Abstract
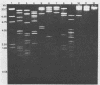
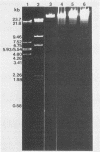
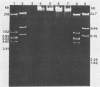
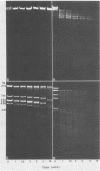
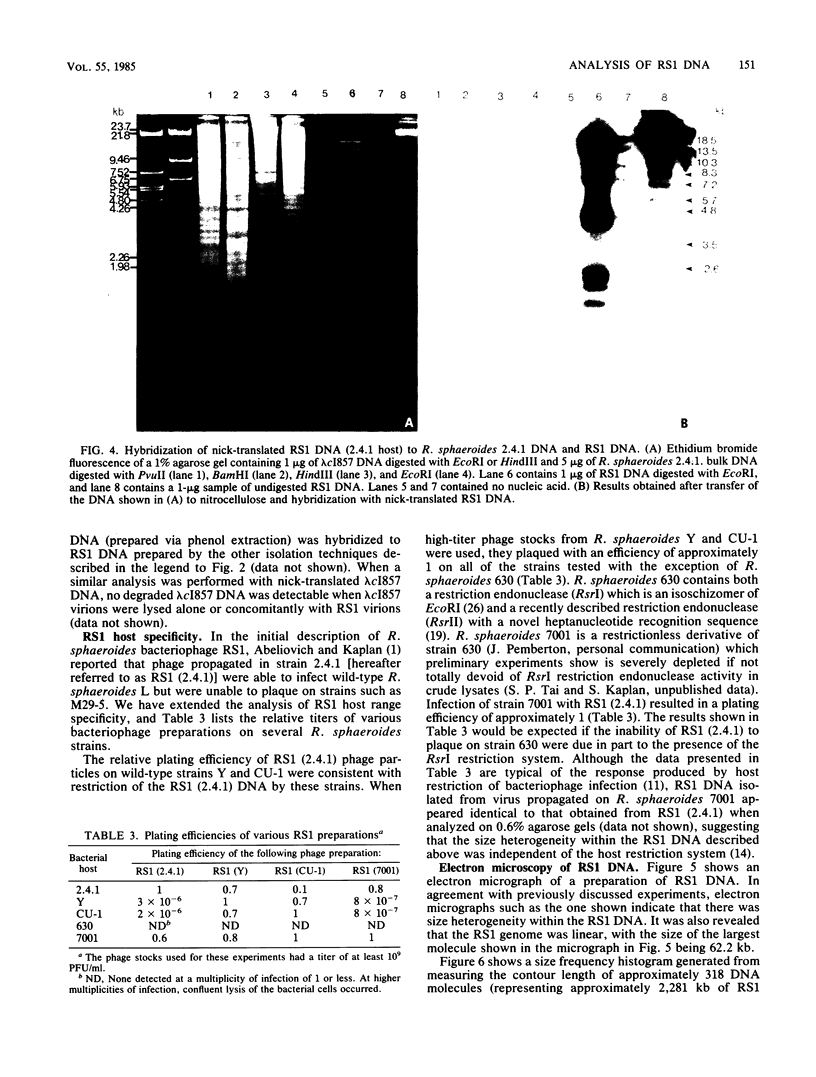
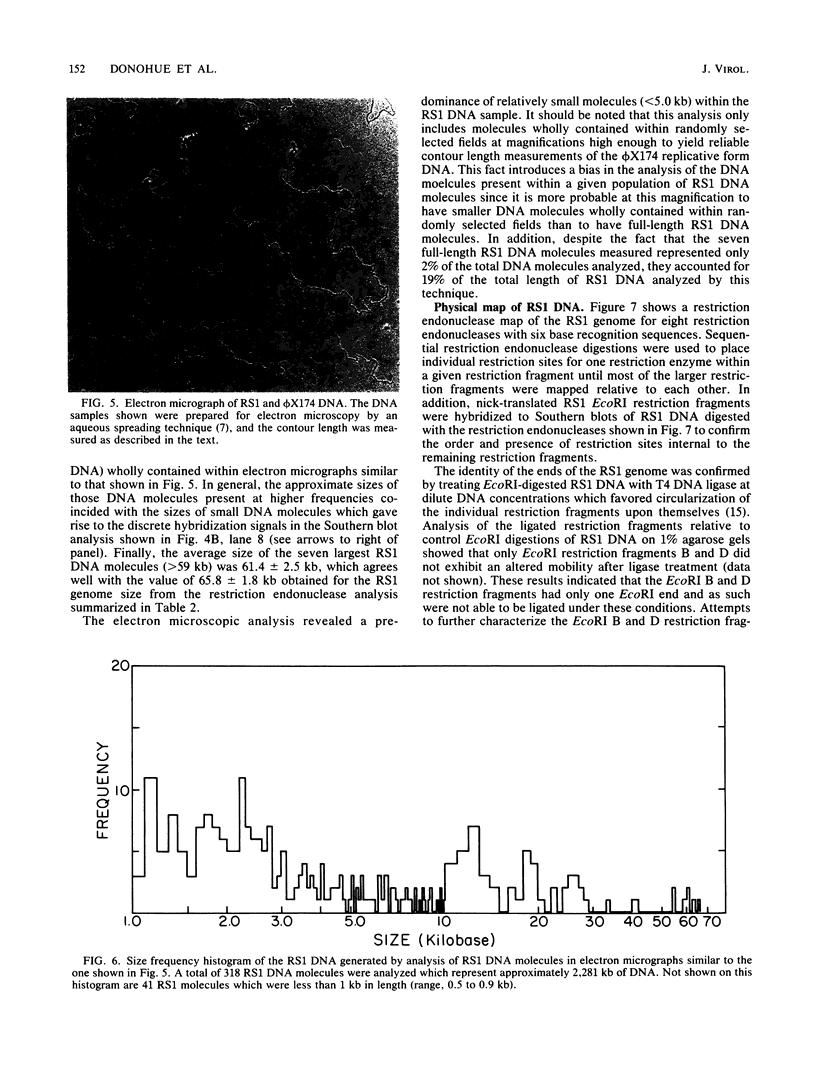
We analyzed, by restriction endonuclease mapping and electron microscopy, the genome of the lytic Rhodopseudomonas sphaeroides-specific bacteriophage RS1 and characterized it as a linear molecule of approximately 60 to 65 kilobases. When the DNA from purified phage particles was examined by several independent methods, considerable size heterogeneity was apparent in the RS1 DNA. This size heterogeneity was concluded to be of biological origin, was independent of the specific host strain used to propagate virus, and was not due to the presence of host DNA within or nonspecifically associated with purified virions. In addition, treatment of RS1 DNA with either BAL 31 nuclease or DNA polymerase I Klenow fragment revealed that several distinct regions exist within the viral chromosome which contain free 3' hydroxyl groups. A restriction endonuclease map of the RS1 genome was constructed by using the restriction endonucleases EcoRI, ClaI, KpnI, BamHI, MluI, SmaI, and BclI; thereby allowing the positioning of some 40 restriction sites within the viral genome. The results are discussed in terms of the significance and the possible biological origin of the unique features discovered within the phage RS1 DNA.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeliovich A., Kaplan S. Bacteriophages of Rhodopseudomonas spheroides: isolation and characterization of a Rhodopseudomonas spheroides bacteriophage. J Virol. 1974 Jun;13(6):1392–1399. doi: 10.1128/jvi.13.6.1392-1399.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Carlson K., Wiberg J. S. In vivo cleavage of cytosine-containing bacteriophage T4 DNA to genetically distinct, discretely sized fragments. J Virol. 1983 Oct;48(1):18–30. doi: 10.1128/jvi.48.1.18-30.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J., Kaplan S. The in vitro transcription-translation of DNA and RNA templates by extracts of Rhodopseudomonas sphaeroides. Optimization and comparison of template specificity with Escherichia coli extracts and in vivo synthesis. J Biol Chem. 1982 Dec 25;257(24):15110–15121. [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Reznikoff W. S. Identification and restriction endonuclease mapping of the threonine operon regulatory region. J Mol Biol. 1978 Dec 5;126(2):241–258. doi: 10.1016/0022-2836(78)90361-3. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Ostrander D. A., Hodnett J. L., Legerski R. J., Robberson D. L. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975 Sep;2(9):1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. P., Gray H. B., Jr Extracellular nucleases of Alteromonas espejiana BAL 31.IV. The single strand-specific deoxyriboendonuclease activity as a probe for regions of altered secondary structure in negatively and positively supercoiled closed circular DNA. Nucleic Acids Res. 1979 Jan;6(1):331–357. doi: 10.1093/nar/6.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn S. P., Cohen L. K., Gardner J. F., Kaplan S. Characterization of a site-specific restriction endonuclease from Rhodopseudomonas sphaeroides. J Bacteriol. 1979 May;138(2):505–509. doi: 10.1128/jb.138.2.505-509.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. D., Chory J., Kaplan S. Cloning and characterization of the gene product of the form II ribulose-1,5-bisphosphate carboxylase gene of Rhodopseudomonas sphaeroides. J Bacteriol. 1985 Jan;161(1):469–472. doi: 10.1128/jb.161.1.469-472.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor C. D., Metcalf E., Wrighton C. J., Harris T. J., Saunders J. R. RsrII--a novel restriction endonuclease with a heptanucleotide recognition site. Nucleic Acids Res. 1984 Sep 11;12(17):6701–6708. doi: 10.1093/nar/12.17.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M., Cooke S., Bowen A. R. Gene transfer mechanisms among members of the genus Rhodopseudomonas. Ann Microbiol (Paris) 1983 Jul-Aug;134B(1):195–204. doi: 10.1016/s0769-2609(83)80105-7. [DOI] [PubMed] [Google Scholar]
- Poindexter J. S. The caulobacters: ubiquitous unusual bacteria. Microbiol Rev. 1981 Mar;45(1):123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. A bacteriophage T5 mutant with an increased frequency of single-chain interruptions. J Virol. 1984 Aug;51(2):553–555. doi: 10.1128/jvi.51.2.553-555.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r135–r167. [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E., Zusman D. R. RNA polymerase of Myxococcus xanthus: purification and selective transcription in vitro with bacteriophage templates. J Bacteriol. 1982 Jul;151(1):89–105. doi: 10.1128/jb.151.1.89-105.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUSSMAN R., JACOB F. [On a thermosensitive repression system in the Escherichia coli lambda bacteriophage]. C R Hebd Seances Acad Sci. 1962 Feb 19;254:1517–1519. [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Wu R., Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 1971 May 14;57(3):491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]