Abstract
The spindle checkpoint arrests the cell cycle at metaphase in the presence of defects in the mitotic spindle or in the attachment of chromosomes to the spindle. When spindle assembly is disrupted, the budding yeast mad and bub mutants fail to arrest and rapidly lose viability. We have cloned the MAD2 gene, which encodes a protein of 196 amino acids that remains at a constant level during the cell cycle. Gel filtration and co-immunoprecipitation analyses reveal that Mad2p tightly associates with another spindle checkpoint component, Mad1p. This association is independent of cell cycle stage and the presence or absence of other known checkpoint proteins. In addition, Mad2p binds to all of the different phosphorylated isoforms of Mad1p that can be resolved on SDS-PAGE. Deletion and mutational analysis of both proteins indicate that association of Mad2p with Mad1p is critical for checkpoint function and for hyperphosphorylation of Mad1p.
INTRODUCTION
Cell cycle progression is a highly ordered and tightly regulated process. For example, mitosis occurs only after DNA synthesis has completed, and chromosome segregation does not begin until all the chromosomes have been correctly aligned on the mitotic spindle. These regulatory linkages are due to cell cycle checkpoints (Hartwell and Weinert, 1989; Elledge, 1996; Rudner and Murray, 1996), mechanisms that arrest the cell cycle if the preceding events have not been completed or if damage has occurred. Defects in checkpoints compromise the faithful transmission of genetic information and have been shown to play an important role in tumor progression (Hartwell and Kastan, 1994; Cahill et al., 1998).
Mitosis in most eukaryotes is regulated by a cyclin-dependent kinase, which is activated by association with the mitotic cyclins, and is encoded by CDC28 in the budding yeast Saccharomyces cerevisiae and the Cdc2 gene of other organisms. Activation of Cdc28 protein kinase leads to mitotic spindle formation. Proteolysis of the anaphase inhibitor Pds1p induces chromatids to separate and move to opposite spindle poles (Cohen-Fix et al., 1996), and the destruction of Clb2p and Ase1p are required for cells to exit from mitosis (Surana et al., 1993; Juang et al. 1997).
Formation of an intact mitotic spindle and attachment of all sister chromatids to the spindle before anaphase occurs is crucial to proper chromosome segregation. Defects in spindle assembly or chromosome attachment prevent the onset of anaphase by activating the spindle checkpoint. Several components of the checkpoint have been identified through budding yeast genetics. Mutations in the MAD (mitotic arrest-deficient) (Li and Murray, 1991) and BUB (budding uninhibited by benzimidazole) (Hoyt et al., 1991) genes abolish this cell cycle arrest and allow cells to enter anaphase in the absence of a functional spindle, leading to cell death and massive chromosome mis-segregation (Hoyt et al., 1991; Li and Murray, 1991). Although the MAD and BUB genes are not essential for cell viability, mutations in these genes increase the chromosome loss rate even in the absence of spindle defects, suggesting that they regulate the metaphase to anaphase transition during normal cell cycles (Hoyt et al., 1991; Li and Murray, 1991).
Many of the Mad and Bub proteins have now been identified and characterized (for review, see Rudner and Murray, 1996). Mad1p is a nuclear protein whose phosphorylation increases greatly upon spindle depolymerization and rises transiently during normal mitosis (Hardwick and Murray, 1995). Genetic and biochemical evidence suggests that Mad1p is phosphorylated by Mps1p whose function is also required for the checkpoint (Hardwick et al., 1996; Weiss and Winey, 1996).
Homologues of spindle checkpoint components have been identified in fission yeast (Kim et al., 1998) and vertebrates (for review, see Hardwick, 1998). MAD2 homologues in the frog Xenopus laevis (XMAD2) (Chen et al., 1996) and humans (HMAD2) (Li and Benezra, 1996) are essential for checkpoint function in frog egg extracts and in cultured human cells, respectively (Chen et al., 1996; Li and Benezra, 1996). Unlike budding yeast, vertebrate cells appear to require the checkpoint even when there is no perturbation of spindle assembly (Gorbsky et al., 1998). Kinetochores that are not attached to microtubules recruit the vertebrate homologues of Mad2 (Chen et al., 1996; Li and Benezra, 1996), Mad1, Mad3, Bub1, and Bub3 (Taylor and McKeon, 1997; Chan et al., 1998; Taylor et al., 1998), and a small fraction of the kinetochores in Taxol-treated cells recruit Mad2 (Waters et al., 1998). The Mad2 and Mad3 proteins bind to and are thought to inhibit the activity of Cdc20p (Fang et al., 1998; Hwang et al., 1998; Kim et al., 1998), a substoichiometric component of the anaphase-promoting complex (Fang et al., 1998), the large complex that initiates anaphase by catalyzing the ubiquitination of cyclin B and proteins that regulate sister chromatid cohesion (King et al., 1995; Sudakin et al., 1995; Cohen-Fix et al., 1996; Zachariae and Nasmyth, 1996). The conservation of the spindle checkpoint proteins in eukaryotes indicates that the checkpoint is an important regulator of cell division and that its mechanism has been conserved throughout evolution.
We report the isolation of the budding yeast MAD2 gene and the characterization of the association between Mad1p and Mad2p that is essential for the function of the spindle checkpoint.
MATERIALS AND METHODS
Yeast Strains and Media
Table 1 lists the strains used in this work, all of which are derivatives of W303 except the three original mad1 alleles, which are in the A364a background, and MAY 2072, which is in the S288c background. Yeast media, growth conditions, stock solutions, and molecular techniques were as previously described (Guthrie and Fink, 1991; Hardwick and Murray, 1995).
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| KH 34 | MATaura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| RHC 1 | MATamad2-1, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| RHC 15.1 | MATamad2Δ::URA3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| BEN 24 | MATamad1-1, ura3-52, leu2, his3, trp1-1, rad9Δ::LEU2 |
| BEN 27 | MATamad1-2, ura3-52, leu2, his3, trp1-1, rad9Δ::LEU2 |
| BEN 79 | MATamad1-3, ura3-52, leu2, his3, trp1-1, rad9Δ::LEU2 |
| KH 144 | MATα mad1Δ.2::URA3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| KH 173 | MATamad3Δ.2::URA3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| KH 127 | MATabub1Δ::HIS3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| KH 128 | MATabub2Δ::URA3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| MAY 2072 | MATabub3Δ::LEU2, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| RHC 88 | MATaURA3::mad2-N5, mad2-1, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| RHC 89 | MATaURA3::mad2-C5, mad2-1, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| RHC 91 | MATaURA3::mad2-N10, mad2-1, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| RHC 93 | MATaURA3::mad2-C10, mad2-1, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| KH 153 | MATaURA3::GAL-MPS1, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
Cloning of MAD2 and mad2 Gene Disruption
A 2.6-kb HindIII–SalI genomic fragment that resides upstream of the translational initiation codon of BET4 was subcloned into the cognate sites in the vector pRS316 (Sikorski and Hieter, 1989). This plasmid pRC2 was able to complement the benomyl-sensitive phenotype of mad2-1 mutant. An ORF of 196 amino acids was identified in this region by DNA sequencing from both ends of the HindIII–SalI fragment.
To generate the mad2::URA3 disruption plasmid pRC10.1, a 1.2-kb fragment containing the URA3 gene was used to replace the fragment between the ApaI site located 20 base pairs upstream of the MAD2 translation initiation codon and the ScaI site that resides in amino acid position 148.
Preparation of Recombinant Mad2 Protein and Mad2 Antibodies
The coding region of MAD2 flanked by EcoRI sites was generated by PCR and cloned into pGEX1 at the EcoRI site. This GST fusion construct was transformed into Escherichia coli strain DH5α, and its expression was induced with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside for 2 h at 37°C. Cells were pelleted and resuspended in PBS (2.7 mM KCl, 137 mM NaCl, 1.5 mM KH2PO4, 4.3 mM Na2HPO4, pH 7.2), and repelleted. The cell pellet was resuspended in PBS containing 0.5% Triton X-100, 1 mM EGTA, 1 mM EDTA, 1 mM PMSF, and 200 μg/ml lysozyme, and sonicated briefly. The lysate was spun at 15 krpm in a Sorvall (Newton, CT) SS-34 rotor for 30 min. The supernatant was loaded onto a 4-ml glutathione-agarose column. The column was washed with 40 ml of PBS, and the GST-Mad2 fusion protein was eluted with 5 mM reduced glutathione in 50 mM Tris, pH 8.0, and 0.5 mM DTT. Purified protein was dialyzed into 50 mM HEPES, pH 7.6, 50 mM KCl, and 50% glycerol. The purified protein was used to raise antisera in rabbits (Babco, Berkeley, California). To affinity purify antibodies, the rabbit serum was passed over a 50-ml column of GST protein coupled to Affi-Gel 10 (Bio-Rad, Hercules, California) to remove anti-GST antibodies, before being loaded over a 3-ml column of GST-Mad2 protein coupled to Affi-Gel 10. Elution of anti-Mad2 antibodies was performed as described (Chen et al., 1996).
Construction of mad2 Deletions
The HindIII–XhoI fragment containing the MAD2 gene (Figure 1A) was subcloned into the corresponding sites in the vector pRS316 to give rise to the plasmid pRC4. The 3′-untranslated region was amplified by PCR, which also converted the EcoRI site following the stop codon to HindIII. This fragment was subcloned into pRS316 at HindIII–XhoI sites, giving rise to pRC66. All the deletion mutants were made by PCR amplification and cloned at the HindIII site of pRC66. The BamHI–XhoI fragments containing various deletions were subcloned into pRS306 (Sikorski and Hieter, 1989). To integrate the plasmids into yeast at URA3 locus, the plasmids were cut at StuI in the URA3 gene.
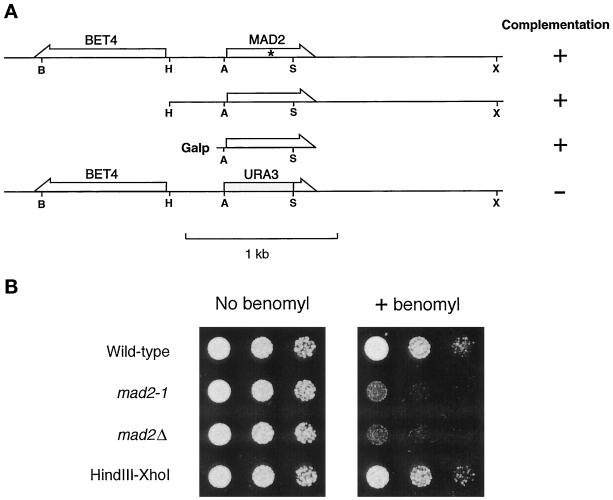
Figure 1.
Identification of MAD2. (A) Relative position of MAD2 and BET4 and the ability of various constructs to rescue mad2-1. The mutation in mad2-1 is marked with an asterisk. The positions of the following restriction enzyme recognition sites are indicated: B, BamHI; H, HindIII; A, ApaI; S, ScaI; X, XhoI. (B) Benomyl sensitivity of mad2 mutants. Cells were spotted onto either a YPD plate (left panel) or a YPD plate containing 7.5 μg/ml benomyl (right panel). Cells were diluted 10-fold from the corresponding spot on the left. Yeast strains are indicated on the left. The HindIII–XhoI fragment upstream of BET4 fully rescued the benomyl sensitivity of mad2-1 when carried on a CEN plasmid.
Construction of mad1 Mutants, Deletions, Truncations, and Allele Sequencing
Three mutations were engineered into the MAD1 sequence by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit and Pfu DNA polymerase according to manufacturer’s instructions (Stratagene, La Jolla, CA). A KpnI site was introduced just before the first methionine of Mad1p using two primers, CTTAAAATCGAGAGGTAATAGGGTACCATGGATGTGAGAGCG -GCATTG and its reverse complement. Two NotI sites were engineered at either side of the asparagine-rich stretch using the primers CCGGATAATCTCTTCAGGAGCGGCCGCTATGTTATTTTTGGT -TC with its reverse complement to introduce a site at position 974 of the coding sequence and GAACCAAAAATAACATAGCGGCCGCCCCTGAAGAGATTATCCGG with its reverse complement to introduce a site at 1109. The other N- and C-terminal Mad1p deletion constructs and the two-hybrid constructs were made by PCR amplification (using VENT polymerase; New England Biolabs, Beverly, MA), followed by subcloning and sequencing of the resulting constructs. pKH601 fuses full-length Mad1p (residues 1-749) to the GAL4 DNA binding domain of pAS1-CYH2; pKH602 fuses residues 313-749; pKH603 fuses residues 529-749; pKH604 fuses residues 1-318; pKH605 fuses residues 593-749; pKH609 fuses residues 529-718; pKH610 fuses 529-649.
The sequences of the mutations in the three original mad1 alleles were determined by PCR amplification of the genomic loci followed by cycle sequencing of the PCR products (Applied Biosystems, Foster City, CA). Each allele was sequenced multiple times on both strands.
Immunoblotting, Immunoprecipitation, and Gel Filtration
Yeast extracts were made, and immunoblotting was performed as previously described (Hardwick and Murray, 1995). The affinity-purified anti-Mad2p antibody was used at a dilution of 1:500 in PBS containing 2% BSA and 0.2% Tween 20, the anti-Mad1p antibody at 1:2000 in Blotto (Harlow and Lane, 1988), and the anti-HA antibody (16B12, Babco) at 1:500 in Blotto.
For immunoprecipitations, yeast extracts were made by bead beating in lysis buffer (50 mM HEPES, pH 7.6, 25 mM KCl, 50 mM NaF, 1 mM MgCl2, 1 mM EGTA, 0.1% Na-deoxycholate, 1 mM PMSF, 0.5 mM DTT, and 10 μg/ml leupeptin, pepstatin, and chymostatin) as previously described (Hardwick and Murray, 1995), except that in some cases the anti-Mad1p antibody was directly coupled to the protein A-agarose (Harlow and Lane, 1988) before use. Gel filtration using a Pharmacia (Piscataway, NJ) Superose 6 fast performance liquid chromatography column was carried out as described (Hardwick and Murray, 1995).
Transfection in COS Cells
For expression in COS7 cells, the coding regions of MAD1, MAD2, or MPS1 were subcloned into the vector SRα (Takebe et al., 1988) at the EcoRI site. The sequence encoding the myc epitope was inserted at the amino terminus of MPS1 for detection with the anti-myc antibody 9E10. The plasmids were purified twice by standard cesium chloride gradient (Maniatis et al., 1982).
COS cells were maintained in Dulbecco’s modified Eagle’s medium plus 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Transfection was performed with standard calcium phosphate precipitation as described (Chen et al., 1996).
RESULTS
Identification and Characterization of MAD2
The spindle checkpoint gene MAD2 in the budding yeast S. cerevisiae was originally identified (Li and Murray, 1991) as the ORF YJL031C, which encodes a subunit of an essential prenyltransferase (Li et al., 1993) and has been renamed BET4. However, sequencing this gene recovered from the original mad2-1 strain failed to identify any mutation. In addition, a genomic DNA fragment outside of the prenyltransferase coding region (HindIII–XhoI region in Figure 1A) fully rescued the benomyl sensitivity of mad2-1 (Figure 1; see correction in Li et al., 1994), suggesting that this fragment encoded the bona fide MAD2 gene. This was confirmed by sequencing a 196-amino acid ORF (YJL030W), recovered from wild-type cells and from the mad2-1 mutant. This analysis shows that the mad2-1 mutation lies within YJL030W converting Trp94 into a stop codon. Deleting most of the coding region of YJL030W produced viable strains that have phenotypes similar to that of mad2-1 (Figure 1B), and expression of the coding region of YJL030W from a galactose-inducible promoter rescued the benomyl sensitivity of mad2-1 in a galactose-dependent manner (Figure 1A). These observations unequivocally show that YJL030W is the bona fide MAD2 gene.
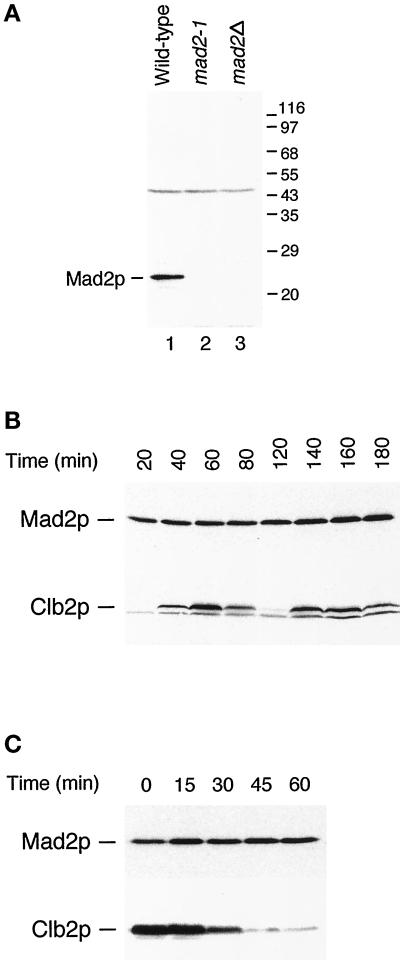
To characterize Mad2p, we generated antiserum against recombinant GST-Mad2. The affinity-purified antibodies recognized Mad2p specifically on immunoblots (Figure 2A, lane 1), and the protein was missing in the mad2Δ strain, as expected (Figure 2A, lane 3). We did not detect the truncated form of Mad2p, which has a predicted molecular mass of 13 kDa, in the mad2-1 strain, indicating that the truncated protein is unstable or that the antibody recognizes epitopes in the C-terminal half. We studied the protein by following its level during a synchronous cell cycle (Figure 2B). Although the level of Clb2p, a mitotic cyclin, showed the expected oscillation, there was no change in either the abundance or the gel mobility of Mad2p during the cell cycle.
Figure 2.
(A) Specificity of anti-Mad2p antibody. The antibody recognizes a 24-kDa protein in wild-type cells (lane 1) that is not detectable in mad2-1 (lane 2) and mad2Δ (lane 3) strains. The migration of molecular size standards is indicated on the right. (B) Mad2p level and its mobility on SDS-PAGE stay constant throughout the cell cycle. Cells arrested at G1 with α-factor were released from the arrest for the time indicated. Cell lysates were immunoblotted with an anti-Mad2p antibody (upper panel) or with an anti-Clb2p antibody. (C) Mad2p levels and gel mobility remain unchanged at the metaphase to anaphase transition. Cells arrested at mitosis with benomyl and nocodazole were released from the arrest for the time indicated on top. Cell lysates were immunoblotted with an anti-Mad2p antibody (upper panel) or with an anti-Clb2p antibody (lower panel).
We examined the effect of activating the spindle checkpoint on Mad2p (Figure 2C). Cells were arrested in mitosis by depolymerizing their spindles with benomyl and nocodazole and then allowed to recover from their arrest. The level of Clb2p fell as cells exited mitosis, but there was no change in either the abundance or the gel mobility of Mad2p. Analyzing the behavior of Mad2p on two-dimensional gels showed a single spot whose mobility was unaffected by activation of the spindle checkpoint (our unpublished data). These results suggest that the function of Mad2p is not regulated by post-translational modification, although we cannot exclude the possibility that only a very small fraction of the Mad2p molecules are modified.
Mad2p and Mad1p Bind Tightly to Each Other In Vivo
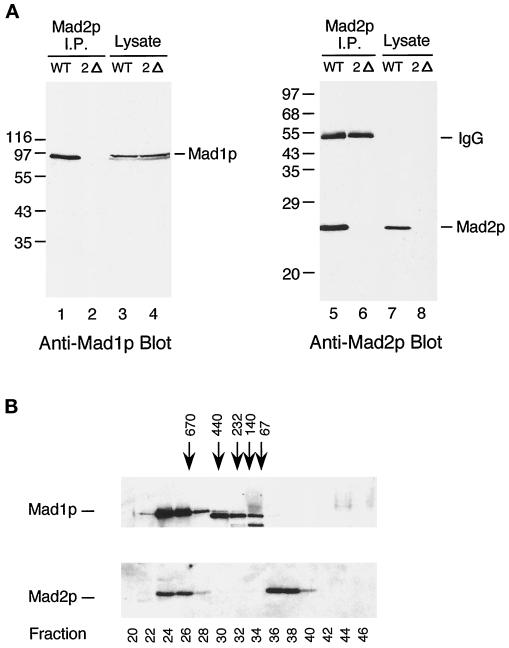
The spindle checkpoint component Mad1p is a nuclear phosphoprotein, which becomes hyperphosphorylated in cells treated with benomyl and in mitotic cells (Hardwick and Murray, 1995). Hyperphosphorylation of Mad1p is not seen in cells containing mutations in BUB1, BUB3, or MPS1 and is dramatically reduced in mad2 mutants, indicating that they likely regulate phosphorylation of Mad1p directly or indirectly (Hardwick and Murray, 1995). We tested whether Mad1p and Mad2p interact with each other by examining whether they could be co-immunoprecipitated from cells. Figure 3A shows that anti-Mad2p immunoprecipitates contained Mad1p. To analyze this Mad2p–Mad1p complex in more detail, whole yeast cell extracts were fractionated by gel filtration (Figure 3B). This experiment showed that there were two pools of Mad2p, and that one co-fractionated with Mad1p in fractions 24-26, thereby predicting a complex larger than 670 kDa. The other pool was in fractions 36-38, running at the size expected for monomeric Mad2p. All of the Mad1p cofractionated with Mad2p. The prominent band in fractions 30-34 is a background band that cross-reacts with the anti-Mad1p antibody. This experiment suggests that all of Mad1p is present in a large protein complex, but we do not know whether some or all of the complex contains Mad2p.
Figure 3.
Mad2p associates with Mad1p in vivo. (A) Mad2p coimmunoprecipitates with Mad1p from wild-type but not from mad2 mutant cells. Cell lysates (lanes 3, 4, 7, and 8) or Mad2p immunoprecipitates (lanes 1, 2, 5, and 6) prepared from wild-type (WT) or mad2Δ (2Δ) strains were immunoblotted with an anti-Mad1p (lanes 1–4) or an anti-Mad2p (lanes 5–8) antibody. The migration of molecular size standards is indicated on the left. The 55-kDa band in lanes 5 and 6 is IgG heavy chain. (B) Gel filtration analysis reveals two discrete pools of Mad2p, one of which co-fractionates with Mad1p. Fractions from Superose 6 column were immunoblotted with an anti-Mad1p (upper panel) or an anti-Mad2p (lower panel) antibody. The bulk of Mad1p is in fractions 24–28, whereas Mad2p fractionates into two separate pools of fractions 24-26 and 36-38. The fractionation of size standards is indicated on top. The fraction number is indicated on the bottom. The prominent band in lanes 30–34 is a background band that cross-reacts with the anti-Mad1p antibody.
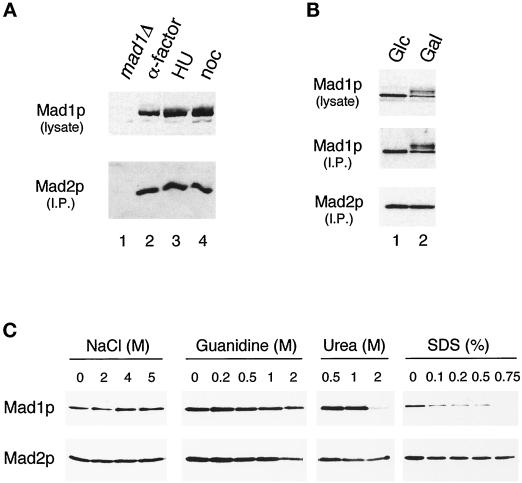
We asked whether the Mad1p–Mad2p interaction is regulated during the cell cycle. Mad1p was immunoprecipitated from yeast cells that were arrested in G1 with α factor, in S phase with hydroxyurea, or in M phase with nocodazole, and the immunoprecipitates were probed with an anti-Mad2p antibody. Figure 4A shows that the levels of Mad2p present in Mad1p immunoprecipitates were similar under all conditions, indicating that the interaction was constant throughout the cell cycle. In addition, the phosphorylation of Mad1p that is observed in mitosis, particularly when the checkpoint is activated with nocodazole, does not appear to affect the Mad2p interaction. To confirm that phosphorylation of Mad1p has no effect on its association with Mad2p in vivo, we compared their interaction in exponentially growing cells and in cells overexpressing the Mps1 protein kinase. We have previously shown that overexpression of this protein kinase is sufficient to activate the spindle checkpoint, and that it leads to a dramatic hyperphosphorylation of Mad1p (Hardwick et al., 1996). Figure 4B shows that all the different phosphorylation isoforms of Mad1p were found in the Mad2p immunoprecipitates isolated from cells overexpressing Mps1p. These results indicate that the association between Mad1p and Mad2p is independent of the phosphorylation state of Mad1p. This result was confirmed by gel filtration studies: a number of extracts were made from checkpoint-activated cells (using either nocodazole or overexpressed MPS1) and then fractionated with a sizing column. In all cases similar pools of Mad2p were found, one in a low-molecular-weight fraction and a second in a larger Mad1p-containing fraction (our unpublished data).
Figure 4.
Regulation of the Mad1–Mad2p complex. (A) The Mad1p–Mad2p complex is similar in cells arrested at G1, S, and M phases. Strains were grown to log phase and then arrested for 3 h in G1 (with α-factor; lane 2), in S phase (with hydroxyurea; lane 3), or in mitosis (with nocodazole; lane 4) before harvesting. Mad1p was immunoprecipitated from the extracts and then immunoblotted with an anti-Mad2p antibody (lower panel). The upper panel is an immunoblot of the Mad1p present in the lysates. In lane 1 a mad1Δ strain is used as a control; this strain was also treated with nocodazole. (B) Mad1p–Mad2p complex formation is independent of the phosphorylation state of Mad1p. All species of Mad1p co-immunoprecipitate with Mad2p in cells overexpressing Mps1p. Mad2p was immunoprecipitated and immunoblotted with an anti-Mad1p (upper panel) or an anti-Mad2p (lower panel) antibody. Lane 1, cells containing MPS1 under the control of galactose-inducible promoter were repressed for Mps1 expression by culturing in media containing glucose (Glc); lane 2, the same strain of cells was grown in galactose (Gal) to induce Mps1p overexpression. (C) Mad1p and Mad2p form a tight complex. Extracts from cells expressing hexahistidine-tagged Mad2p were applied to nickel-nitrilotriacetic acid beads. The gels show the proteins that remain on the beads after washing with the indicated concentrations of sodium chloride, guanidine hydrochloride, urea, or SDS. Samples were immunoblotted with an anti-Mad1p (upper panel) or an anti-Mad2p (lower panel) antibody.
To study the strength of the interaction between Mad1p and Mad2p, we used a variety of washing conditions during the isolation of Mad2p to determine what condition disrupted the association. Hexahistidine-tagged Mad2p was isolated from exponentially growing cells with nickel beads, and the Mad2p-bound beads were washed with various concentrations of sodium chloride, urea, guanidine hydrochloride, or SDS. We found that the Mad1p–Mad2p complex was stable in solutions containing up to 5 M sodium chloride, 1 M urea, and 1 M guanidine hydrochloride (Figure 4C). Even though more than half of the Mad1p–Mad2p complex was disrupted by 0.1% SDS, some of the complex was stable in up to 0.5% SDS (Figure 4C). Gel filtration analysis carried out in the presence of 1 M NaCl confirmed the stability of the Mad1–Mad2p complex (our unpublished data). These results show that Mad1p and Mad2p form a tight complex in yeast cells.
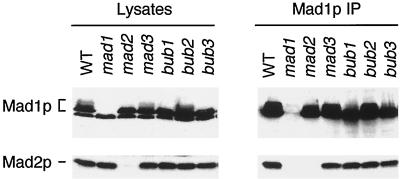
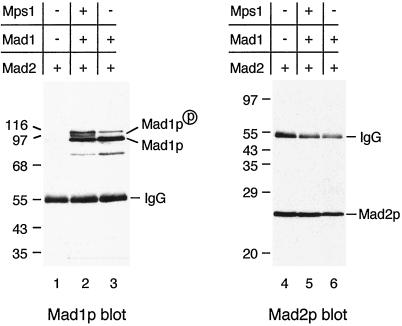
Co-immunoprecipitation of Mad1p and Mad2p from yeast cells suggests that these proteins are assembled into a complex. However, it is possible that the interaction between these two proteins is mediated through another cellular component. To test this possibility, we asked whether any other spindle checkpoint proteins were required for the assembly of the Mad1p–Mad2p complex. Deletion of the BUB1, BUB3, and MAD3 genes or a point mutation in BUB2 had no effect on the Mad1p–Mad2p complex (Figure 5). The interaction was also intact in a temperature-sensitive mps1 strain grown at nonpermissive temperature (our unpublished data). These data suggest that the assembly of Mad1p–Mad2p complex is independent of other known spindle checkpoint proteins. In an attempt to rule out the possibility that other, unknown, proteins were required for complex formation, we determined whether Mad1p and Mad2p bound to each other when they were expressed in mammalian cells. When the two proteins were transiently expressed in COS7 cells by co-transfection, Mad1p was found in Mad2p immunoprecipitates (Figure 6). This result shows that Mad1p and Mad2p can form a complex in the absence of any other yeast protein, and that they likely interact with each other directly. Similar to yeast cells overexpressing Mps1p, co-transfection of MPS1 and MAD1 in COS7 cells also enhanced Mad1p phosphorylation, and all isoforms of Mad1p bound to Mad2p (Figure 6).
Figure 5.
The association of Mad1p and Mad2p is independent of Mad3p, Bub1p, Bub2p, and Bub3p. Cell lysates or Mad1p immunoprecipitates prepared from nocodazole-treated wild-type (WT) or mutant strains, as indicated on top, were immunoblotted with an anti-Mad1p (upper panel) or an anti-Mad2p (lower panel) antibody. All mad or bub mutant strains were deletions, except for bub2-1.
Figure 6.
Mad1p and Mad2p interact in the absence of other yeast proteins. Mad1p and Mad2p co-immunoprecipitate from COS cells co-transfected with MAD2 and MAD1. MAD2 was transfected into COS cells alone (lanes 1 and 4) or co-transfected with MAD1 (lanes 3 and 6) or with MAD1 and MPS1 (lanes 2 and 5) as indicated. Mad2p was immunoprecipitated from cell lysates and immunoblotted with an anti-Mad1p (lanes 1–3) or an anti-Mad2p (lanes 4–6) antibody. Both the unphosphorylated Mad1p and the Mps1-induced phosphorylated form co-immunoprecipitated with Mad2p. The migration of molecular size standards is indicated.
Analysis of Binding Regions in Mad1p and Mad2p
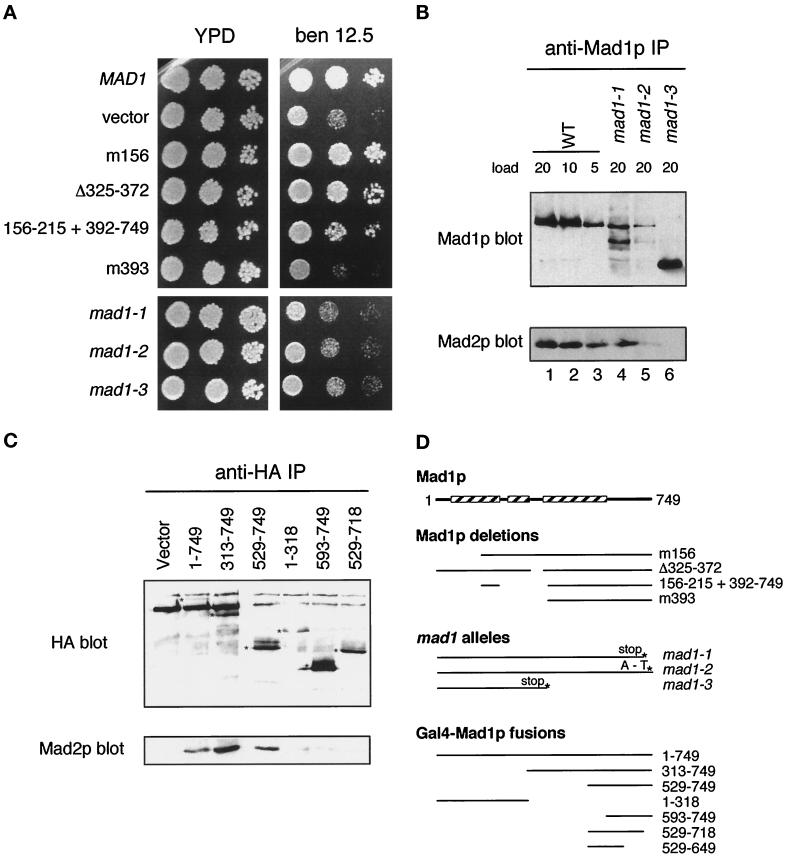
We wanted to find the basis of the interaction between Mad1p and Mad2p and to determine the importance of the interaction for the spindle checkpoint. To map the Mad2p-binding region in Mad1p, the three original mad1 alleles (Li and Murray, 1991) were sequenced (Table 2). The mad1-3 allele is a stop codon at amino acid 380 and leads to a truncated protein that does not bind to Mad2p (Figure 7B). The mad1-1 and mad1-2 alleles map to the C terminus of the protein and remove the last 33 amino acids (mad1-1) of Mad1p or change alanine (736) to threonine (mad1-2). The phenotype of all three mutants was indistinguishable from that of mad1Δ (Figure 7A), suggesting that the C terminus of Mad1p is critical for its function. The level of Mad1p protein was reduced in mad1-1 and mad1-2 cells relative to wild-type cells (Figure 7B). Immunoprecipitation experiments showed that the levels of Mad2p that could be co-immunoprecipitated with Mad1p were reduced. Approximately 25% of the wild-type level of the Mad1p–Mad2p complex appeared to be present in mad1-1 extracts, and Mad2p was barely detectable in the mad1-2 immunoprecipitate (Figure 7B, lane 5).
Table 2.
Sequence of mad1 alleles
| DNA sequence | Protein sequence | |
|---|---|---|
| mad1-1 | TGG > TAG | W (717) > stop |
| mad1-2 | GCA > ACA | A (736) > T |
| mad1-3 | TGG > TGA | W (380) > stop |
Figure 7.
The Mad2p binding domain in Mad1p. (A) mad1 constructs were assayed for their ability to complement the benomyl sensitivity of a mad1Δ strain and are compared with mad1-1, 2, and 3. Yeast strains were spotted onto plates at three dilutions and grown at 24°C. Benomyl was used at 12.5 μg/ml. (B) mad1 mutant proteins fail to co-immunoprecipitate efficiently with Mad2p. Mad1p immunoprecipitates prepared from wild-type (WT) and mutant (mad1-1, mad1-2, and mad1-3) extracts as indicated on top were immunoblotted with an anti-Mad1p (upper panel) or an anti-Mad2p (lower panel) antibody. The numbers indicate the volume (microliters) loaded of the immunoprecipitates. (C) The indicated MAD1-GAL4 DNA binding domain fusion constructs containing a hemagglutinin (HA) epitope tag were assayed for Mad2p interaction by immunoprecipitation. 16B12 (anti-HA) immunoprecipitates were immunoblotted and probed with 16B12 antibody (upper panel) or anti-Mad2p antibody (lower panel). The position of the different fusion proteins is marked with an asterisk on the anti-HA blot. (D) Summary of the different mad1 mutants, deletions, and fusion protein constructs. The boxed regions indicate the portions of Mad1p that are predicted to form a coiled coil (Hardwick and Murray, 1995).
To further map the interaction between Mad1p and Mad2p, we constructed deletion mutations in the two proteins and tested their ability to bind to their partners and their function in the spindle checkpoint. Analyzing their ability to rescue the benomyl sensitivity of a mad1Δ strain (Figure 7A) shows that up to 155 amino acids could be deleted from the N terminus of Mad1p without affecting its ability to bind to Mad2p or to complement a mad1 mutant. In addition a large, central, non–coiled-coil region from residues 216-391 was also dispensable. This region includes a highly asparagine-rich region (34 of 39 residues are asparagine or aspartate), which is not found in Mad1p homologues in other organisms. A Mad1 protein starting at methionine 393 was nonfunctional; however, a similar fusion protein with the additional residues 156-215 did rescue the benomyl sensitivity of a mad1 mutant (Figure 7A). This suggests that the region of Mad1p between amino acids 156 and 215 is structurally or functionally important. We also produced a C-terminal Mad1 truncation lacking the last 147 amino acids and found that it was unable to complement a mad1Δ strain (our unpublished data).
A series of MAD1 constructs were made fusing regions of Mad1p to the GAL4 DNA binding domain (in pAS1-CYH2) and tested for their interaction with the endogenous Mad2p in a mad1Δ strain by co-immunoprecipitation (Figure 7C). This experiment confirms the importance of the C terminus of Mad1p for its Mad2p interaction: the smallest fusion protein capable of binding to Mad2p contained residues 529-749 (pKH603), and deleting the last 35 amino acids (pKH609) abolished that ability.
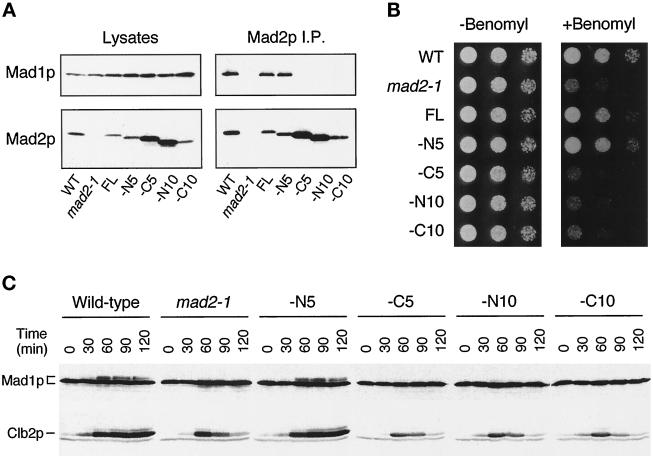
Small deletions were generated in MAD2, and the proteins were expressed in cells to determine their ability to bind Mad1p and to rescue the benomyl sensitivity of the mad2-1 mutant. Mad2p missing the N-terminal 5 amino acids could still bind to Mad1p, whereas deletion of the N-terminal 10 amino acids abolished the interaction (Figure 8A). Removal of 5 or 10 amino acids from the C terminus of Mad2p also diminished the binding (Figure 8A). Interestingly, among the four deletion mutants we generated, only the one without the N-terminal 5 amino acids could rescue the benomyl sensitivity of mad2-1 (Figure 8B). Once again, our results show a correlation between the activity of Mad1p and Mad2p in the spindle checkpoint and their ability to form a stable complex and suggest that the formation of the Mad1p–Mad2p complex is important for checkpoint function.
Figure 8.
The ability of Mad2p to rescue mad2-1 correlates with its ability to bind Mad1p. Cells used in the experiments are strains of wild-type (WT), mad2-1, mad2-1 containing a full-length MAD2 gene (FL), or a MAD2 gene that lacks regions encoding the N-terminal 5 (−N5, RHC88), C-terminal 5 (−C5, RHC89), N-terminal 10 (−N10, RHC91), or C-terminal 10 (−C10, RHC93) amino acids. (A) Co-immunoprecipitation between Mad1p and various Mad2p molecules. Lysates or Mad2p immunoprecipitated from exponentially growing cells were immunoblotted for either Mad1p (upper panels) or Mad2p (lower panels). (B) Benomyl sensitivity of various strains. Cells were spotted onto either YPD plates (left panel) or YPD plates containing 7.5 μg/ml benomyl (right panel). Cells were diluted 10-fold from the corresponding spot on the left. (C) Phosphorylation of Mad1p in various strains. Cells were first arrested at early G1 with α-factor and then released from the arrest into YPD containing 30 μg/ml benomyl and 10 μg/ml nocodazole. Aliquots of cells were taken every 30 min as indicated. Cell lysates were prepared and immunoblotted for Mad1p (upper panel) or Clb2p (lower panel).
Because phosphorylation of Mad1p is greatly reduced in a mad2-1 mutant (Hardwick and Murray, 1995), it is possible that Mad2p, by binding to Mad1p, may facilitate Mad1p phosphorylation. We tested this hypothesis by examining Mad1p phosphorylation in mad2-1 mutant cells containing various truncated forms of Mad2p. In a synchronized cell cycle, Mad1p became hyperphosphorylated in wild-type cells and in cells expressing Mad2p missing the N-terminal 5 amino acids. Mad1p hyperphosphorylation was not observed in cells that expressed Mad2 proteins lacking the N-terminal 10 amino acids or the C-terminal 5 or 10 amino acids (Figure 8C), all of which failed to bind Mad1p (Figure 8A). This result shows a correlation between the assembly of Mad1p–Mad2p complex and Mad1p hyperphosphorylation, indicating that a possible function of the Mad1p–Mad2p complex in the spindle checkpoint is to allow efficient phosphorylation of Mad1p.
DISCUSSION
We have investigated the budding yeast spindle checkpoint component Mad2p. Sequence analysis indicates that it encodes a 23-kDa protein with homology to human, Xenopus, and fission yeast proteins. Mad1p is tightly bound to Mad2p, and this interaction requires almost all of Mad2p and the C-terminal third of Mad1p. Consistent with Xmad2 in Xenopus egg extracts (Chen et al., 1998), the yeast Mad2p also exists in two different pools, a Mad1p-bound pool and a Mad1p-free pool.
MAD1 and MAD2 Encode Conserved Checkpoint Components
Since the mad1 and mad2 mutants were first identified in 1991 (Li and Murray, 1991), their homologues have been cloned from a wide variety of organisms, including human, mouse, frog, and yeast. Sequence comparisons reveal that both proteins have regions of primary sequence conservation, yet to date no homologues have been shown to rescue mad2 mutants. In the case of Mad2p the whole protein appears to be conserved. Most of the protein (residues 8-193) forms a domain that was defined by comparison of the protein sequence of Hop1p, Rev7p and Mad2p, three yeast proteins that participate in a variety of protein–protein interactions, and has been dubbed the HORMA domain (Aravind and Koonin, 1998). Our analysis of Mad2p–Mad1p binding supports the idea that this entire domain is necessary for protein–protein interaction. Mad2p deletions that removed 10 residues from the N terminus or 5 residues from the C terminus, both of which disrupted Mad1p binding and abolished checkpoint function, also removed residues from the proposed HORMA domain (Figure 8).
Mad1p is less well conserved. The bulk of this protein is predicted to be coiled-coil, with a C-terminal globular domain. The level of conservation is higher toward the C terminus, and we have shown through co-immunoprecipitation studies that it is the last 30% of Mad1p (residues 528-749) that is critical for its Mad2p interaction. In studies on the human homologue of Mad1p (TXBP181; Jin et al., 1998), it was found that residues 465-584 are sufficient for the interaction of the human Mad1p and Mad2p in a two-hybrid assay. In our hands a similar region of yeast Mad1p (pKH610 contains residues 529-649; our unpublished data) failed to bind efficiently to Mad2p by co-immunoprecipitation. Although this could reflect real differences in functional domains between the yeast and human proteins, we are unable to rule out effects from fusion constructs and their stability on these results.
The extreme C terminus of Mad1p is clearly critical for its function. Removing the last 33 amino acids of Mad1p (in mad1-1) or a single amino acid change (A736 → T in mad1-2) 13 residues from the C terminus of Mad1p is sufficient to abolish its checkpoint function. Because both the mad1-1 and mad1-2 mutations affect the stability of Mad1p, it is possible that this explains their reduced ability to bind to Mad2p and act in the spindle checkpoint. However, the importance of the C terminus was confirmed in our Gal4-Mad1 co-immunoprecipitation studies, in which a fusion containing residues 529-749 (pKH603) of Mad1p bound Mad2p, but another containing residues 529-718 (pKH609) did not (Figure 7C).
The rest of Mad1p is much more forgiving: almost the entire N-terminal half can be deleted without any apparent effect, including the asparagine-rich domain, which might form a flexible hinge within a coiled-coil rod but is not conserved in other Mad1 homologues. It has previously been reported that Mad1p, Mad2p, and Mad3p can all be co-immunoprecipitated with Cdc20p (Hwang et al., 1998). Further studies will be necessary to determine whether other regions of the Mad1 protein are necessary for other protein–protein interactions.
Regulation of the Mad1p–Mad2p Complex
We find that co-transfection of MAD1 and MAD2 constructs into animal tissue culture cells leads to the production of a stable Mad1p–Mad2p complex, indicating that no other yeast proteins are necessary for its formation or maintenance. The Mad1p–Mad2p complex isolated from yeast is very stable in vitro, and formation of the complex in vivo appears to be independent of the cell cycle or checkpoint status. These molecules interact at both a mitotic arrest induced by microtubule disruption and at metaphase arrest induced by a cdc23 mutation (our unpublished data), indicating that kinetochore attachment has no apparent effect on the Mad1p–Mad2p interaction. However, we cannot rule out the possibility that unattached kinetochores may regulate a small fraction of the complex or have a subtle effect on the affinity between these molecules. In addition, all of the different phosphorylated isoforms of Mad1p that can be resolved on SDS-PAGE were found complexed with Mad2p, indicating that complex formation is not regulated by such phosphorylation. Our previous work has shown that in cells lacking Mad2p the level of Mad1p hyperphosphorylation is dramatically reduced, suggesting that complex formation improves the ability of Mad1p to act as a substrate for its kinase(s). This notion is supported by our observation that phosphorylation of Mad1p is also reduced in cells expressing truncated Mad2p molecules that fail to bind to Mad1p. In addition, all checkpoint-defective alleles of mad1 produce proteins that do not get phosphorylated (Hardwick and Murray, 1995; Brady and Hardwick, unpublished data). It has recently been shown that overexpression of a dominant BUB1 allele can lead to checkpoint activation without any apparent phosphorylation of Mad1p (Farr and Hoyt, 1998). The functional significance of Mad1p hyperphosphorylation remains unclear and will require the mapping of the Mad1p phosphorylation sites and their mutational analysis. Analysis of HsMad1 indicated that it is phosphorylated on serine during S, G2, and M phases (Jin et al., 1998).
Coimmunoprecipitation studies in Xenopus egg extracts suggest that all of Xmad1 is bound to Xmad2 and that only a fraction of Xmad2 is present in the complex (Chen et al., 1998), indicating that Xmad1 may be the limiting factor in the complex formation. Consistent with the Xenopus proteins, we now show that yeast Mad2p also exists in two different pools, a Mad1p-bound and a Mad1p-free pool and that all of Mad1p co-fractionates with Mad2p by gel filtration chromatography. However, it requires future studies to determine whether all of Mad1p is indeed in the complex containing Mad2p and whether Mad1p and/or another component is the limiting factor for the complex formation.
Possible Functions of the Mad1p–Mad2p Complex
Conservation of the Mad1p–Mad2p interaction in yeast, frog (Chen et al., 1998), and human (Jin et al., 1998) indicates the importance of this complex. The frog homologue of Mad1p, Xmad1, has been shown to recruit Xmad2 to unattached kinetochores (Chen et al., 1998). We have attempted to localize Mad2p in yeast cells; however, we have been unable to detect the protein with our polyclonal anti-Mad2p antibody or with an anti-myc epitope antibody when the myc-Mad2p fusion protein was expressed to the endogenous level (our unpublished data). When overexpressed, both Mad2p and GFP-Mad2p fusion protein are distributed throughout the whole cell (our unpublished data). Nevertheless, the conservation of the Mad1p–Mad2p complex during evolution suggests that the proteins likely function similarily in different organisms. We now show that the ability of Mad2p to bind to Mad1p appears to play an important role in Mad1p phosphorylation. Taken together, these results indicate that the functions of Mad1p and Mad2p are likely dependent on each other and that they regulate each other through direct interaction. Mad1p affects the ability of Mad2p and Mad3p to interact stably with the checkpoint effector Cdc20p (Hwang et al., 1998). It remains unclear precisely how the Mad proteins inhibit the function of Cdc20p. Recent in vitro studies have shown that a tetramerized form of recombinant human Mad2 protein is sufficient to inhibit the action of human Cdc20 if they are incubated together before incubation with the anaphase-promoting complex (APC) (Fang et al., 1998). Perhaps Mad1p plays a role in the formation of Mad2p multimers at unattached kinetochores, in which case the hyperphosphorylation of Mad1p may promote this activity.
Mad1p–Mad2p is one of several complexes known to be formed by spindle checkpoint components, although the precise roles that the formation and interaction of these complexes play in the checkpoint is currently unclear. Both the localization and the activity of checkpoint components could be regulated by complex formation. As mentioned above, in Xenopus Xmad1 recruits Xmad2 to kinetochores (Chen et al., 1998), and in mammalian cells the Bub3 protein binds to unattached kinetochores and appears to recruit both Bub1 (Taylor et al., 1998) and a protein that has homology to Mad3 and Bub1 (Chan et al., 1998; Taylor et al., 1998). In budding yeast Bub1p binds to and phosphorylates Bub3p, and it has been suggested that the formation of this complex affects the kinase activity of Bub1p (Roberts et al., 1994). The Mad1p–Mad2p complex could regulate both the localization and/or the activity of other spindle checkpoint components by providing a structural framework for the assembly of Mad and Bub protein complexes at kinetochores that lack bound microtubules. This could regulate their ability to interact with the APC and its associated regulators such as Cdc20p. In so doing the Mad1p–Mad2p complex would play a crucial role in the inhibition of APC activity by the spindle checkpoint.
ACKNOWLEDGMENTS
We thank all our lab members for their advice and encouragement. This work was supported by grants from National Institutes of Health and the Human Frontiers in Science Program (to A.W.M.), from National Institutes of Health (to R.-H.C.), and the Wellcome Trust (D.M.B. and K.G.H.). K.G.H. was a Special Research Fellow of the Leukemia Society of America. R.-H.C. was a Helen Hay Whitney postdoctoral fellow.
REFERENCES
- Aravind L, Koonin EV. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23:284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Juo P-C, Curran T, Blenis J. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene. 1996;12:1493–1502. [PubMed] [Google Scholar]
- Chen R-H, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein xmad1 recruits xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ, Chen RH, Murray AW. Microinjection of antibody to mad2 protein into mammalian cells in mitosis induces premature anaphase. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego: Academic Press; 1991. [Google Scholar]
- Hardwick K, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG. The spindle checkpoint. Trends Genet. 1998;14:1–4. doi: 10.1016/S0168-9525(97)01340-1. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Trotis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jin DY, Spencer F, Jeang KT. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Li R, Chen R-H, Murray AW. Feedback control of mitosis in budding yeast (correction) Cell. 1994;79:388. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li R, Havel C, Watson JA, Murray AW. The mitotic feedback control gene MAD2 encodes the α subunit of a prenyl transferase. Nature. 1993;336:82–84. doi: 10.1038/366082a0. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Roberts RT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hersko J, Luca F, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitination ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of bub3 is required for kinetochore localization of bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B type cyclins in yeast. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]