the cell cycle (Kurjan, 1993). In the “halo” assay (Dietzel and Kurjan, 1987a), pheromone secreted from a patch of cells arrests the growth of an underlying lawn of cells of the opposite mating type, resulting in a clear zone immediately surrounding the patch of cells. Response of the MATα lawn to a-factor requires the a-factor receptor, Ste3p (Figure 4A). Expression of Bbr1 in a MATα ste3 strain resulted in a halo surrounding the MATa cells expressing Bbp2(4). Controls indicated that Bbp2(4) expression by the MATa strain and Bbr1 expression by the MATα strain were necessary for halo formation. Therefore, response to the Schizophyllum pheromone Bbp2(4) through the Schizophyllum Bbr1 receptor was sufficient to promote cell-cycle arrest.
Figure 4.
A compatible pheromone–receptor pair from Schizophyllum elicits cell-cycle arrest and mating. (A) Cell-cycle arrest in response to pheromone was tested by the halo assay. Master plates with duplicate patches of MATa cells (W303–1A) containing either the control vector pPGK (upper two patches in each panel) or pPGK-bbp2(4) (lower two patches) were replica plated on MATα lawns to test growth arrest by secreted pheromone. The lawns used to test response were STE3 sst2 (Tn44–1B) containing pPGK (left panel), ste3 sst2 (SDK47) containing pPGK (middle panel), and ste3 sst2 (SDK47) containing pPGK-bbr1. (B) Mating tests were performed between MATa and MATα cells by pairing complementing auxotrophs (his4 versus his3) and assaying for growth of diploids on SD(his−) medium. Rows: the MATa MFA1 MFA2 strain (SM1058) containing pPGK (top), the MATa mfa1 mfa2 strain (SM2331) containing pPGK (middle), and the MATa mfa1 mfa2 strain containing pPGK-bbp2(4). Columns: The MATα STE3 strain (Tn44–1B) containing pPGK (left), the MATα ste3 strain (SDK47) containing pPGK (middle), the MATα ste3 strain (SDK47) containing pPGK-bbr1 (right).
Another response to pheromone in S. cerevisiae is a morphological change in which the cells produce a mating projection. No morphological change was seen in MATα cells (SDK47) expressing Schizophyllum receptor Bbr1 when treated for ≤12 h with supernatant containing Schizophyllum pheromone Bbp2(4). In control experiments, MATα STE3 cells (TN44–1B) exposed to culture supernatants containing a-factor showed morphological changes associated with pheromone response within 2 h. Previous dose response analysis for yeast pheromone response indicated that morphological changes require ∼100-fold higher concentrations of pheromone than are required for cell-cycle arrest or transcriptional induction (Moore, 1983); the absence of morphological changes via the Schizophyllum pheromone–receptor pair may reflect these differing dosage requirements.
The various responses to pheromone in S. cerevisiae lead to the mating reaction in which cells, and then nuclei, fuse to form the MATa/MATα diploid. In mating assays (Figure 4B), production of Bbp2(4) by MATa mfa1 mfa2 cells (SM2331) allowed mating with MATα ste3 cells (SDK47) expressing Bbr1, whereas paired combinations that included control strains that did not express either Bbp2(4) or Bbr1 were defective in mating. Therefore, expression of the Bbp2(4)–Bbr1 pheromone–receptor pair in S. cerevisiae cells of opposite mating type can initiate transcriptional induction, cell-cycle arrest, and mating.
The Schizophyllum Pheromone–Receptor Pair Signals through Components of the Yeast Pheromone-Response Pathway
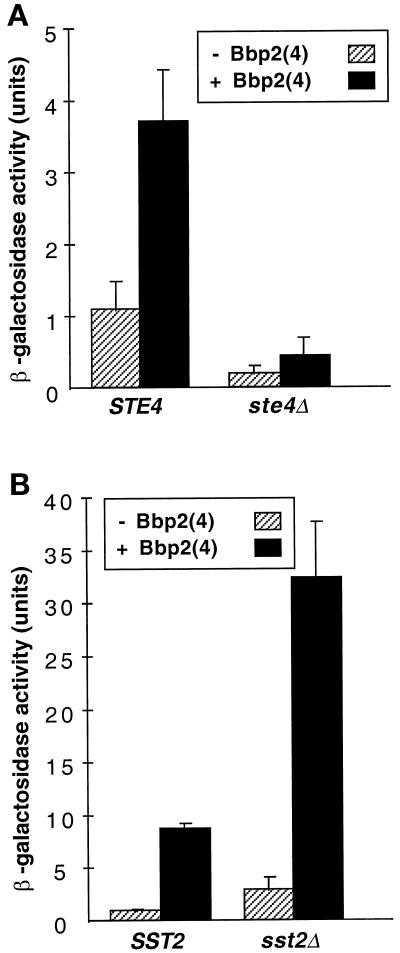
Three aspects of the pheromone-response pathway were tested to determine whether the signal generated from the interaction of the compatible Schizophyllum pheromone–receptor pair Bbp2(4)–Bbr1 follows the same path as a signal generated by a pheromone-activated yeast receptor. The yeast pheromone receptors interact with a trimeric G protein composed of Gpa1p (α), Ste4p (β), and Ste18p (γ) (Dietzel and Kurjan, 1987b; Miyajima et al., 1987; Jahng et al., 1988; Whiteway et al., 1989). The Gβγ dimer acts as a positive component to activate the downstream pathway, whereas the Gα subunit binds to Gβγ to inhibit signaling by the dimer. Hence, ste4 null mutants, which lack the β subunit, are defective in response to pheromone and mating (Whiteway et al., 1989). Culture supernatants containing Bbp2(4) did not induce the FUS1-lacZ reporter gene in a MATα ste4 null mutant expressing Bbr1, but did show about a fourfold induction in the isogenic MATα STE4 strain (Figure 5A), indicating that the Schizophyllum pheromone–receptor pair acts through the G protein of the pheromone-response pathway.
Figure 5.
Components of the pheromone-response pathway govern signaling from the Schizophyllum pheromone–receptor pair. The MATα strains tested for response contained the pheromone-inducible FUS1-lacZ reporter plasmid and pPGK-bbr1. Cell culture supernatants were from a MATa strain (W303–1A) that contained either pPGK (stripes) or pPGK-bbr2(4) (black). Reporter gene assays were done as in Figure 2. (A) Comparison of responses by MATα ste3 STE4 (SDK45) and MATα ste3 ste4 (SDK48) strains indicate that Ste4p, the β-subunit of the yeast G protein, is required for signaling via activated Bbr1. (B) Comparison of responses by MATα ste3 SST2 (SDK45) and MATα ste3 sst2 (SDK47) strains indicate that sst2, a mutation that enhances sensitivity to yeast pheromone, also increases sensitivity to Schizophyllum pheromone.
The S. cerevisiae SST2 gene product is involved in recovery from pheromone-induced cell-cycle arrest (Dietzel and Kurjan, 1987a). sst2 Mutants show a greatly increased sensitivity to pheromone and an increase in both the basal and α-factor–induced expression of pheromone-inducible genes (Chan and Otte, 1982). A similar increase in FUS1-lacZ expression was observed in response to Bbp2(4) in the MATα ste3 sst2 mutant expressing Bbr1 (Figure 5B). Cell-cycle arrest in response to secreted yeast pheromones can be detected by the halo assay in sst2 mutants, but not in a wild-type SST2 strain. Whereas Bbp2(4) secretion resulted in halo formation on a lawn of Bbr1-expressing MATα sst2 cells, no halo was observed on a MATα SST2 lawn (our unpublished data). The sst2 mutation therefore increased the sensitivity of cells expressing Schizophyllum receptor Bbr1 to the pheromone Bbp2(4).
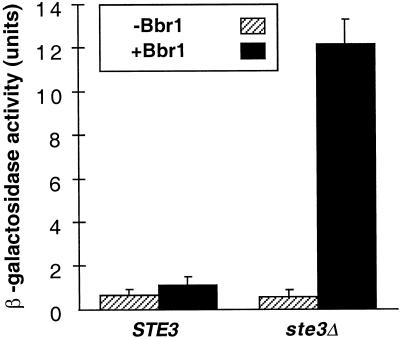
Initial assays in this system were done in a ste3 mutant to preclude activation of the pheromone-response pathway through the a-factor receptor, Ste3p, by a-factor present in the culture supernatants (Figure 2). Production of Bbp2(4) in an a-factor–deficient strain (MATa mfa1 mfa2) permitted a test for any effect of coexpression of receptors Ste3p and Bbr1 on signaling. An inhibitory effect of Ste3p upon signaling by α-factor receptor, Ste2p, was previously described for strains that simultaneously express these two receptors (Bender and Sprague, 1989; Hirsch and Cross, 1993). Induction of FUS1-lacZ by Bbp2(4) through Bbr1 was ∼15-fold over background levels in the ste3 strain as compared with an approximate twofold induction in an isogenic STE3 strain (Figure 6). These results indicated that Ste3p inhibited pheromone-responsive signaling through Bbr1. Signaling by a-factor through Ste3p, however, was not significantly affected by the expression of Bbr1 (our unpublished data).
Figure 6.
The yeast Ste3p receptor inhibits response to activated Schizophyllum Bbr1. Cell culture supernatants of the MATa strain (W303–1A) containing pPGK-bbp2(4) were used for all treatments. Responses by the MATα STE3 (W303–1B) and MATα ste3 (SDK45) strains containing either pPGK (striped) or pPGK-bbr1 (black) were compared. Reporter gene assays were done as in Figure 2.
Functional Expression of Bbp2(4) in S. cerevisiae Requires Only Some of the Components Required for a-Factor Processing, Modification, and Secretion
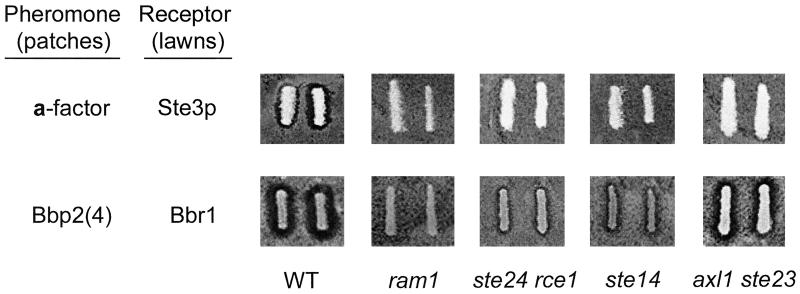
a-Factor secretion is independent of the classical secretory pathway. The a-factor precursors undergo a series of modification and processing steps, followed by secretion of mature lipopeptide by a transmembrane pump (Michaelis et al., 1992). To determine whether biosynthesis and secretion of active Bbp2(4) involve the same steps required for maturation of a-factor, we used mutant strains defective in several aspects of a-factor production. Each of these mutant strains, containing pPGK-bbp2(4), was used in halo assays to test for secreted Bbp2(4) activity.
The a-factor precursor is farnesylated on a cysteine residue that is part of the C-terminal CaaX sequence (Anderegg et al., 1988; Michaelis and Herskowitz, 1988). The CaaX motif is found at the C termini of all predicted Schizophyllum pheromones, suggesting that prenylation of these molecules is likely. The S. cerevisiae RAM1 and RAM2 genes encode the two subunits of the farnesyltransferase responsible for prenylation of a-factor (He et al., 1991). The ram1 mutant containing pPGK-bbp2(4) did not produce a halo on the lawn expressing Bbr1, whereas the wild-type RAM1 strain containing pPGK-bbp2(4) did produce a halo (Figure 7). A similar defect was seen in a nonisogenic ram2 mutant (our unpublished data). These results suggest that the Bbp2(4) precursor is farnesylated in S. cerevisiae by Ram1p/Ram2p.
Figure 7.
Active, secreted Bbp2(4) requires only some of the a-factor processing and modification machinery. Halo assays, in duplicate, were done as in Figure 4A. Top row: Secreted a-factor was monitored using halo assays on MATα sst2 STE3 (Tn44–1B) lawns. MATa cell patches tested for a-factor production were the wild-type parental strain (“WT”, SM1058) and isogenic mutants defective in farnesylation (ram1, LHK1), C-terminal processing (ste24 rce1, SM3614), carboxyl methylation (ste14, SM1188), and N-terminal processing (axl1 ste23, SM2744), all containing pPGK. Bottom row: A MATa mfa1 mfa2 mutant strain (SM2331) containing pPGK-bbp2(4) (“WT”) served as the control for Bbp2(4) production. The ram1, ste24 rce1, ste14, and axl1 ste23 mutant strains described above, but containing pPGK-bbp2(4), were tested for Bbp2(4) production. Halo assays were done on MATα sst2 ste3 (SDK47) lawns containing pPGK-bbr1. Quantifications of Bbp2(4) activity in the ste24 rce1 and ste14 mutants were done by FUS1-lacZ reporter gene assays (see text for details). The same mutant strains containing pPGK, tested on MATα sst2 ste3 (SDK47) lawns containing pPGK-bbr1, showed no halo effect (our unpublished data).
After farnesylation of the cysteine residue, the C-terminal aaX residues of the a-factor precursor are removed by the functionally redundant Ste24p and Rce1p proteases (Boyartchuk et al., 1997; Tam et al., 1998). The ste24 rce1 double mutant expressing Bbp2(4) produced a much smaller halo than the isogenic wild-type strain expressing Bbp2(4) (Figure 7). β-Galactosidase assays with the ste24 rce1 double mutant revealed that the Bbp2(4) activity was <2% of the activity of an isogenic STE24 RCE1 strain (our unpublished data). These results suggest that one or both proteases are nearly essential for production of active Bbp2(4).
After C-terminal proteolysis, the farnesylated cysteine residue of the a-factor precursor is carboxyl methylated by Ste14p (Sapperstein et al., 1994). The ste14 mutant expressing Bbp2(4) produced a much smaller halo than the isogenic wild-typeSTE14 strain expressing Bbp2(4) (Figure 7), and β-galactosidase assays showed that Bbp2(4) activity secreted by the mutant strain was 5–15% of the level obtained from the isogenic STE14 strain (our unpublished data). These results indicate that Ste14p activity plays an important, but not an essential, role in production of active Bbp2(4).
N-terminal processing of a-factor precursor involves two steps. Ste24p acts to carry out the initial N-terminal trimming in addition to its role in C-terminal processing (Fujimura-Kamada et al., 1997; Tam et al., 1998). The reduced halo size (Figure 7) and the extremely low levels of β-galactosidase produced by the ste24 rce1 mutant suggest that Ste24p may be required for Bbp2(4) activity. Processing of the mature N terminus of a-factor involves the functionally redundant Axl1p and Ste23p proteases (Adames et al., 1995; Chen et al., 1997b). The axl1 ste23 mutant expressing Bbp2(4) showed halos of similar size to those of the wild-type control (Figure 7), indicating that Axl1p and Ste23p are not required for production of active Bbp2(4).
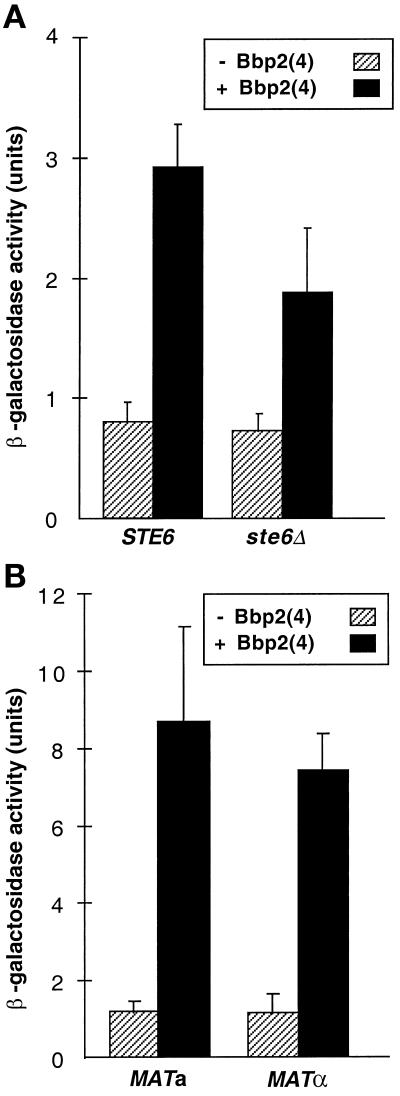
Secretion of the mature a-factor lipopeptide occurs through the transporter Ste6p, which is a family member of the ATP-binding cassette transporters (Kuchler et al., 1989; McGrath and Varshavsky, 1989). The level of active Bbp2(4) produced by ste6 mutants was about half of the level produced by the wild-type STE6 strain, as assayed by β-galactosidase activity (Figure 8A). Halo assays also indicated that the ste6 mutation did not significantly affect Bbp2(4) secretion (our unpublished data). Bbp2(4) can therefore be secreted independently of Ste6p.
Figure 8.
Bbp2(4) processing and secretion are independent of transporter Ste6p and all other a-specific gene products. The MATα ste3 strain (SDK45) containing the pheromone-inducible FUS1-lacZ reporter plasmid and pPGK-bbr1 was tested for response. Reporter gene assays were done as in Figure 2. (A) The effects of cell culture supernatants from the MATa STE6 strain (W303–1A) and the isogenic ste6 strain (RAK32) containing either pPGK (stripes) or pPGK-bbp2(4) (black) were compared. (B) The effects of cell culture supernatants from MATa (W303–1A) and MATα (W303–1B) cells containing either pPGK (stripes) or pPGK-bbp2(4) (black) were compared.
In all previous experiments, Bbp2(4) was expressed in MATa cells because it was thought that steps involved in processing, modification, and secretion of Bbp2(4) might be similar to steps used for a-factor production. However, STE6 is the only MATa-specific gene known to play a role in a-factor production (Wilson and Herskowitz, 1984). Because Ste6p was not essential for Bbp2(4) secretion by MATa cells, we tested whether MATα cells could secrete functional Bbp2(4). Levels of induction of the FUS1-lacZ reporter gene were similar using supernatants from either MATα cells containing pPGK-bbp2(4) or MATa cells containing pPGK-bbp2(4) (Figure 8B). This result is consistent with a Ste6p-independent transport mechanism and indicated that no MATa-specific gene products are required for Bbp2(4) production.
The Specificity of Two Schizophyllum Pheromone–Receptor Pairs Is Reproduced in Yeast
In Schizophyllum, interaction between compatible pheromones and receptors encoded within different B-locus specificities is required for activation of the B-regulated pathway of development. Pheromones and receptors encoded within the same specificity of the B locus are always incompatible and do not activate this pathway (Figure 1A; Vaillancourt et al., 1997; Fowler et al., 1998). We expressed a second pheromone–receptor pair in yeast to determine whether similar compatibility/incompatibility relationships prevail in the reconstituted yeast system. The Bbp1(1) pheromone was expressed in the MATa mfa1 mfa2 strain, and the Bbr2 receptor was expressed in the MATα ste3 strain. This compatible combination induced β-galactosidase activity by approximately threefold (our unpublished data) and also allowed mating (Figure 9). Equally important, pheromone–receptor pairs derived from the same Bβ specificity (e.g., Bbp1(1)/Bbr1 or Bbp2(4)/Bbr2) did not induce the FUS1-lacZ reporter or allow mating. The specificity observed in Schizophyllum was therefore maintained in the reconstituted system in S. cerevisiae.
Figure 9.
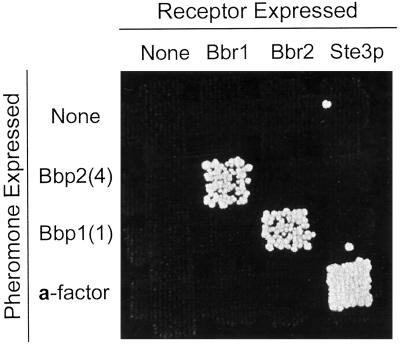
Schizophyllum pheromones and receptors maintain their specificity in yeast. Mating was tested as in Figure 4B. Rows: the MATa mfa1 mfa2 strain (SM2331) containing either pPGK (top), pPGK-bbp2(4) (second from top), or pPGK-bbp1(1) (third from top), and the MATa MFA1 MFA2 strain (SM1058) containing pPGK (bottom). Columns: the MATα ste3 sst2 strain (SDK47) containing either pPGK (left), pPGK-bbr1 (second from left), or pPGK-bbr2 (third from left), and the MATα STE3 sst2 strain (Tn44–1B) containing pPGK (right).
DISCUSSION
This study demonstrates that interactions of pheromones and receptors encoded by the B mating-type genes of S. commune can be reconstituted in S. cerevisiae to activate the pheromone-response pathway and mating in yeast. This reconstitution indicates that the seven-transmembrane receptors of Schizophyllum can localize appropriately in the plasma membrane in order to allow activation by extracellular pheromone and subsequent coupling to the yeast G protein. The extracellular production of Schizophyllum pheromones by yeast indicates that the lipopeptide precursors can be processed, modified, and secreted to produce active pheromones. A previous study in which a rat somatostatin receptor was able to activate the response pathway through the yeast G protein provided a precedent for coupling between the yeast G protein and a heterologous seven-transmembrane receptor (Price et al., 1995). There was no precedent, however, for the secretion of active heterologous lipopeptide pheromones by S. cerevisiae. Secretion of the yeast lipopeptide pheromone, a-factor, is independent of the classical secretory pathway and involves a complex set of modification and processing steps. This study indicates that Schizophyllum pheromones are modified by farnesylation and demonstrates that active heterologous lipopeptides are secreted by yeast.
Secretion of Active Schizophyllum Lipopeptide Pheromones by S. cerevisiae
Similarities of the predicted Schizophyllum lipopeptide pheromone precursors to the a-factor precursors suggested the possibility of common processes in the biosyntheses of a-factor and the active forms of Schizophyllum pheromones by S. cerevisiae. The two a-factor precursors, which differ by a single amino acid within the mature peptide sequence, undergo farnesylation, C-terminal processing, carboxyl methylation, and N-terminal processing. Mature a-factor is then secreted by a specific ATP-binding cassette transporter, Ste6p.
We investigated whether Schizophyllum pheromone Bbp2(4) production and secretion in the yeast system used the same machinery as a-factor production and secretion by analyzing Bbp2(4) expression in mutants defective for each of the steps involved in a-factor production. Interestingly, farnesylation was the only step absolutely required for processing of the Schizophyllum pheromone: mutants defective in a-factor farnesylation (ram1 and ram2) were also defective in production of active Bbp2(4), suggesting that Bbp2(4), secreted by S. cerevisiae, is a lipopeptide and is modified with a farnesyl moiety. Analysis of other mutants showed that two proteases involved in production of the mature a-factor N terminus (Axl1p and Ste23p) are not involved in active Bbp2(4) production. A mutation eliminating the a-factor carboxyl methyltransferase (Ste14p) resulted in ∼90% decrease in Bbp2(4) activity, indicating that although the decrease in activity is large, an active form of Bbp2(4) can be produced independently of this protein (Figure 7 and our unpublished data). A double-mutant strain eliminating the C-terminal protease, Rce1p, and the C- and N-terminal protease, Ste24p, showed a small halo (Figure 7) that represents <2% of the activity of a wild-type strain, as quantified by β-galactosidase assays. One or both of these proteases are nearly essential for Bbp2(4) activity. These results suggest either that the Schizophyllum pheromone is processed in a way comparable to a-factor but may be able to utilize other enzymes or that the secretion of active Schizophyllum pheromone does not require all of the processing and modification steps required by a-factor.
Bbp2(4) secretion was independent of the ATP-dependent a-factor transporter, Ste6p, as shown by ste6 mutant studies and by analysis of Bbp2(4) secretion in MATα cells, which do not express STE6 (Figure 8). S. cerevisiae contains ∼30 genes predicted to encode other proteins with homology to ATP-binding cassette transporters, and Bbp2(4) export may involve one of these alternative transporters (Taglicht and Michaelis, 1998). Another small lipopeptide product known to be transported in a Ste6p-independent manner is a-factor-related peptide (AFRP), a heptapeptide derived from the C termini of the a-factor precursors (Chen et al., 1997a). Unlike a-factor and Bbp2(4), AFRP does not have pheromone activity. AFRP and Bbp2(4) both require farnesylation for production, but not the N-terminal protease Axl1p. In contrast, methylation by Ste14p is important for robust production of Bbp2(4) activity but appears to be unimportant to AFRP maturation and export. During N-terminal processing of AFRP, the yeast cell appears to use a length-specific protease that measures from the C terminus of the a-factor precursors to the point of N-terminal cleavage, rather than a protease with sequence-specific recognition. It will be interesting to know whether yeast cells use a length-specific mechanism or a site-specific protease(s) to achieve the mature size of Bbp2(4), or whether the N terminus of the Bbp2(4) precursor is processed at all.
Activation of the Pheromone-Response Pathway by Schizophyllum Receptors
The activated yeast pheromone receptors signal through the heterotrimeric G protein, composed of Gpa1p (α), Ste4p (β), and Ste18p (γ)(Dietzel and Kurjan, 1987b; Miyajima et al., 1987; Jahng et al., 1988; Whiteway et al., 1989). After G protein activation in S. cerevisiae, the Gβγ dimer (Ste4p/Ste18p) transmits the signal to the downstream pathway. Elimination of Ste4p function therefore abolishes pheromone response and mating (Whiteway et al., 1989). Similarly, signaling by activated Bbr1 was blocked in a ste4 null mutant (Figure 5A), indicating that signaling by the Schizophyllum receptor acts through the yeast G protein. This finding is consistent with the results of two previous investigations in which it was demonstrated that heterologous receptor–G protein couplings could be used to transmit signals in yeast. Expression of the Candida albicans GPA1 homologue in S. cerevisiae allowed mating, indicating that the S. cerevisiae pheromone receptors could interact with C. albicans Gpa1p (Sadhu et al., 1992). A rat somatostatin receptor expressed in S. cerevisiae and activated by exogenous somatostatin induced the pheromone-response pathway through the S. cerevisiae G protein, demonstrating a fruitful interaction between this distantly related receptor and the yeast G protein (Price et al., 1995).
In yeast, Sst2p acts in desensitization to pheromone through Gpa1p (Dohlman and Thorner, 1997); sst2 mutants show greatly increased sensitivity to pheromone and a defect in recovery from pheromone-induced cell-cycle arrest (Chan and Otte, 1982; Dietzel and Kurjan, 1987a). The sst2 mutation similarly resulted in increased sensitivity of cells expressing Bbr1 to the secreted Bbp2(4) pheromone. This increased response was evidenced not only by assays of a pheromone-inducible reporter gene (Figure 5B) but in halo assays as well (our unpublished data). These data strengthen the conclusion that the Schizophyllum receptor signals through the S. cerevisiae G protein.
Interestingly, expression of the a-factor receptor, Ste3p, and Bbr1 in the same strain inhibited signaling through Bbr1 (Figure 6). This inhibition was unidirectional: Bbr1 expression did not block signaling through Ste3p (our unpublished data). This result suggests that attempts to couple heterologous receptors to the S. cerevisiae pathway may be more successful if done in a strain lacking endogenous pheromone receptor. The inhibition phenomenon resembles a previous observation that Ste3p inhibits α-factor–induced signaling through Ste2p (Bender and Sprague, 1989; Hirsch and Cross, 1993), but that Ste2p does not inhibit signaling through Ste3p. Recent results indicate that Ste3p inhibition of the Ste2p signal acts at the level of the Gβ subunit, Ste4p, and that a MATa-specific protein is involved in the inhibition process (Kim et al., 1999). A MATa-specific protein cannot be involved in Ste3p inhibition of Bbr1 signaling, however, because our assays tested signaling in a MATα strain.
Overall, the experiments demonstrate that activation of the pheromone- response pathway in yeast by Schizophyllum pheromone–receptor interactions must depend on secreted pheromones, since culture supernatants from pheromone-producing cells induce a response. The probable location of the receptors is within the plasma membrane of the responding cells, since the receptors couple with the plasma membrane-associated yeast G protein.
Advantages of the Heterologous Expression System for Future Studies of Schizophyllum Pheromone–Receptor Interactions
The pheromone-stimulated pathway of sexual development in homobasidiomycetes, e.g., Schizophyllum commune and Coprinus cinereus, differs in important ways from other fungal mating systems that communicate through secreted pheromones and G protein-linked receptors (reviewed by Vaillancourt and Raper, 1996). In the hemiascomycetes, e.g., S. cerevisiae and Schizosaccharomyces pombe, activation of the pheromone-signaling pathway is necessary for the conjugation of cells of opposite mating type, which is followed by nuclear fusion to establish diploidy. Pheromone signaling is required also for cell conjugation in the dimorphic hemibasidiomycetes, e.g., Ustilago maydis, Rhodosporidium toruloides, and Tremella sp. In the strictly filamentous homobasidiomycetes, hyphal fusion between two strains is independent of mating type, and self- versus non-self-recognition occurs only after cell fusion. Non-self-recognition (compatibility) is necessary for continued sexual development, and nuclear fusion occurs in specialized cells of the fruiting body well after hyphal fusion. These differences in the pheromone-stimulated pathway of sexual development in homobasidiomycetes suggested no obvious requirement for extracellular secretion of signaling molecules or for mate recognition through cell-surface receptors. The mechanism by which this recognition process acts in the sexual development of this class of fungi is not understood. Production of Schizophyllum pheromones in yeast may allow these pheromones to be concentrated using methods developed for yeast a-factor (Strazdis and MacKay, 1983; Chen et al., 1997a). Isolation and concentration of Schizophyllum pheromone secreted by yeast would facilitate studies on the biochemical nature of these pheromones and the mechanism by which they activate compatible receptors to initiate the B pathway of development in Schizophyllum. As a start toward this goal, we have concentrated Bbp2(4) and have shown that it elicits a dose-dependent halo response in yeast cells that express Bbr1 (our unpublished data).
Functional expression of Schizophyllum pheromones and receptors that can activate the yeast pheromone-response pathway provides a powerful system for addressing the molecular nature of specificity between compatible pheromones and receptors. For example, how do several pheromones of quite different sequences activate the same Schizophyllum receptor, and how can one Schizophyllum pheromone activate several different receptors? To be useful for these studies, the specificity of Schizophyllum pheromone–receptor interactions must be reproducible in this yeast system. We have shown that two pheromone–receptor pairs reproduce their natural specificity in yeast; Bbp1(1) activates Bbr2 but not Bbr1, and Bbp2(4) activates Bbr1 but not Bbr2 (Figure 9). An extrapolation from analyses to date suggest that about half of the estimated 300 or more possible pairings of Bβ pheromones and receptors extant in nature activate the B-regulated pathway of development. A comparable number of active and inactive combinations are postulated for the series of Bα pheromones and receptors. In addition, a number of mutant variants of both pheromones and receptors are known to alter specificity of interaction (Raper and Raper, 1973; Fowler et al., 1998). Comparisons of predicted amino acid sequences among mutant and natural variants show that both minor and major differences in either type of molecule can result in changes in the spectrum of partners that are used to trigger the identical pathway of sexual development. How is this possible? Exploitation of the heterologous yeast sytem for extensive and rapid screening of the effects of amino acid substitutions on Schizophyllum pheromone–receptor interactions will help us to answer this question. The information obtained from such studies should complement studies of other complex systems of ligand–seven-transmembrane receptor interactions such as those involved in mammalian olfaction, pheromone sensing, and taste (Buck and Axel, 1991; Dulac and Axel, 1995; Belluscio et al., 1999; Hoon et al., 1999; Rodriguez et al., 1999). This body of information may lead to a general understanding of the basic structure–function relationships between ligands and receptors.
ACKNOWLEDGMENTS
The authors thank Dr. Marjatta Raudaskoski for the generous gift of cDNA clones, Dr. Susan Michaelis for providing yeast strains, and Laura Hill-Eubanks for excellent technical support. The helpful suggestions of Drs. Eunice Froeliger, Mary Tierney, Joyce Heckman, and Murry Stein during the manuscript preparation are also gratefully acknowledged. This work was supported by research grant MCB9513513 from the National Science Foundation to C.A.R.
REFERENCES
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Anderegg RJ, Betz R, Carr SA, Crabb JW, Duntze W. Structure of the Saccharomyces cerevisiae mating hormone a-factor. J Biol Chem. 1988;263:18236–18240. [PubMed] [Google Scholar]
- Belluscio L, Koentges G, Axel R, Dulac C. A map of pheromone receptor activation in the mammalian brain. Cell. 1999;97:209–220. doi: 10.1016/s0092-8674(00)80731-x. [DOI] [PubMed] [Google Scholar]
- Bender A, Sprague GF., Jr Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics. 1989;121:463–476. doi: 10.1093/genetics/121.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Otte CA. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a-factor and α-factor pheromones. Mol Cell Biol. 1982;2:11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D-C, Yang B-C, Kuo T-T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- Chen P, Choi JD, Wang R, Cotter RJ, Michaelis S. A novel a-factor-related peptide of Saccharomyces cerevisiae that exits the cell by a Ste6p-independent mechanism. Mol Biol Cell. 1997a;8:1273–1291. doi: 10.1091/mbc.8.7.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Sapperstein S, Choi JD, Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol. 1997b;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Kurjan J. Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol Cell Biol. 1997;17:3429–3439. doi: 10.1128/mcb.17.6.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. “Marker swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- DeVries S, Hoge H, Bisseling T. Isolation of total and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoort RA, Verma DPS, editors. Plant Molecular Biology Manual. Dortrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–13. [Google Scholar]
- Dietzel C, Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol Cell Biol. 1987a;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel C, Kurjan J. The yeast SCG1 gene: a Gα-like protein implicated in the a- and α -factor response pathway. Cell. 1987b;50:1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Fowler TJ, Mitton MF, Raper CA. Gene mutations affecting specificity of pheromone/receptor mating interactions in Schizophyllum commune. In: Van Griensven LJLD, Visser J, editors. Proceedings of the Fourth Meeting on the Genetics and Cellular Biology of Basidiomycetes. Horst, The Netherlands: Mushroom Experimental Station; 1998. pp. 130–134. [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen DC, McCaffrey G, Sprague GF., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a-factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci USA. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Chen P, Chen S-Y, Vancura KL, Michaelis S, Powers S. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc Natl Acad Sci USA. 1991;88:11373–11377. doi: 10.1073/pnas.88.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JP, Cross FR. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated Gα protein. Genetics. 1993;135:943–953. doi: 10.1093/genetics/135.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJP, Zuker CS. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- Jahng K-Y, Ferguson J, Reed SI. Mutations in a gene encoding the a subunit of a Saccharomyces cerevisiae G protein indicate a role in mating pheromone signaling. Mol Cell Biol. 1988;8:2484–2493. doi: 10.1128/mcb.8.6.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y-S, Kane J, Kurjan J, Stadel JM, Tipper DJ. Effects of expression of mammalian Gα and hybrid mammalian-yeast Gα proteins on the yeast pheromone response signal transduction pathway. Mol Cell Biol. 1990;10:2582–2590. doi: 10.1128/mcb.10.6.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Couve A, Hirsch JP. Receptor inhibition of pheromone signaling is mediated by the Ste4p Gβ subunit. Mol Cell Biol. 1999;19:441–449. doi: 10.1128/mcb.19.1.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K, Dolhman HG, Thorner J, Caron MG, Lefkowitz RJ. Control of yeast mating signal transduction by a mammalian β2-adrenergic receptor and Gsα subunit. Science. 1990;250:121–123. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- Koltin Y, Flexer AS. Alteration of nuclear distribution in B-mutant strains of Schizophyllum commune. J Cell Sci. 1969;4:739–749. doi: 10.1242/jcs.4.3.739. [DOI] [PubMed] [Google Scholar]
- Koltin Y, Raper JR, Simchen G. Genetic structure of the incompatibility factors of Schizophyllum commune: the B factor. Proc Natl Acad Sci USA. 1967;47:55–63. doi: 10.1073/pnas.57.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Sterne RE, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J. The pheromone response pathway in Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:147–179. doi: 10.1146/annurev.ge.27.120193.001051. [DOI] [PubMed] [Google Scholar]
- Marcus S, Caldwell GA, Miller D, Xue CB, Naider F, Becker JM. Significance of C-terminal cysteine modifications to the biological activity of the Saccharomyces cerevisiae a-factor mating pheromone. Mol Cell Biol. 1991;11:3603–3612. doi: 10.1128/mcb.11.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989;340:400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- Michaelis S, Chen P, Berkower C, Sapperstein S, Kistler A. Biogenesis of yeast a-factor involves prenylation, methylation and a novel export mechanism. Antonie van Leeuwenhoek. 1992;61:115–117. doi: 10.1007/BF00580617. [DOI] [PubMed] [Google Scholar]
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987;50:1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- Moore SA. Comparison of dose-responsive curves for alpha factor-induced agglutinin, cell division arrest, and projection formation of Saccharomyces cerevisiae MATa yeast cells. J Biol Chem. 1983;258:13849–13856. [PubMed] [Google Scholar]
- Price LA, Kajkowski EM, Hadcock JR, Ozenberger BA, Pausch MH. Functional coupling of a mammalian somatostatin receptor to the yeast pheromone response pathway. Mol Cell Biol. 1995;15:6188–6195. doi: 10.1128/mcb.15.11.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper JR. Genetics of Sexuality in Higher Fungi. New York: Ronald Press; 1966. [Google Scholar]
- Raper JR, Baxter MG, Ellingboe AH. The genetic structure of the incompatibility factors of Schizophyllum commune: the A factor. Proc Natl Acad Sci USA. 1960;44:889–900. doi: 10.1073/pnas.46.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper JR, Hoffman RM. Schizophyllum commune. In: King RC, editor. Handbook of Genetics. New York: Plenum Press; 1974. pp. 597–626. [Google Scholar]
- Raper CA, Raper JR. Mutational analysis of a regulatory gene for morphogenesis in Schizophyllum. Proc Natl Acad Sci USA. 1973;69:1426–1431. doi: 10.1073/pnas.70.5.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudaskoski M. The relationship between B-mating-type genes and nuclear migration in Schizophyllum commune. Fungal Genet Biol. 1998;24:207–227. doi: 10.1006/fgbi.1998.1069. [DOI] [PubMed] [Google Scholar]
- Raudaskoski M, Färdig M, Uuskallio M. In: The structure of pheromone and receptor gene transcripts in Bα1 and Bβ1 mating-type loci of Schizophyllum commune. In: Proceedings of the Fourth Meeting on the Genetics and Cellular Biology of Basidiomycetes. Van Griensven LJLD, Visser J, editors. Horst, The Netherlands: Mushroom Experimental Station; 1998. pp. 119–124. [Google Scholar]
- Reynolds A, Lundblad V, Dorris D, Keaveney M. Yeast vector and assays for expression of cloned genes. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. pp. 13.6.1–13.6.6. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Feinstein P, Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell, 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Sadhu C, Hoekstra D, McEachern MJ, Reed SI, Hicks JB. A G-protein α subunit from asexual Candida albicans functions in the mating signal transduction pathway of Saccharomyces cerevisiae and is regulated by the a1-α2 repressor. Mol Cell Biol. 1992;12:1977–1985. doi: 10.1128/mcb.12.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol Cell Biol. 1994;14:1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR, Rine J. Protein prenylation: genes, enzymes, targets, and functions. Annu Rev Genet. 1992;30:209–237. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- Sprague GF, Jr, Thorner J. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1992. pp. 657–744. [Google Scholar]
- Stamberg J, Koltin Y. The organization of the incompatibility factors in higher fungi: the effects of structure and symmetry of breeding. Heredity. 1972;30:15–26. [Google Scholar]
- Strazdis JR, MacKay VL. Induction of yeast mating pheromone a-factor by α cells. Nature. 1983;305:543–545. doi: 10.1038/305543a0. [DOI] [PubMed] [Google Scholar]
- Taglicht D, Michaelis S. Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease. Methods Enzymol. 1998;292:130–162. doi: 10.1016/s0076-6879(98)92012-2. [DOI] [PubMed] [Google Scholar]
- Tam A, Nouvet FJ, Fujimura-Kamada K, Slunt H, Sisodia SS, Michaelis S. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco D, Lundblad V. Preparation of yeast media. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. pp. 13.1.1–13.1.7. [DOI] [PubMed] [Google Scholar]
- Trueheart J, Fink GR. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc Natl Acad Sci USA. 1989;86:9916–9920. doi: 10.1073/pnas.86.24.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt LJ, Raper CA. Pheromones and pheromone receptors as mating-type determinants in basidiomycetes. In: Setlow JK, editor. Genetic Engineering. Vol. 18. New York: Plenum Press; 1996. pp. 219–247. [DOI] [PubMed] [Google Scholar]
- Vaillancourt LJ, Raudaskoski M, Specht CA, Raper CA. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics. 1997;146:541–551. doi: 10.1093/genetics/146.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J, Vaillancourt LJ, Hegner J, Lengeler KB, Laddison KJ, Specht CA, Raper CA, Kothe E. The mating-type locus Bα1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 1995;14:5271–5278. doi: 10.1002/j.1460-2075.1995.tb00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels J, Marchant R. Enzymatic degradation of septa in wall preparations from a monokaryon and a dikaryon of Schizophyllum commune. J Gen Microbiol. 1974;83:359–368. [Google Scholar]
- Whiteway M, Hougan L, Dignard D, Thomas DY, Bell L, Saari GC, Grant FJ, O’Hara P, MacKay VL. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Wilson K, Herskowitz I. Negative regulation of STE6 gene expression by the α 2 product of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2420–2427. doi: 10.1128/mcb.4.11.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]