Abstract
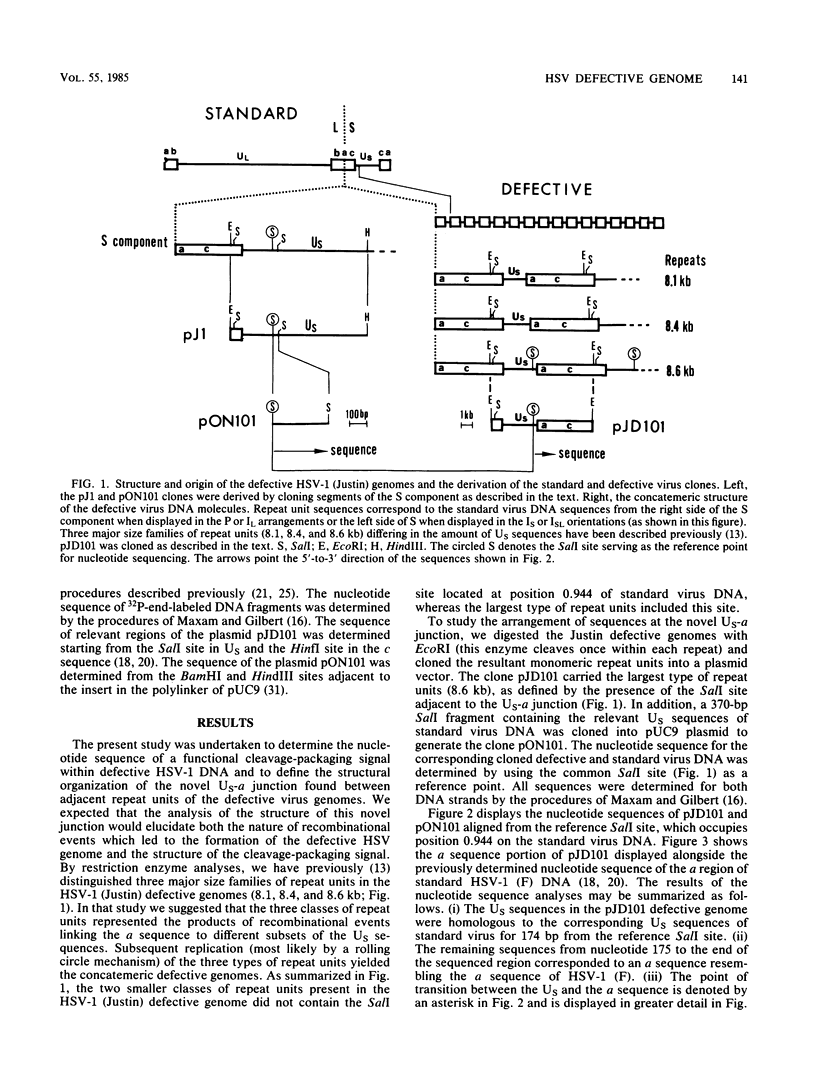
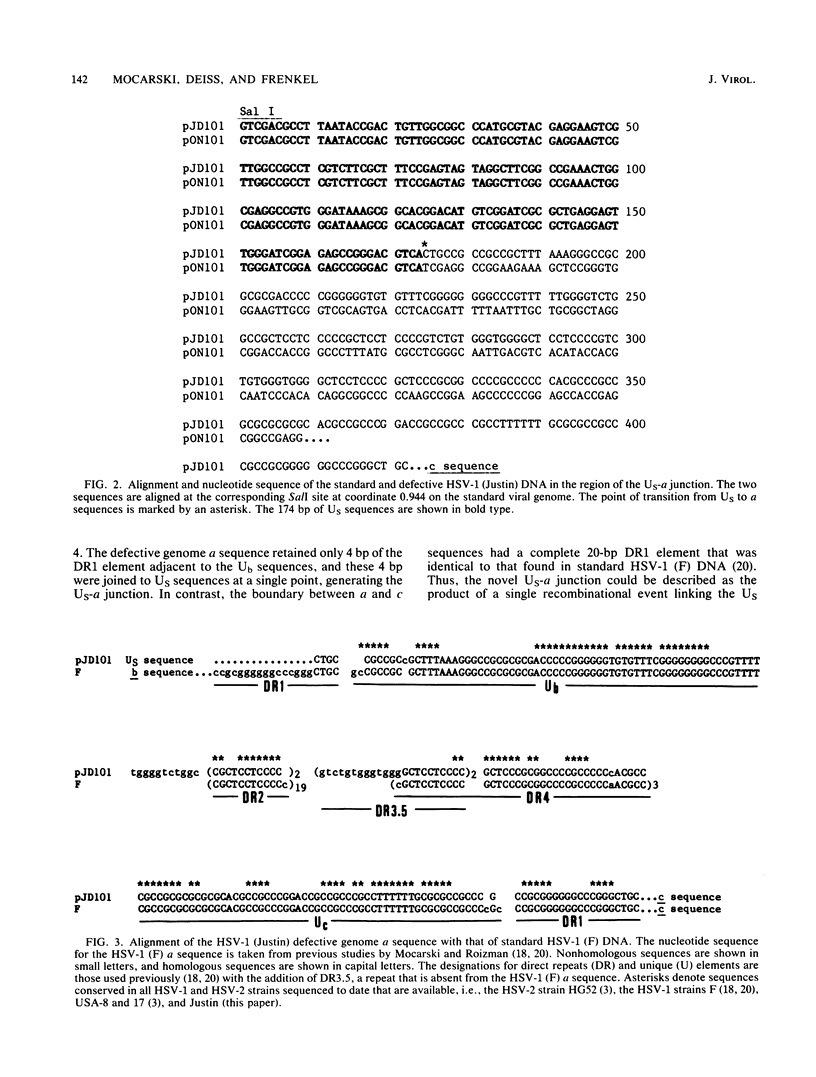
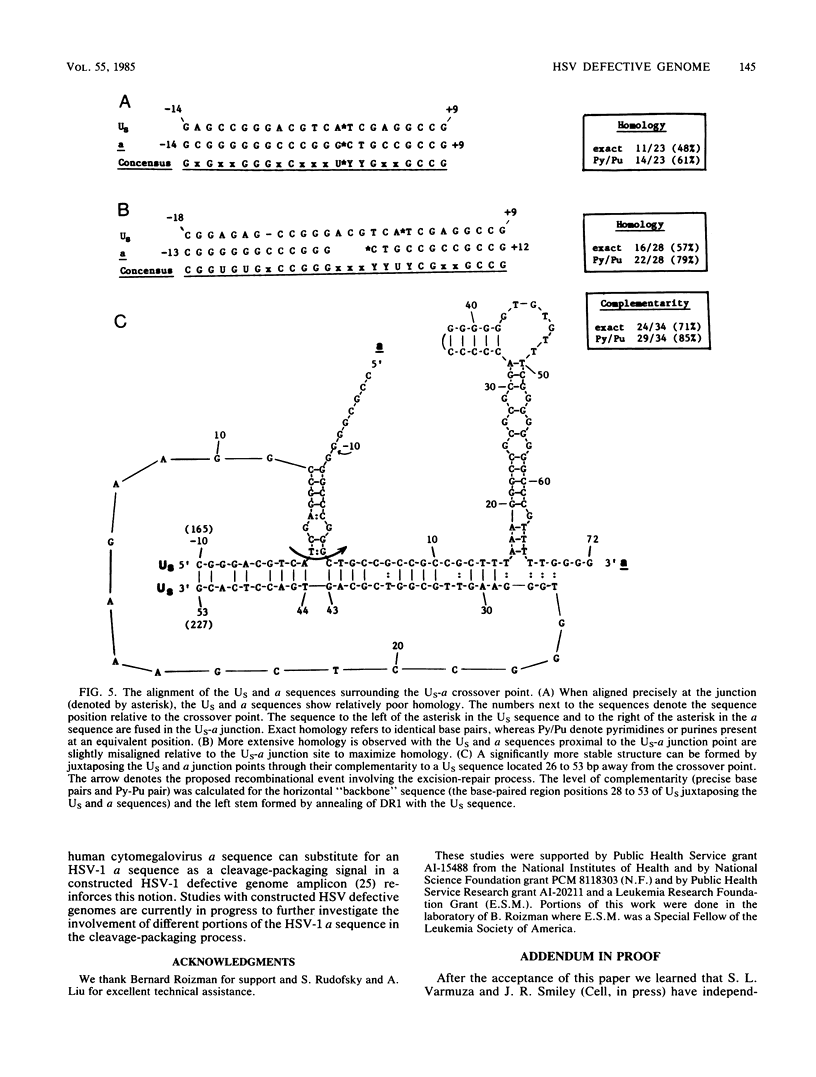
Defective genomes generated during serial propagation of herpes simplex virus type 1 (Justin) consist of tandem reiterations of sequences that are colinear with a portion of the S component of the standard viral genome. We determined the structure of the novel US-a junction, at which the US sequences of one repeat unit join the a sequences of the adjacent repeat unit. Comparison of the nucleotide sequence at this junction with the nucleotide sequence of the corresponding US region of the standard virus genome indicated that the defective genome repeat unit arose by a single recombinational event between an L-S junction a sequence and the US region. The recombinational process might have been mediated by limited sequence homology. The sequences retained within the US-a junction further define the signal for cleavage and packaging of viral DNA.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnett J. W., Eppstein D. A., Chan H. W. Class I defective herpes simplex virus DNA as a molecular cloning vehicle in eucaryotic cells. J Virol. 1983 Nov;48(2):384–395. doi: 10.1128/jvi.48.2.384-395.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Denniston K. J., Madden M. J., Enquist L. W., Vande Woude G. Characterization of coliphage lambda hybrids carrying DNA fragments from Herpes simplex virus type 1 defective interfering particles. Gene. 1981 Dec;15(4):365–378. doi: 10.1016/0378-1119(81)90180-3. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Jacob R. J., Honess R. W., Hayward G. S., Locker H., Roizman B. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J Virol. 1975 Jul;16(1):153–167. doi: 10.1128/jvi.16.1.153-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkeĺ N., Locker H., Batterson W., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. VI. Defective DNA originates from the S component. J Virol. 1976 Nov;20(2):527–531. doi: 10.1128/jvi.20.2.527-531.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaerner H. C., Maichle I. B., Ott A., Schröder C. H. Origin of two different classes of defective HSV-1 Angelotti DNA. Nucleic Acids Res. 1979 Apr;6(4):1467–1478. doi: 10.1093/nar/6.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf C., Strauss G., Ott-Hartmann A., Schatten R., Kaerner H. C. Herpes simplex virus defective genomes: structure of HSV-1 ANG defective DNA of class II and encoded polypeptides. J Gen Virol. 1983 Nov;64(Pt 11):2455–2470. doi: 10.1099/0022-1317-64-11-2455. [DOI] [PubMed] [Google Scholar]
- Kwong A. D., Frenkel N. Herpes simplex virus amplicon: effect of size on replication of constructed defective genomes containing eucaryotic DNA sequences. J Virol. 1984 Sep;51(3):595–603. doi: 10.1128/jvi.51.3.595-603.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979 Nov;32(2):429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N., Halliburton I. Structure and expression of class II defective herpes simplex virus genomes encoding infected cell polypeptide number 8. J Virol. 1982 Aug;43(2):574–593. doi: 10.1128/jvi.43.2.574-593.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin). J Virol. 1979 Mar;29(3):1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Post L. E., Conley A. J., Mocarski E. S., Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Fong B. S., Leung W. C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981 Aug;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C., Davison A. J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983 Dec 10;11(23):8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro J. C., Filpula D., Friedmann T., Spector D. H. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J Virol. 1984 Nov;52(2):541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]



