Abstract
Attachment of ubiquitin to cellular proteins frequently targets them to the 26S proteasome for degradation. In addition, ubiquitination of cell surface proteins stimulates their endocytosis and eventual degradation in the vacuole or lysosome. In the yeast Saccharomyces cerevisiae, ubiquitin is a long-lived protein, so it must be efficiently recycled from the proteolytic intermediates to which it becomes linked. We identified previously a yeast deubiquitinating enzyme, Doa4, that plays a central role in ubiquitin-dependent proteolysis by the proteasome. Biochemical and genetic data suggest that Doa4 action is closely linked to that of the proteasome. Here we provide evidence that Doa4 is required for recycling ubiquitin from ubiquitinated substrates targeted to the proteasome and, surprisingly, to the vacuole as well. In the doa4Δ mutant, ubiquitin is strongly depleted under certain conditions, most notably as cells approach stationary phase. Ubiquitin depletion precedes a striking loss of cell viability in stationary phase doa4Δ cells. This loss of viability and several other defects of doa4Δ cells are rescued by provision of additional ubiquitin. Ubiquitin becomes depleted in the mutant because it is degraded much more rapidly than in wild-type cells. Aberrant ubiquitin degradation can be partially suppressed by mutation of the proteasome or by inactivation of vacuolar proteolysis or endocytosis. We propose that Doa4 helps recycle ubiquitin from both proteasome-bound ubiquitinated intermediates and membrane proteins destined for destruction in the vacuole.
INTRODUCTION
Protein degradation by the ubiquitin–proteasome pathway is required for altering levels of key regulatory proteins as well as for clearing misfolded and damaged proteins from the cell. Substrates for this pathway function in processes as diverse as cell cycle progression, antigen presentation, cell fate specification, the stress response, and DNA repair. Ubiquitin is ligated to proteins through an isopeptide bond between the C-terminal Gly of ubiquitin and a Lys side chain on the substrate (Hochstrasser, 1995, 1996; Pickart, 1997; Hershko and Ciechanover, 1998). Assembly of a polyubiquitin chain(s) on the substrate is generally necessary for targeting to the 26S proteasome. In addition to its well-established role in proteasome-dependent degradation, protein ubiquitination has been shown to stimulate the internalization of cell surface proteins (Kolling and Hollenberg, 1994; Hein et al., 1995; Hicke and Riezman, 1996). Rather than being degraded by the proteasome, these ubiquitinated proteins are destroyed by vacuolar proteases. Exactly how ubiquitin serves as an internalization signal is not known.
Ubiquitin can be recovered from ubiquitin–protein conjugates by the action of members of a family of thiol proteases referred to as deubiquitinating enzymes (Dubs) (Wilkinson and Hochstrasser, 1998). Dubs are also responsible for generating ubiquitin from its C-terminally extended precursor forms. Dubs can be grouped into two distinct classes that share no obvious homology. The ubiquitin C-terminal hydrolases are a set of generally small enzymes, most of which are specialized for cleaving peptides and other small adducts from the C terminus of ubiquitin (Larsen et al., 1998). Members of the second class of Dubs are referred to as ubiquitin-specific processing proteases. These enzymes vary in size from ∼40 to 300 kDa, and the only well-conserved regions common to all of them are two short elements containing absolutely conserved Cys and His residues, respectively, which probably constitute part of the active site (Baker et al., 1992; Papa and Hochstrasser, 1993; Wilkinson and Hochstrasser, 1998). Sequence analyses in Saccharomyces cerevisiae have revealed that this class of enzymes is remarkably large, consisting of 16 members (Hochstrasser, 1996). Although deubiquitinating activity for many of the Dubs has been demonstrated in vitro, their precise functions in ubiquitin-dependent processes are not well understood.
Some Dubs may negatively regulate protein degradation by removing ubiquitin from substrates before the substrates can be targeted to or destroyed by the proteasome (Huang et al., 1995; Lam et al., 1997; Chung et al., 1998). Other Dubs positively regulate proteolysis. Yeast Ubp14 and its mammalian homologue isopeptidase T stimulate substrate degradation by the 26S proteasome both in vitro (Hadari et al., 1992) and in vivo (Amerik et al., 1997). Ubp14 and its orthologs appear to facilitate proteolysis by specifically disassembling unanchored ubiquitin chains that accumulate in vivo and can bind to and inhibit the 26S proteasome (Wilkinson et al., 1995; Amerik et al., 1997; Piotrowski et al., 1997).
Protein ubiquitination marks a substrate for eventual degradation by the 26S proteasome, but it is unclear how ubiquitin itself escapes proteolysis. That ubiquitin can be recycled is suggested by in vitro degradation assays using 125I-labeled ubiquitin–lysozyme conjugates (Hough and Rechsteiner, 1986). A Dub that associates with the 26S proteasome would be uniquely suited to prevent ubiquitin degradation by releasing the polyubiquitin chain either just before or at some point during substrate proteolysis. Previous work suggested that Doa4 removes ubiquitin from proteasome-targeted proteolytic intermediates (Papa and Hochstrasser, 1993). More recently, we have shown that a fraction of Doa4 copurifies with the 26S proteasome (Papa et al., 1999). Doa4 was identified in a genetic screen for mutants that stabilize Deg1-β-galactosidase (Deg1-βgal), a fusion protein containing the Deg1 degradation signal from the short-lived MATα2 transcriptional repressor (Hochstrasser and Varshavsky, 1990). Deletion of the DOA4 gene leads to multiple defects including the inhibition of degradation of all tested ubiquitin–proteasome pathway substrates (Papa and Hochstrasser, 1993). In addition, anti-ubiquitin immunoblot analysis of extracts from doa4Δ cells in logarithmic growth revealed a striking accumulation of low molecular mass ubiquitin-containing species that cluster most prominently above free ubiquitin and diubiquitin. These ubiquitinated species in doa4Δ cells are believed to be ubiquitinated peptide remnants of proteasome substrates that can bind to and inhibit degradation by 26S proteasomes (Papa and Hochstrasser, 1993).
Here, we show that cells lacking the Doa4 enzyme are significantly depleted for ubiquitin, particularly as they enter stationary phase, and a number of defects of the doa4Δ mutant can be rescued by restoring wild-type ubiquitin levels. Whereas ubiquitin is a long-lived protein in wild-type yeast, it is degraded relatively rapidly in doa4Δ cells, leading to ubiquitin depletion. Ubiquitin degradation in the mutant is dependent on the 26S proteasome and, surprisingly, vacuolar proteases. Additionally, we find that mutations that suppress endocytosis raise ubiquitin levels in the doa4Δ mutant. These findings suggest that Doa4 recycles ubiquitin from ubiquitinated substrates destined for either the 26S proteasome or the vacuole and thereby spares ubiquitin from degradation by the two major cellular proteolytic systems.
MATERIALS AND METHODS
Media
Yeast rich and minimal media were prepared as described, and standard genetic procedures were followed (Ausubel et al., 1989).
Yeast Strain Construction
All strains used in this study are congenic with MHY501 unless otherwise noted. The doa4-Δ1::LEU2 null allele (MHY623) was described previously (Papa and Hochstrasser, 1993). MHY1063 was made by crossing MHY622 to MHY792. MHY1232 and MHY1269, strains bearing the vps24Δ::HIS3 and vps27Δ::LEU2 alleles, respectively, were generated as described (Amerik, Nowak, Swaminathan, and Hochstrasser, unpublished data). MHY1251 and MHY1275 were derived from crosses between MHY623 and MHY1233 and MHY1270, respectively (Amerik et al., unpublished data). To generate MHY1475 (end3-1) and MHY1479 (end3-1 doa4-Δ1::LEU2), we crossed MHY623 to LHY500 (end3-1) (Raths et al., 1993). Spores corresponding to an end3-1 single mutant or an end3-1 doa4-Δ1::LEU2 double mutant were isolated after tetrad dissection of the resultant diploid. The end3-1 single-mutant spore was identified by an inability to grow at 37°C. end3-1 doa4-Δ1::LEU2 double mutants were identified by following segregation of the LEU2 marker and by heat sensitivity at 37°C (the doa4Δ single mutant still grows at near wild-type rates at this temperature). Several double-mutant segregants were analyzed to minimize differences caused by differences in strain backgrounds. To construct MHY1046, we first mated MHY623 to the congenic BBY61 strain (Bartel et al., 1990), and a doa4-Δ1::LEU2 pep4::HIS3 spore was isolated. A PCR-based strategy was used to disrupt the gene encoding PRB1 with the kanMX module (Wach et al., 1994) in the doa4-Δ1::LEU2 pep4::HIS3 to create the final strain MHY1046. MHY1061 was isolated from a cross between MHY1046 and MHY500. MHY1528 (doa4Δ doa3-1 pep4 prb1Δ) was made by crossing MHY784 to MHY1045, identifying the quadruple chromosomal null mutant by marker analysis, and then replacing the wild-type DOA3 plasmid with YCplac22doa3-1.
Pulse-Chase and Immunoblot Analysis
Pulse-chase assays to determine degradation rates were performed as described previously (Chen et al., 1993). Immunoprecipitations were performed with antibodies against α2 (Hochstrasser and Varshavsky, 1990) or Escherichia coli β-galactosidase (Cappel, West Chester, PA). To induce high levels of ubiquitin expression from the CUP1 promoter, we treated cells with 100 μM CuSO4 (J.T. Baker, Phillipsburg, NJ) for ∼3 h (Ellison and Hochstrasser, 1991).
For measuring rates of ubiquitin synthesis, cells grown in minimal medium to an OD600 of ∼1.5 were labeled for 30 min at 30°C using 300 μCi of 35S-Translabel (ICN Pharmaceuticals, Costa Mesa, CA). Radiolabeled yeast cells were lysed by mixing with an equal aliquot of 2% SDS, 90 mM HEPES, pH 7.5, and 30 mM DTT and boiling for 5 min. Ubiquitin was immunoprecipitated using a rabbit antiserum that we raised against ubiquitin by methods described previously (Haas and Bright, 1985).
Extracts for anti-ubiquitin immunoblot analysis were made from cells grown at 30°C in minimal media except where noted. Cells were resuspended in Laemmli SDS gel-loading buffer, boiled for 10 min, and centrifuged at 14,000 × g for 5 min to remove cell debris. Extracts from 0.25 OD600 units of cells were loaded onto 16% Tricine gels (Schägger and von Jagow, 1987) and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Blots were boiled in water for 30 min before incubating with a 1:500 dilution of affinity-purified anti-ubiquitin antibodies in TBST buffer (Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Tween-20) containing 1% nonfat dry milk. Anti-ubiquitin antibodies were affinity-purified against denatured ubiquitin as described previously (Haas and Bright, 1985). Antibody binding was detected using ECL reagents from Amersham (Arlington Heights, IL). For quantitative immunoblot analysis, [125I]protein A (New England Nuclear, Boston, MA) was used for detecting the anti-ubiquitin primary antibody. The linearity of [125I]protein A binding was determined using serial dilutions of yeast cell extracts. To follow 3-phosphoglycerate kinase (Pgk1), anti-ubiquitin immunoblots were stripped by heating for 30 min at 50°C in stripping solution (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.7). A mouse monoclonal antibody against Pgk1 (Molecular Probes, Eugene, OR) was used at a dilution of 1:500. A rabbit anti-mouse IgG antibody (Cappel) at a dilution of 1:1000 was used as a “sandwich antibody” before incubating with [125I]protein A. All antibody incubations were performed in 1% nonfat dry milk in TBST for 1 h. Blots were washed twice for 10 min in TBST between antibody incubations. The data were quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Measurements of Ubiquitin Half-Life
To measure the rate of disappearance of cellular ubiquitin, we added cycloheximide to a final concentration of 50 μg/ml to cells grown in minimal medium to an OD600 of ∼1.5. At the desired time points, extracts from equal aliquots of cells were removed and heated for 10 min at 100°C in lysis buffer (0.0625 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol). Proteins were electrophoresed through 16% Tricine gels and processed for anti-ubiquitin immunoblot analysis as described above. The amount of protein loaded in each lane was normalized after measurements of protein concentration with the bicinchoninic acid reagent (Pierce Chemical, Rockford, IL). Affinity-purified anti-ubiquitin antibody provided by C. Pickart (Johns Hopkins University, Baltimore, MD) was used at a dilution of 1:6000 in TBST with 1% nonfat dry milk, and membranes were incubated for 1 h at room temperature. Antibody binding was detected with [125I]protein A (1 μCi in 10 ml of TBST) after a 1 h incubation at room temperature. Ubiquitin degradation rates were derived from quantitation of these blots using a PhosphorImager and linear least-squares curve fitting of the data. The membranes were subsequently stripped and probed with an anti-Pgk1 antibody as described above. Cell viability was determined by plating appropriate culture dilutions from the 0 and 120 min time points onto minimal medium. Viability was found to decrease by 25–30% after 2 h in cycloheximide for all the strains examined (see Figure 7).
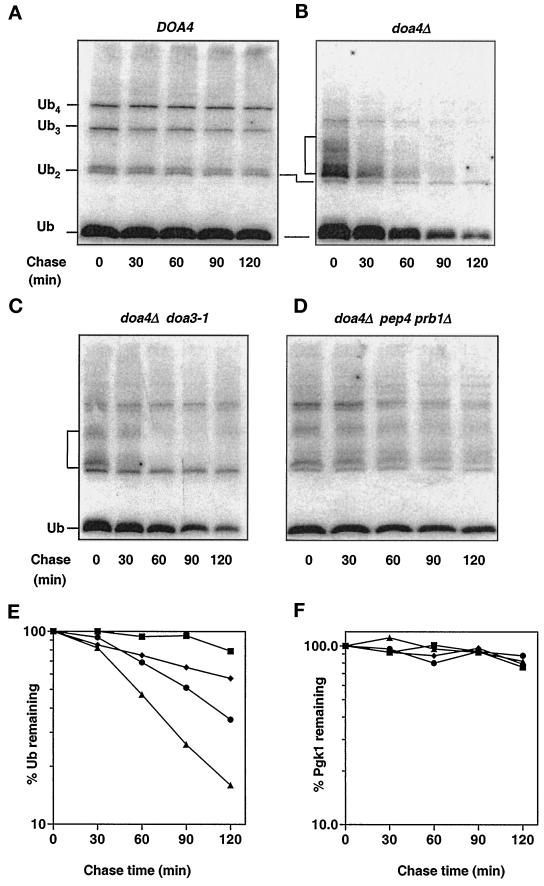
Figure 7.
Analysis of ubiquitin degradation in doa4Δ cells. (A) Rates of ubiquitin turnover determined in wild-type (MHY501) cells in late logarithmic phase (OD600 ∼ 1.5). Cycloheximide was added to a final concentration of 50 μg/ml, and at the indicated time points, extracts from equal aliquots of cells were processed for anti-ubiquitin immunoblotting. Ubiquitin and ubiquitin-containing species were detected using affinity-purified anti-ubiquitin antibodies (from C. Pickart) and [125I]protein A as the secondary antibody. (B) doa4Δ (MHY623) cells analyzed as described in A. (C) doa4Δ doa3-1 (MHY1063) cells analyzed as described in A. (D) doa4Δ pep4 prb1Δ (MHY1046) cells analyzed as described in A. (E) Quantitation of the rates of ubiquitin disappearance in A–D. Rates of ubiquitin turnover were derived from PhosphorImager (Molecular Dynamics) quantitation of the blots. (F) Quantitation of rates of Pgk1 disappearance in A–D. The symbols denote the following strains: wild type (▪), doa4Δ (▴), doa4Δ doa3-1 (●), doa4Δ pep4 prb1Δ (♦).
Analysis of doa4Δ Defects in the Presence of Augmented Ubiquitin Expression
Cells were transformed with the control vector (pES12) or plasmids carrying genes under the control of the CUP1 promoter that encoded ubiquitin (YEp96 [Ellison and Hochstrasser, 1991]), ubiquitin-K48R (YEp110 [Hochstrasser et al., 1991]), or ubiquitin-K63R (pTER103 [Arnason and Ellison, 1994]). The strains were grown to midlogarithmic phase in Trp-dropout medium that selected for plasmid retention. Tenfold serial dilutions of cells were spotted on selective plates supplemented with CdCl2 (30 μM) or canavanine sulfate (0.8 μg/ml). Plates were incubated at 30°C for 3 d. For assessing temperature sensitivity, plates were placed at 38°C for 3–5 d.
To follow viability of doa4Δ cells entering stationary phase, we diluted saturated yeast cultures into selective medium and grew the cultures overnight at 30°C. Viability measurements were begun when cells reached an OD600 of ∼0.5. Cell death was followed by uptake of the fluorescent DNA dye propidium iodide (50 μg/μl; Molecular Probes). Propidium iodide–stained cells were counted using a hemocytometer (Fisher Scientific, Pittsburgh, PA). In separate experiments, cell viability was also determined by plating cells onto solid medium and counting colonies. The results from the propidium iodide uptake and cell-plating assays were in close agreement.
Northern RNA Hybridization Analysis
Total RNA was purified from yeast cells, and Northern RNA hybridization analysis was performed as described (Ausubel et al., 1989). Ten micrograms of RNA from each sample were used for each lane. To detect UBI4 mRNA, a 1.3-kb BstXI-BclI fragment from UBI4 (Finley et al., 1987) was radiolabeled with [α-32P]dATP using a random-primed DNA-labeling kit (Boehringer Mannheim, Indianapolis, IN). The actin probe was a radiolabeled 560-bp ClaI fragment from the yeast ACT1 gene.
Lucifer Yellow Uptake Assays
Yeast overnight cultures grown in minimal medium were rediluted into fresh medium and harvested at an OD600 of ∼0.5. One OD600 unit of cells was collected by centrifugation, resuspended in 90 μl of fresh medium, and incubated with 4 mg/ml Lucifer yellow (Sigma, St. Louis, MO) at 30°C for 2 h (Raths et al., 1993). At the end of the incubation period, cells were washed three to four times with ice-cold 50 mM sodium succinate, pH 5, and 10 mM sodium azide buffer to remove excess Lucifer yellow. Cells were viewed using epifluorescence optics on a Zeiss Axioskop microscope (Thornwood, NY).
RESULTS
Reduced Ubiquitin Levels and Increased Cell Death in Stationary Phase doa4Δ Cells
Logarithmic phase doa4Δ cells accumulate small ubiquitinated species (Papa and Hochstrasser, 1993) (Figure 1A, lane 7). During logarithmic growth, the mutant cells also show an approximately threefold decrease in free ubiquitin levels relative to that in wild-type cells based on quantitative anti-ubiquitin immunoblotting (see Figure 1A). Mutant doa4Δ cells from a culture in early stationary phase/diauxic shift (OD600 ∼ 2.5) had more severely reduced free ubiquitin, having 10-fold less ubiquitin than wild-type cells at the same stage in the growth cycle (Figure 1A, lane 2 vs. lane 8). Compared with cells in logarithmic growth, early stationary phase wild-type cells had at most a slight decrease in free ubiquitin (Figure 1A, lane 1 vs. lane 2). For normalization, levels of the glycolytic enzyme Pgk1 were followed by quantitative immunoblotting and were found to vary by <50% in either wild-type or mutant cells (Figure 1A).
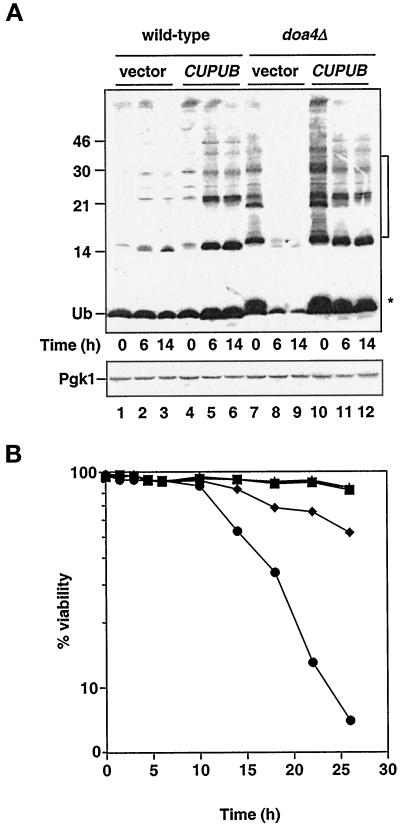
Figure 1.
Characteristics of stationary phase doa4Δ cells. (A) Analysis of ubiquitin levels in wild-type and doa4Δ cells at distinct stages in the growth cycle. Lanes 1–3, wild type (vector); lanes 4–6, wild type (YEp96); lanes 7–9, doa4Δ (vector); lanes 10–12, doa4Δ (YEp96). At the indicated time points, extracts for immunoblot analysis were made from the same yeast cultures that were used in B. Anti-ubiquitin antibody binding was detected by enhanced chemiluminescence, whereas anti-Pgk1 antibody binding was followed by [125I]protein A. The * and bracket indicate the positions of ubiquitinated species that accumulate in doa4Δ cells. The slight increase in ubiquitin (Ub) levels in wild-type cells carrying YEp96 at 6 h is most likely caused by upregulation of the CUP1 promoter in response to nutrient starvation (Tamai et al., 1994). The position of size standards (in kilodaltons) is indicated on the left. (B) Viability of wild-type and doa4Δ cells in the presence and absence of exogenously supplied ubiquitin. Rates of survival of wild-type (MHY501) and congenic doa4Δ (MHY623) cells carrying pES12 (vector) or YEp96 (CUP1UB) were measured by propidium iodide staining. Viability measurements were begun when yeast cultures were in logarithmic growth at an OD600 of ∼0.5, ∼10 h before growth levels off. The symbols denote the following strains: wild type (vector; ▪), wild type (CUP1UB; ▴), doa4Δ (vector; ●), and doa4Δ (CUP1UB; ♦).
Because doa4Δ cells are considerably depleted for ubiquitin in stationary phase, we asked whether the mutant cells could maintain viability. Survival of doa4Δ cells in logarithmic growth was indistinguishable from that of wild-type cells (Figure 1B). However, mutant cells in stationary phase experienced a striking loss of viability, with cell survival falling to ∼5% by 26 h (Figure 1B). The decrease in ubiquitin levels in doa4Δ cells occurred well before the increase in cell death. By 6 h, mutant cells exhibited a pronounced depletion of free ubiquitin, whereas cell viability remained close to that of wild-type (Figure 1).
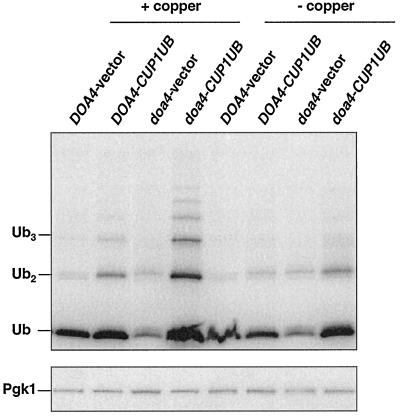
We then determined whether providing mutant cells with additional ubiquitin would enhance their survival in stationary phase. Ubiquitin was expressed from a high-copy allele under the control of the copper-inducible CUP1 promoter (YEp96 [Ellison and Hochstrasser, 1991]). Without addition of copper, the YEp96 plasmid allowed sufficient expression to restore ubiquitin to wild-type levels (see Figure 5), so copper was not added to the medium in these experiments. Under these conditions, doa4Δ cell viability was restored to ∼50% of that noted with wild-type cells (Figure 1B), suggesting that ubiquitin depletion could account for most but not all of the decreased survival of stationary phase mutant cells. Provision of ubiquitin to stationary phase doa4Δ cells restored free ubiquitin levels; however, the small ubiquitinated species that are characteristic of exponentially growing doa4Δ cells accumulated only to low levels (Figure 1A, lanes 11 and 12), suggesting that reduction of these species in stationary phase mutant cells is not simply a consequence of reduced ubiquitin levels. One possible explanation is that these ubiquitinated species are rapidly turned over in stationary phase doa4Δ cells, thus preventing accumulation to high levels.
Figure 5.
Levels of ubiquitin in doa4Δ cells carrying the ubiquitin-encoding YEp96 multicopy plasmid. Anti-ubiquitin immunoblot analysis of logarithmic phase extracts from wild-type (MHY501) and doa4Δ (MHY623) cells cotransformed with plasmid-borne Deg1-Ura3 and either pES12 (vector) or YEp96 (CUP1UB). The same transformants were used for the pulse-chase analysis in Figure 4, B–D. [125I]protein A was used to detect ubiquitin and Pgk1 levels.
To establish that decreased ubiquitin levels in doa4Δ cells were caused by loss of the deubiquitinating activity of Doa4, we examined ubiquitin levels in mutant cells expressing hemagglutinin (HA) epitope-tagged doa4C571S, a protein that has no deubiquitinating activity in vitro (Papa and Hochstrasser, 1993). HA-doa4C571S levels were similar to those of wild-type HA-Doa4 by anti-HA immunoblot analysis. Expression of the active site mutant failed to restore ubiquitin in stationary phase doa4Δ cells to wild-type levels (Figure 2, lane 5 vs. lane 6). From these data, we conclude that doa4Δ cells are unable to maintain normal ubiquitin levels and that the deubiquitinating activity of Doa4 is necessary for intracellular ubiquitin homeostasis.
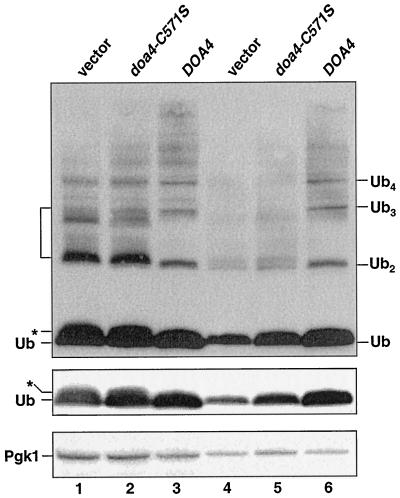
Figure 2.
The deubiquitinating activity of Doa4 is required to maintain ubiquitin levels in stationary phase. Anti-ubiquitin immunoblots (ECL detection) of doa4Δ (MHY623) cells carrying YCplac33-based plasmids expressing either the HA-tagged active site mutant, doa4C571S, or wild-type HA-Doa4. Lanes 1–3 and 4–6 show extracts from logarithmic phase cells (OD600 ∼ 1.0) and early stationary phase cells (OD600 ∼ 2.5), respectively. Positions of putative ubiquitin-peptide species in doa4Δ cells are indicated by the * and bracket; free ubiquitin and unanchored ubiquitin chain positions are also marked. The blot (middle) showing free ubiquitin alone is a shorter exposure of the top immunoblot. As a loading control, Pgk1 levels (bottom blot) were followed, with [125I]protein A as the secondary antibody. Very weak suppression of ubiquitin depletion is detected in cells expressing doa4C571S; the mechanistic basis of this suppression is unknown.
Effect of Ubiquitin Supplementation on the doa4Δ Stress and Proteolytic Defects
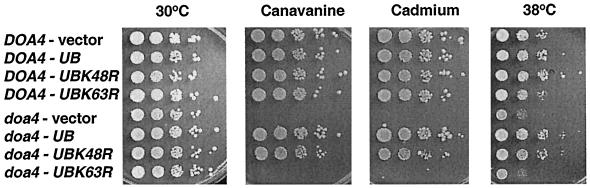
Mutations in genes encoding proteins that function in ubiquitin-dependent proteolysis often cause cells to become sensitive to various stress conditions (Jungmann et al., 1993; Chen and Hochstrasser, 1995; Amerik et al., 1997). Deletion of the DOA4 gene leads to multiple phenotypic abnormalities such as sensitivity to heat, the amino acid analogue canavanine, or the heavy metal cadmium (Papa and Hochstrasser, 1993). Provision of additional ubiquitin to doa4Δ cells (YEp96, no copper addition) completely rescued the stress sensitivities of these cells (Figure 3), suggesting that limited ubiquitin pools account for these particular defects of the mutant.
Figure 3.
Suppression of the heat, canavanine, and cadmium sensitivities of doa4Δ cells by ubiquitin expression. Tenfold serial dilutions of wild-type (MHY501) or doa4Δ (MHY623) cells transformed with pES12 (vector), YEp96 (CUP1-UB), YEp90 (CUP1-UBK48R), and pTER103 (CUP1-UBK63R) were spotted onto selective plates containing 0.8 μg/ml canavanine sulfate or 30 μM CdCl2. No copper was added. Plates were incubated at either 30 or 38°C for 3–5 d.
Of the seven Lys residues in ubiquitin, Lys-48, Lys-63, and Lys-29 can function in yeast as ubiquitin addition sites during the formation of polyubiquitin chains (Arnason and Ellison, 1994; Spence et al., 1995). Mutation of Lys-48 to Arg is lethal; the Lys-29 and Lys-63 mutations are not. There are no known phenotypic defects associated with the ubiquitin-K29R mutation, but K63-linked ubiquitin polymers have been implicated in tolerance to stress and DNA damage (Arnason and Ellison, 1994; Spence et al., 1995). In agreement with previous evidence, the ubiquitin-K63R mutant failed to suppress the stress sensitivity of doa4Δ cells (Figure 3). In contrast, the ubiquitin-K48R derivative supported nearly wild-type growth of mutant cells exposed to high temperature, cadmium, or canavanine.
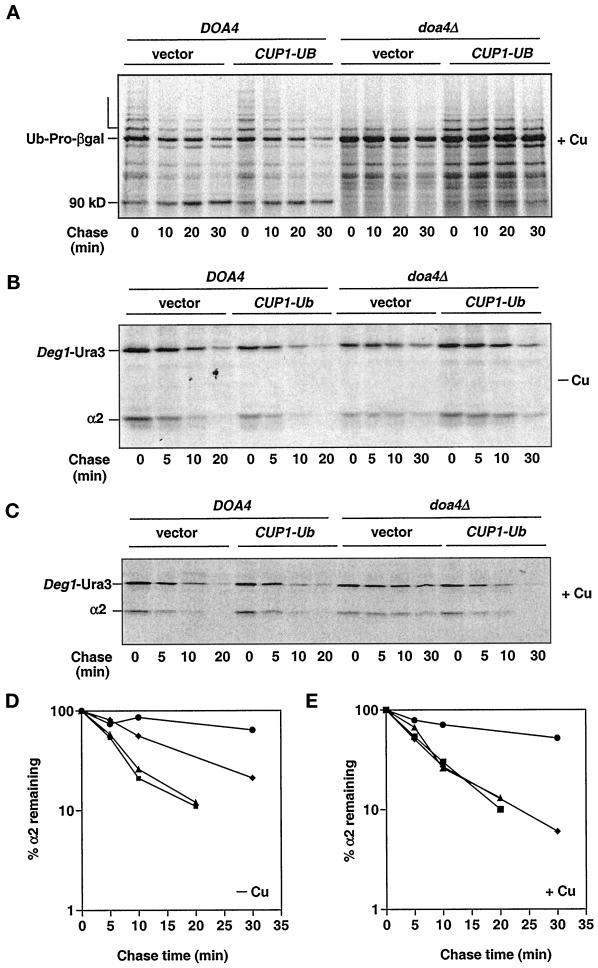
The observation that many of the cellular defects of the doa4Δ mutant can be at least partially suppressed by restoration of normal ubiquitin levels led us to examine in more detail whether a reduced ubiquitin pool may contribute to the proteolytic defects in doa4Δ cells. In wild-type cells, polyubiquitinated forms of the Ub-Pro-βgal test substrate accumulated transiently and subsequently disappeared (Bachmair et al., 1986) (Figure 4A, open bracket). As shown previously, these species were greatly diminished in doa4Δ cells, and Ub-Pro-βgal was a long-lived protein (Papa and Hochstrasser, 1993). Providing additional ubiquitin well in excess of wild-type levels (YEp96 + copper) enhanced the polyubiquitinated forms of Ub-Pro-βgal, suggesting that ubiquitin levels were limiting for modification of Ub-Pro-βgal in doa4Δ cells (Figure 4A). Nevertheless, Ub-Pro-βgal continued to be long-lived both in the presence and absence of added copper. Similarly, only very weak suppression of the degradation defect of Leu-βgal, a substrate for the N-end rule pathway (Bachmair et al., 1986), was observed in doa4Δ cells under the same conditions (after a 45-min chase, the amount of Leu-βgal remaining was reduced by <30%).
Figure 4.
Pulse-chase analysis of Ub-Pro-βgal (A), Deg1-Ura3 (B and C), and α2 (B–E) in doa4Δ cells supplemented with ubiquitin. Turnover rates of Ub-Pro-βgal and Deg1-Ura3 were followed in wild-type (MHY501) and doa4Δ (MHY623) cells carrying plasmids expressing Ub-Pro-βgal from the GAL1 promoter and Deg1-Ura3 from the α2 promoter. Endogenous α2 levels were followed in wild-type (MHY501) and doa4Δ (MHY623) cells transformed with a plasmid carrying Deg1-Ura3. Where indicated, 100 μM CuSO4 was added to cultures ∼3 h before harvesting. For D and E, the symbols denote the following strains: wild type (vector; ▪), wild type (CUP1UB; ▴), doa4Δ (vector; ●), and doa4Δ (CUP1UB; ♦).
The yeast α2 repressor is destroyed in wild-type cells with a half-life of ∼5 min, and deleting DOA4 extends the half-life of α2 at least fourfold (Papa and Hochstrasser, 1993). Restoration of wild-type ubiquitin levels (YEp96, no copper) partially rescued the α2 degradation defect in doa4Δ cells (Figure 4, B and D). Induction of high levels of ubiquitin expression with 100 μM copper resulted in complete rescue of the α2 degradation defect in the mutant (Figure 4, C and E). Under the same conditions, the kinetics of α2 turnover in wild-type cells was unchanged relative to that of the vector control (Hochstrasser et al., 1991). Similarly, degradation of Deg1-Ura3 (Figure 4, B and C) and Deg1-βgal, substrates containing the Deg1 degradation signal of α2, was restored upon ubiquitin overproduction (YEp96 + copper) in the mutant.
These experiments establish that the degradation of substrates such as Ub-Pro-βgal, Leu-βgal, and to some extent α2 is inhibited in doa4Δ cells even with normal amounts of free ubiquitin (Figure 5). Thus, the degradation defect of proteasome-dependent substrates in doa4Δ cells cannot be explained by ubiquitin depletion alone, suggesting an additional block to proteolysis in doa4Δ cells.
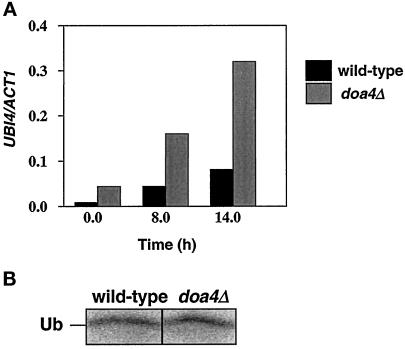
Ubiquitin Synthesis Is Not Reduced in doa4Δ Cells
Because the yeast polyubiquitin gene UBI4 is specifically induced in stationary phase cells (Finley et al., 1987), it was possible that ubiquitin depletion in stationary phase doa4Δ cells resulted from an inability to accumulate UBI4 transcripts. However, as demonstrated in Figure 6A, UBI4 was expressed even earlier in doa4Δ than in wild-type cells, being easily detectable in cells that were still in exponential growth (t = 0 h; OD600 ∼ 0.5). By 8 h (OD600 ∼ 2.5), when ubiquitin protein levels had already declined in doa4Δ cells, UBI4 transcripts accumulated to a level fourfold higher than that observed in wild-type cells and remained elevated as the culture approached stationary phase.
Figure 6.
Analysis of ubiquitin synthesis rates in doa4Δ cells. (A) Northern analysis of UBI4. Total RNA was prepared from wild-type (MHY501) and doa4Δ (MHY623) cells grown in minimal media in early logarithmic phase (t = 0 h) and stationary phase (t = 8 and 14 h). UBI4 transcripts were detected using a 1.3-kb BstXI-BclI fragment from UBI4. The blot was stripped and reprobed with a 560-bp ClaI fragment from the ACT1 gene. (B) Pulse labeling of ubiquitin. Radiolabeled ubiquitin was immunoprecipitated from wild-type (MHY501) and doa4Δ (MHY623) cells in late logarithmic phase (OD600 ∼ 1.5) with anti-ubiquitin antiserum, which does not efficiently immunoprecipitate ubiquitin–protein conjugates. Cells were pulse-labeled with 35S-TransLabel for 30 min.
To ascertain whether reduced ubiquitin levels in the doa4Δ mutant were caused by a decline in synthesis from the remaining ubiquitin genes UBI1–3, we estimated overall rates of ubiquitin protein synthesis in wild-type and mutant cells. Immunoprecipitation of pulse-labeled ubiquitin suggested that synthesis of ubiquitin was similar in wild-type and doa4Δ cells (Figure 6B). Hence, the reduced ubiquitin levels in doa4Δ cells cannot be explained by an inhibition of ubiquitin protein synthesis.
Aberrant Ubiquitin Degradation in doa4Δ Cells
We then investigated the possibility that ubiquitin was being degraded at abnormally high rates in the doa4Δ mutant during progression into stationary phase. To follow the entire cellular pool of conjugated and unconjugated ubiquitin, we added cycloheximide to yeast cultures to block protein synthesis and assayed extracts made at various times thereafter by quantitative anti-ubiquitin immunoblotting. Ubiquitin was very long-lived in wild-type cells, showing little or no degradation during a 2 h chase (Figure 7, A and E). By contrast, ubiquitin disappeared in doa4Δ cells with a half-life of ∼45–60 min (Figure 7, B and E).
Because of the evidence linking Doa4 and the 26S proteasome (Papa et al., 1999), the proteasome was an obvious candidate for a protease that degrades ubiquitin in doa4Δ cells. Failure to release ubiquitin efficiently from a ubiquitinated substrate targeted to the proteasome might cause the entire conjugate to get degraded. Indeed, introduction into doa4Δ cells of the doa3-1 mutation, which affects a catalytic subunit of the proteasome (Chen and Hochstrasser, 1995), extended the half-life of ubiquitin by ∼80% (Figure 7, C and E). The doa3-1 allele results in only a partial loss of function, so complete inactivation of the protease might be expected to have a further stabilizing effect.
To test the specificity of 26S proteasome involvement in ubiquitin degradation, we inactivated the vacuolar proteolytic system in doa4Δ cells by disruption of PEP4 and PRB1. These genes encode vacuolar proteases that are required for maturation and activation of most or all vacuolar hydrolases (Zubenko et al., 1982). Unexpectedly, ubiquitin degradation was significantly suppressed in doa4Δ pep4 prb1Δ triple-mutant cells. The half-life of ubiquitin was ∼2.5 h in these cells (Figure 7, D and E). The control Pgk1 protein was similarly long-lived in wild-type, doa4Δ, doa4Δ doa3-1, and doa4Δ pep4 prb1Δ strains (Figure 7F). Inactivation of proteolysis by the 26S proteasome or the vacuole also reduced levels of the ubiquitinated species characteristic of doa4Δ cells, suggesting that both proteolytic pathways, either directly or indirectly, contribute to the formation of these species. This observation suggests a correlation between generation of the low molecular mass ubiquitinated species and ubiquitin degradation in the mutant.
The doa3-1 and pep4 prb1Δ mutations led to an approximate two- and sixfold increase in ubiquitin levels, respectively, in stationary phase doa4Δ cells. The partial restoration of ubiquitin levels may result from an incomplete stabilization of ubiquitin by mutation of only one of these proteolytic pathways. Indeed, a quadruple mutant, doa4Δ doa3-1 pep4 prb1Δ, which is defective for both proteasomal and vacuolar degradation had wild-type levels of ubiquitin in stationary phase, consistent with distinct contributions of both proteolytic pathways to ubiquitin turnover.
Genetic Interactions between doa4Δ and Components of the Endocytic Pathway
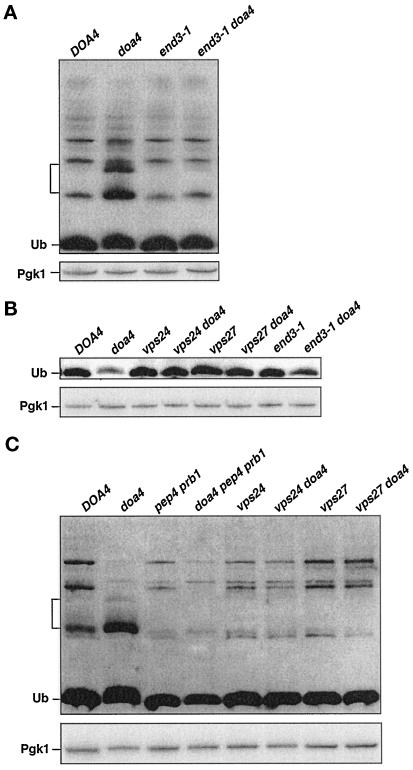
Ubiquitination of cell surface proteins is known to promote their endocytosis and eventual degradation in the vacuole or lysosome (Kolling and Hollenberg, 1994; Hein et al., 1995; Hicke and Riezman, 1996; Govers et al., 1999). Due to the low levels of ubiquitin in doa4Δ cells, internalization of a number of endocytic pathway substrates is inhibited (Galan and Haguenauer-Tsapis, 1997; Terrell et al., 1998), although some membrane proteins are still internalized normally (Loayza and Michaelis, 1998). Because ubiquitin is a metabolically stable protein in wild-type yeast (Figure 7), ubiquitin must also be released before vacuolar degradation of the endocytosed proteins. Vacuolar proteolysis of ubiquitinated membrane proteins that failed to be deubiquitinated in the doa4Δ mutant could explain the participation of vacuolar proteases in ubiquitin degradation. To test this idea, we assessed the effects of blocking endocytosis in the doa4Δ mutant by introducing end3-1, a temperature-sensitive allele of END3 that specifically inhibits the internalization step of endocytosis (Raths et al., 1993). Analysis of the endocytic block using a fluorescent fluid phase marker, Lucifer yellow, indicated that at 30°C, the end3-1 allele in our strain background was primarily but not completely defective for endocytosis, with ∼10% of the cells still showing Lucifer yellow staining in the vacuole compared with nearly 100% of wild-type cells. In logarithmic phase end3-1 doa4Δ cells, the ubiquitinated species characteristic of doa4Δ cells were substantially suppressed (Figure 8A), and in saturated cultures, ubiquitin levels were partially restored (Figure 8B), reflecting a reduction in the rate of ubiquitin degradation relative to that in doa4Δ cells based on cycloheximide-chase experiments. Similar results were found with a second end3-1 doa4Δ mutant derived from the same cross.
Figure 8.
Suppression of doa4Δ defects by mutations in the endocytic pathway. (A) end3-1 suppresses accumulation of low molecular mass ubiquitinated species (bracket) in doa4Δ cells. Anti-ubiquitin immunoblotting (ECL detection) of extracts from cells in logarithmic growth is shown. Wild-type (MHY501), doa4Δ (MHY623), end3-1 (MHY1475), and end3-1 doa4Δ (MHY1479) cells were grown at 23°C. Cultures were shifted to 30°C for ∼2–3 h before extracts for immunoblot analysis were made. (B) Suppression of ubiquitin depletion in stationary phase doa4Δ cells by mutations in VPS24, VPS27, or END3 as assayed by anti-ubiquitin immunoblot analysis (ECL detection) is shown. Extracts were made from cells at an OD600 of ∼2.5. (C) Inhibition of the vacuolar protein–sorting pathway or of vacuolar proteases suppresses the accumulation of the low molecular mass ubiquitinated species (bracket) in doa4Δ cells. Anti-ubiquitin immunoblot analysis (ECL detection) of logarithmic phase cells from wild-type (MHY501), doa4Δ (MHY623), pep4 prb1Δ (MHY1061), doa4Δ pep4 prb1Δ (MHY1046), vps24Δ (MHY1232), vps24Δ doa4Δ (MHY1251), vps27Δ (MHY1269), and vps27Δ doa4Δ (MHY1275) cells is shown. Bottom, blots in A–C were stripped and reprobed with an anti-Pgk1 antibody, using [125I]protein A for antibody detection.
We also analyzed the effects of interfering with a later step in the endocytic pathway. A recent suppressor screen for mutations that restored Deg1-βgal degradation in doa4 mutant cells identified mutations in several vacuolar protein–sorting (VPS) genes, including VPS24 and VPS27 (Amerik, Nowak, Swaminathan, and Hochstrasser, unpublished data). Vps24 and Vps27 regulate membrane traffic from the late endosome to the vacuole (Piper et al., 1995; Babst et al., 1998). In accordance with our findings with the end3-1 doa4Δ double mutant, deletion of either VPS24 or VPS27 suppressed the accumulation of the low molecular mass ubiquitinated species in logarithmic phase doa4Δ cells (Figure 8C) as well as the depletion of ubiquitin levels (Figure 8B) and abnormal ubiquitin degradation in stationary phase cells. Several related mechanisms for the targeting of cytoplasmic proteins to the vacuole are also known. The autophagy pathway takes up a random portion of cytosol for delivery to the vacuole, whereas the cytoplasm-to-vacuole (Cvt) pathway is more substrate selective (Bryant and Stevens, 1998). The endocytic, Cvt, and autophagy pathways all appear to converge at the late endosome (Scott et al., 1997), so the latter two pathways may also contribute to aberrant ubiquitin metabolism in the doa4Δ mutant. However, we found that mutations in the autophagy and Cvt pathways did not suppress formation of the low molecular mass ubiquitinated species in doa4Δ cells (our unpublished data).
In summary, mutations that impeded delivery of ubiquitinated proteins from the cell surface to the vacuole suppressed aberrant ubiquitin proteolysis in doa4Δ cells. These findings suggest that in wild-type cells, but not in the doa4Δ mutant, ubiquitinated endocytosed substrates are normally deubiquitinated before their destruction by vacuolar proteases. In agreement with these findings, levels of high molecular mass ubiquitinated proteins were elevated in purified vacuolar fractions from doa4Δ pep4 prb1Δ cells relative to the doa4Δ or pep4 prb1Δ mutants; however, we were unable to detect free ubiquitin or the low molecular mass ubiquitin conjugates in the same preparations (our unpublished data).
DISCUSSION
In this study, we show that the yeast deubiquitinating enzyme Doa4 is required to regulate ubiquitin levels in vivo. Cells deleted for DOA4 are unable to maintain normal amounts of ubiquitin, and this defect is responsible for the impairment of a subset of ubiquitin-dependent processes in the mutant. Ubiquitin depletion results from aberrant ubiquitin degradation, which depends on a functional proteasome, an observation consistent with the previously characterized role for Doa4 in proteasome-dependent proteolysis. However, the present work has also uncovered an unexpected contribution by vacuolar proteases to aberrant ubiquitin degradation in doa4Δ cells. As discussed below, these findings have implications for cellular ubiquitin homeostasis and point to a central role for Doa4 in recycling ubiquitin from proteins destined for degradation by the 26S proteasome or by the vacuole.
Ubiquitin Levels and the Proteolytic Defect in doa4Δ Cells
Mutant doa4Δ cells are compromised for degradation of all tested substrates of the 26S proteasome. Substrates such as Ub-Pro-βgal show reduced ubiquitination in the doa4Δ mutant, and provision of additional ubiquitin enhances Ub-Pro-βgal ubiquitination. Mutant doa4Δ cells have reduced levels of ubiquitin, but even with normal or strongly elevated ubiquitin levels, degradation of substrates such as Ub-Pro-βgal or the N-end rule Leu-βgal substrate is not restored, indicating that the doa4Δ mutant is also defective for a postubiquitination step in the ubiquitin–proteasome pathway, presumably proteolysis by the 26S proteasome (Papa et al., 1999).
The reduced ubiquitin levels in doa4Δ cells do limit degradation of other tested proteasomal substrates. Raising ubiquitin to wild-type levels in the doa4Δ mutant partially restores the degradation of Deg1-Ura3, Deg1-βgal, and α2, suggesting that the mutant is defective for ubiquitination of these substrates. Strong ubiquitin overproduction in the mutant almost completely suppresses the defective degradation of these proteins. Levels of ubiquitinated α2 are known to increase significantly in wild-type cells under these conditions (Hochstrasser et al., 1991); such enhanced ubiquitination might make α2 a more effective substrate for a compromised 26S proteasome in doa4Δ cells, thereby restoring wild-type degradation kinetics. Together, the data with the different substrates indicate that the doa4Δ degradation defect is due to reduced ubiquitination of substrates because of ubiquitin depletion and/or to inhibition of a postubiquitination event. The extent to which the impairment of one or the other of these two processes affects proteolysis in the mutant cells is substrate dependent.
Provision of additional ubiquitin also rescues the cellular growth defects of doa4Δ cells to varying degrees. Whereas providing these cells with ubiquitin only partially suppresses the reduced viability in stationary phase, the other stress abnormalities are almost completely suppressed. Although the mechanistic basis for the stress sensitivities exhibited by mutants in the ubiquitin–proteasome pathway is not well understood, a generally accepted notion is that exposure of cells to agents such as heat, cadmium, or canavanine leads to the accumulation of damaged and aberrantly folded proteins that must be degraded. Tolerance of canavanine or cadmium may also depend on downregulation of certain cell surface proteins by ubiquitin-dependent internalization. For example, canavanine and cadmium treatment may lead to ubiquitination and endocytosis of the Arg permease and transporters for divalent cations, respectively. Ubiquitin-dependent internalization is strongly inhibited in the doa4Δ mutant, and for a number of proteins, the endocytic defect is a consequence of reduced ubiquitin levels (Galan and Haguenauer-Tsapis, 1997; Medintz et al., 1998; Terrell et al., 1998). Restoring ubiquitin levels may reduce canavanine and cadmium sensitivity by suppressing the endocytic defect in doa4Δ cells.
Ubiquitin Homeostasis in doa4Δ Cells
Under conditions where ubiquitin levels have already declined, doa4Δ mutant cells accumulate UBI4 transcripts and display no reduction in the synthesis of ubiquitin protein relative to that in wild-type cells. Extraction of doa4Δ cells under conditions that efficiently extract large ubiquitin–protein conjugates revealed a strong reduction in these species as well. These experiments indicate that in stationary phase there is neither a strong reduction in ubiquitin synthesis rates nor a redistribution of ubiquitin into high molecular mass conjugates in doa4Δ cells relative to wild-type cells. On the basis of these results, we conclude that proteolysis of ubiquitin is the major reason for ubiquitin depletion in stationary phase doa4Δ strains. For the logarithmically growing mutant, the rate of ubiquitin degradation has been more difficult to assess. Degradation may be somewhat slower than in stationary phase, although the rate measurements are potentially complicated by deconjugation of the low molecular mass ubiquitinated species, which are present at higher concentrations in these cells than in late logarithmic and early stationary phase cells. A greater depletion of ubiquitin in stationary phase doa4Δ cells relative to logarithmic phase cells may result from a normal reduction in ubiquitin expression because of the reduced expression of the UBI1–3 ubiquitin genes (Özkaynak et al., 1987).
Analysis of ubiquitin and ubiquitin-conjugate profiles in a variety of yeast proteasome pathway mutants suggests the existence of a homeostatic mechanism that maintains free ubiquitin levels within a certain range. An increase in ubiquitin-conjugate species without a corresponding decrease in free ubiquitin is seen in many of these mutants. For example, ubp14Δ cells accumulate unanchored polyubiquitin chains, and doa3-1 mutants amass ubiquitin–protein conjugates (Chen and Hochstrasser, 1995; Amerik et al., 1997). These mutants are presumably able to maintain wild-type levels of free ubiquitin by increasing rates of ubiquitin synthesis. Higher ubiquitin levels may enhance substrate ubiquitination, thereby facilitating degradation by a crippled 26S proteasome. The exact mechanism(s) leading to ubiquitin induction is unknown. Inhibition of proteasome function has been reported to increase expression of cellular chaperones and induce thermotolerance in yeast (Bush et al., 1997; Mathew et al., 1998). Activation of a general stress response pathway in cells inhibited for proteasome-mediated proteolysis may increase ubiquitin expression. In doa4Δ cells, the feedback mechanism that augments ubiquitin synthesis may be defective, and/or the rate of ubiquitin degradation may exceed the capacity of this compensatory mechanism.
Mechanism of Ubiquitin Degradation
As noted above, previous work indicated that Doa4 action is closely linked to the 26S proteasome. Failure to remove ubiquitin from proteasome-bound intermediates may result in the ubiquitin portion of these proteolytic intermediates being unfolded and translocated into the catalytic chamber of the proteasome along with the substrate moiety. Consistent with this hypothesis, we find that ubiquitin is relatively short-lived in doa4Δ cells and that mutation of a catalytic subunit of the proteasome in the doa4Δ background not only reduces levels of the putative ubiquitin–peptide species, as seen previously (Papa et al., 1999), but also partially stabilizes ubiquitin.
Our studies have also revealed an unexpected role for vacuolar proteolysis in the formation of the doa4Δ cell–specific ubiquitin-linked species and in ubiquitin turnover. An end3-1 mutation, which inhibits cell membrane endocytosis, partially inhibits accumulation of these ubiquitin conjugates as well and causes an elevation of ubiquitin levels in doa4Δ cells. These results suggest that a fraction of the small ubiquitinated species in doa4Δ cells are derived from ubiquitinated cell surface proteins, which are subsequently proteolyzed by vacuolar proteases. Perturbation of either endocytosis or endosome-to-vacuole trafficking blocks the delivery of these substrates to the site of degradation (i.e., the vacuole) and may thereby stabilize ubiquitin and suppress its depletion in the doa4Δ mutant.
Direct participation of Doa4 in releasing ubiquitin from proteasome substrates is suggested by the association of this enzyme with the 26S proteasome (Papa et al., 1999). The genetic interactions between doa4Δ and vps24Δ, vps27Δ and end3-1 described in this study cannot distinguish between a direct or indirect role for Doa4 in cleaving ubiquitinated, endocytosed proteins. It is also not clear whether destruction of ubiquitin by the proteasome and by the vacuole in doa4Δ cells always occurs via independent mechanisms or whether the two proteolytic pathways sometimes converge. The proteasome may also contribute to the degradation of ubiquitin conjugated to membrane proteins that are targeted to the vacuole. Degradation of Ste6, the a-factor transporter, has been reported to depend on both the proteasome and vacuolar proteases (Loayza and Michaelis, 1998). Given that Ste6 localizes to the vacuolar membrane in a doa4Δ mutant (Loayza and Michaelis, 1998), the ubiquitinated cytosolic domain of Ste6 would remain accessible to degradation by proteasomes. Destruction of ubiquitin attached to such substrates may require an interdependence between lumenal vacuolar proteases and the cytosolic proteasome, whereas ubiquitin on membrane proteins that localize to the vacuole interior in doa4Δ cells should be degraded in a proteasome-independent manner. The experiments comparing ubiquitin depletion in stationary phase doa4Δ cells impaired for vacuolar proteolysis, proteasomal proteolysis, or both indicate that the two proteolytic pathways define two distinguishable mechanisms for ubiquitin degradation.
Analysis of the Dub family of enzymes suggests that these enzymes can be highly substrate specific. For example mammalian isopeptidase T and yeast Ubp14 selectively release ubiquitin from unanchored polyubiquitin chains (Wilkinson et al., 1995; Amerik et al., 1997). From the present work, Doa4 appears to act in both the proteasome and vacuolar proteolytic pathways, suggesting that this enzyme should be able to deubiquitinate a wide spectrum of substrates. Because the yeast genome encodes 16 additional Dubs, it is surprising that Doa4 should have a central function in such disparate pathways. Identification and characterization of the low molecular mass ubiquitinated species that accumulate in the doa4Δ mutant should clarify whether Doa4 works directly in both pathways and might shed light on other cellular processes that depend on Doa4.
ACKNOWLEDGMENTS
We thank C. Pickart for the anti-ubiquitin antibody, L. Hicke for the end3-1 strain, and J. Laney for comments on the manuscript. This work was supported by National Institutes of Health grant GM-53756 to M.H.
REFERENCES
- Amerik AY, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason T, Ellison MJ. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1989. [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;10:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M. Biogenesis, structure, and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Chung YC, Reddy TBK, Zhou K, Firtel RA. A novel, putative MEK kinase controls developmental timing and spatial patterning in Dictyostelium and is regulated by ubiquitin-mediated protein degradation. Genes Dev. 1998;12:3564–3578. doi: 10.1101/gad.12.22.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison MJ, Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- Finley D, Özkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Galan J, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers R, Broeke T, Kerkhof P, Schwartz AL, Strous GJ. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 1999;18:28–36. doi: 10.1093/emboj/18.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Bright PM. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- Hadari T, Warms JVB, Rose IA, Hershko A. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J Biol Chem. 1992;267:719–727. [PubMed] [Google Scholar]
- Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, Andre B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur1 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Ellison MJ, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Hough R, Rechsteiner M. Ubiquitin-lysozyme conjugates: purification and susceptibility to proteolysis. J Biol Chem. 1986;261:2391–2399. [PubMed] [Google Scholar]
- Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- Jungmann J, Reins H-A, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- Kolling R, Hollenberg CP. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz I, Jiang H, Michels CA. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J Biol Chem. 1998;273:4454–4462. doi: 10.1074/jbc.273.51.34454. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Özkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Papa FR, Amerik AY, Hochstrasser M. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J Biol Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-SDS-PAGE for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai KT, Liu X, Silar P, Sosinowsk IT, Thiele DJ. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signaling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD, Hochstrasser M. Deubiquitinating enzymes. In: Peters JM, Finley D, Harris JR, editors. Ubiquitin and Biology of the Cell. New York: Plenum Press; 1998. pp. 99–120. [Google Scholar]
- Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Park FJ, Jones E. Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics. 1982;102:679–690. doi: 10.1093/genetics/102.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]