Abstract
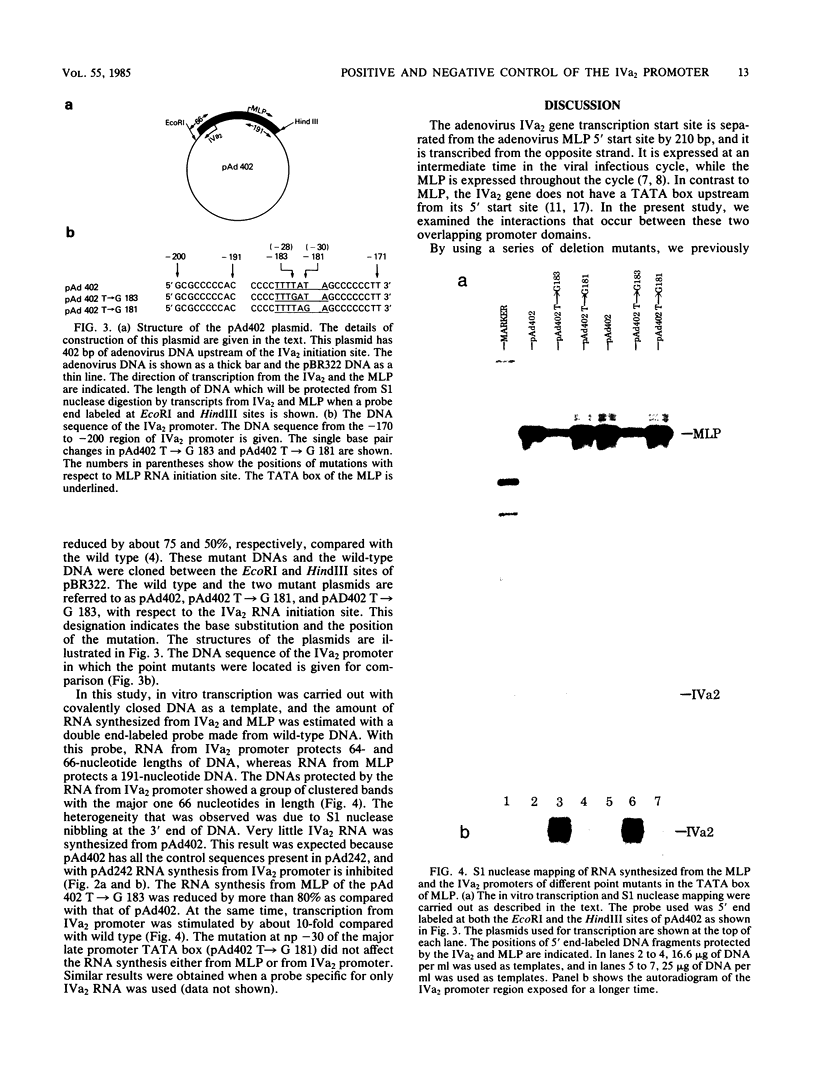
The RNA initiation sites of the adenovirus IVa2 and major late promoters (MLP) are separated by 210 base pairs and transcribed from opposite DNA strands. We had previously shown that they contained overlapping promoter sequences (V. Natarajan, M. J. Madden, and N. P. Salzman, Proc. Natl. Acad. Sci. U.S.A. 81:6290-6294, 1984). The transcription efficiencies of these two promoters were studied in vitro with templates of covalently closed circular DNAs that contained various deletion and point mutants. The distal control region of the IVa2 promoter that is located at nucleotide position (np) -152 to -242 from the RNA initiation site consists of at least two domains. The first distal domain, present between np -152 and -179, is necessary for efficient transcription of the IVa2 promoter, and it overlaps with sequences that have been shown to be necessary for efficient transcription of MLP. This region may serve as the entry site for the transcription machinery. The second distal domain consists of sequences present between np -211 and -242. These sequences are contained at the 5' end in the MLP transcript, and they inhibit transcription from the IVa2 promoter. However, these sequences are not necessary for transcription of the MLP with a covalently closed template but are needed for transcription with a linear template. The TATA box that is located at np -180 to -186 between these two domains has a critical role for efficient transcription of the MLP. A point mutation that reduces transcription from MLP by more than 80% stimulates transcription from IVa2 promoter by 10-fold. This finding is consistent with the proposal that MLP and IVa2 promoters share an entry site for transcription machinery and compete for its use.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brady J., Radonovich M., Vodkin M., Natarajan V., Thoren M., Das G., Janik J., Salzman N. P. Site-specific base substitution and deletion mutations that enhance or suppress transcription of the SV40 major late RNA. Cell. 1982 Dec;31(3 Pt 2):625–633. doi: 10.1016/0092-8674(82)90318-x. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Hernandez N., Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983 Sep 22;305(5932):336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Hickman J., Bishop J. A 45-kb DNA domain with two divergently orientated genes is the unit of organisation of the murine major urinary protein genes. EMBO J. 1984 Sep;3(9):2055–2064. doi: 10.1002/j.1460-2075.1984.tb02091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concino M. F., Lee R. F., Merryweather J. P., Weinmann R. The adenovirus major late promoter TATA box and initiation site are both necessary for transcription in vitro. Nucleic Acids Res. 1984 Oct 11;12(19):7423–7433. doi: 10.1093/nar/12.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concino M., Goldman R. A., Caruthers M. H., Weinmann R. Point mutations of the adenovirus major late promoter with different transcriptional efficiencies in vitro. J Biol Chem. 1983 Jul 10;258(13):8493–8496. [PubMed] [Google Scholar]
- Crossland L. D., Raskas H. J. Identification of adenovirus genes that require template replication for expression. J Virol. 1983 Jun;46(3):737–748. doi: 10.1128/jvi.46.3.737-748.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Earley J. M., 3rd, Roebuck K. A., Stumph W. E. Three linked chicken U1 RNA genes have limited flanking DNA sequence homologies that reveal potential regulatory signals. Nucleic Acids Res. 1984 Oct 11;12(19):7411–7421. doi: 10.1093/nar/12.19.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Baker C. C., Manley J. L., Ziff E. B., Sharp P. A. In vitro transcription of adenovirus. J Virol. 1981 Dec;40(3):703–719. doi: 10.1128/jvi.40.3.703-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen R., Sassone-Corsi P., Corden J., Gaub M. P., Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R., Manley J. L. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984 Jul 10;259(13):8513–8521. [PubMed] [Google Scholar]
- Lee D. C., Roeder R. G. Transcription of adenovirus type 2 genes in a cell-free system: apparent heterogeneity of initiation at some promoters. Mol Cell Biol. 1981 Jul;1(7):635–651. doi: 10.1128/mcb.1.7.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L. Analysis of the expression of genes encoding animal mRNA by in vitro techniques. Prog Nucleic Acid Res Mol Biol. 1983;30:195–244. doi: 10.1016/s0079-6603(08)60687-x. [DOI] [PubMed] [Google Scholar]
- Natarajan V., Madden M. J., Salzman N. P. Preferential stimulation of transcription from simian virus 40 late and adeno IVa2 promoters in a HeLa cell extract. J Biol Chem. 1983 Dec 10;258(23):14652–14655. [PubMed] [Google Scholar]
- Natarajan V., Madden M. J., Salzman N. P. Proximal and distal domains that control in vitro transcription of the adenovirus IVa2 gene. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6290–6294. doi: 10.1073/pnas.81.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Santoro C., Costanzo F., Ciliberto G. Inhibition of eukaryotic tRNA transcription by potential Z-DNA sequences. EMBO J. 1984 Jul;3(7):1553–1559. doi: 10.1002/j.1460-2075.1984.tb02010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984 Sep 6;311(5981):81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]