Abstract
Blot hybridization of thymocyte DNA from AKR/J mice was used to detect new proviral junction fragments as markers of clonality at different stages of viral leukemogenesis and to detect DNA rearrangements at the c-myc locus due to proviral insertion. Clonal populations of thymocytes were observed in mink cell focus-forming virus-injected mice as early as 35 days postinjection, at a stage distinguishable from frank leukemia by flow cytometric analysis and transplantation bioassay. Specific proviral integrations in the c-myc locus were detected in 15% of these early clones and in up to 65% of late-developing thymomas and frank leukemias. Thus, in this system c-myc activation appears to be a common mechanism in T-cell leukemogenesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boccara M., Souyri M., Magarian C., Stavnezer E., Fleissner E. Evidence for a new form of retroviral env transcript in leukemic and normal mouse lymphoid cells. J Virol. 1983 Oct;48(1):102–109. doi: 10.1128/jvi.48.1.102-109.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Cloyd M. W., Linemeyer D. L., Lander M. R., Rands E., Lowy D. R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature. 1982 Jan 7;295(5844):25–31. doi: 10.1038/295025a0. [DOI] [PubMed] [Google Scholar]
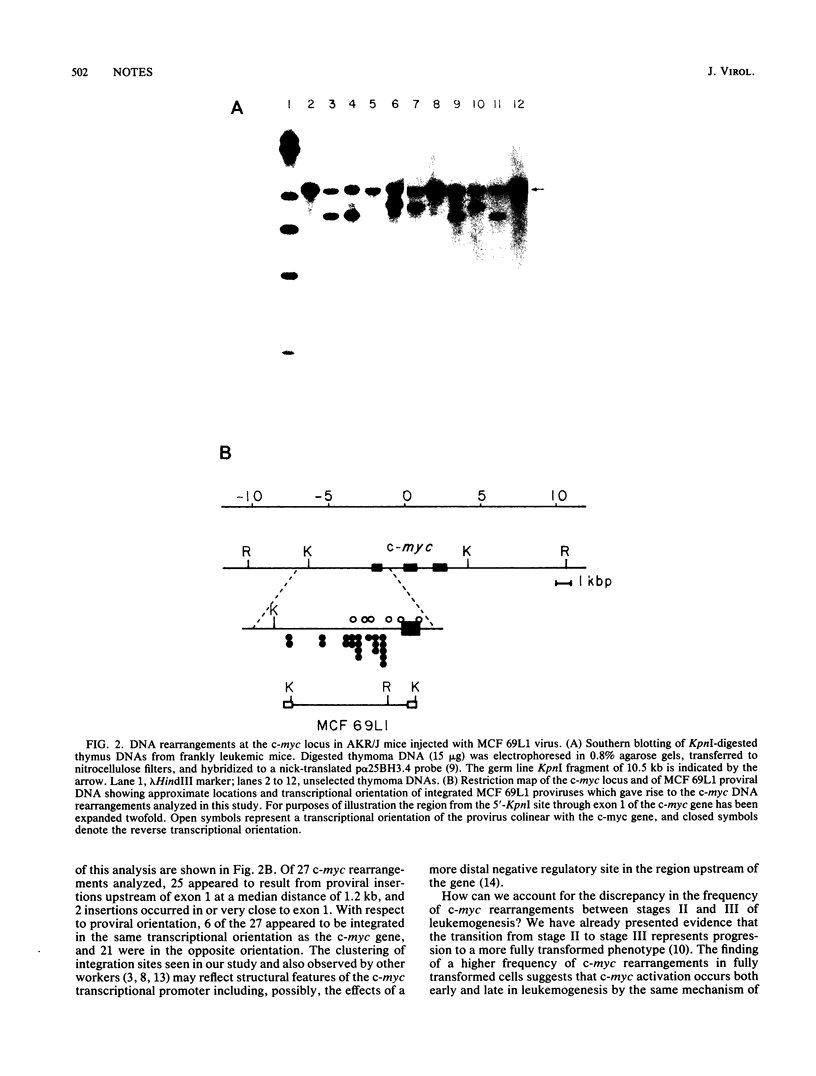
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Famulari N. G. Murine leukemia viruses with recombinant env genes: a discussion of their role in leukemogenesis. Curr Top Microbiol Immunol. 1983;103:75–108. doi: 10.1007/978-3-642-68943-7_4. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Herr W., Gilbert W. Somatically acquired recombinant murine leukemia proviruses in thymic leukemias of AKR/J mice. J Virol. 1983 Apr;46(1):70–82. doi: 10.1128/jvi.46.1.70-82.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Holland C. A., Hartley J. W., Hopkins N. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Harris L. J., Stanton L. W., Erikson J., Watt R., Croce C. M. Transcriptionally active c-myc oncogene is contained within NIARD, a DNA sequence associated with chromosome translocations in B-cell neoplasia. Proc Natl Acad Sci U S A. 1983 Jan;80(2):519–523. doi: 10.1073/pnas.80.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P. V., Woller R., Chu A. Stages in development of mink cell focus-inducing (MCF) virus-accelerated leukemia in AKR mice. J Exp Med. 1984 Sep 1;160(3):914–934. doi: 10.1084/jem.160.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint W., Quax W., van der Putten H., Berns A. Characterization of AKR murine leukemia virus sequences in AKR mouse substrains and structure of integrated recombinant genomes in tumor tissues. J Virol. 1981 Jul;39(1):1–10. doi: 10.1128/jvi.39.1.1-10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]