Abstract
Previous work has revealed a cytoplasmic pool of flagellar precursor proteins capable of contributing to the assembly of new flagella, but how and where these components assemble is unknown. We tested Chlamydomonas outer-dynein arm subunit stability and assembly in the cytoplasm of wild-type cells and 11 outer dynein arm assembly mutant strains (oda1-oda11) by Western blotting of cytoplasmic extracts, or immunoprecipitates from these extracts, with five outer-row dynein subunit-specific antibodies. Western blots reveal that at least three oda mutants (oda6, oda7, and oda9) alter the level of a subunit that is not the mutant gene product. Immunoprecipitation shows that large preassembled flagellar complexes containing all five tested subunits (three heavy chains and two intermediate chains) exist within wild-type cytoplasm. When the preassembly of these subunits was examined in oda strains, we observed three patterns: complete coassembly (oda 1, 3, 5, 8, and 10), partial coassembly (oda7 and oda11), and no coassembly (oda2, 6, and 9) of the four tested subunits with HCβ. Our data, together with previous studies, suggest that flagellar outer-dynein arms preassemble into a complete Mr ≃ 2 × 106 dynein arm that resides in a cytoplasmic precursor pool before transport into the flagellar compartment.
INTRODUCTION
Chlamydomonas flagella are made up of at least 150 different proteins (Piperno et al., 1977) and show an intricate structural arrangement, with complex components such as inner and outer dynein arms, radial spokes, and a ring of nine doublet microtubules surrounding two central singlet microtubules (Ringo, 1967; reviewed in Dutcher, 1995). At the flagellar base or transition zone (Randall et al., 1967), flagellar components appear to be functionally separated from the cell body (Musgrave et al., 1986; Jarvik and Suhan, 1991). Since neither DNA nor ribosomes have been found within flagella (Ringo, 1967; Johnson and Rosenbaum, 1990, 1993), all flagellar proteins must be transported from the cytoplasm for assembly into the complex flagellar structure. How these proteins are selected and transported through the transition zone is unknown (reviewed by Dentler, 1981).
Studies of many flagellated and ciliated cells have revealed a cytoplasmic pool of precursor protein ca-pable of contributing to the assembly of new flagella or cilia under conditions where synthesis of new proteins is inhibited (Rosenbaum and Child, 1967; Rosenbaum et al., 1969; Child and Apter, 1969; Stephens, 1977). In Chlamydomonas this pool is sufficient for assembly of two new half-length flagella (Rosenbaum et al., 1969). In spite of these studies, several questions about flagellar organelle assembly remain little understood, including the location of flagellar precursor proteins in the cytoplasm, how they are transported to the flagellar compartment, whether most proteins are transported as individual subunits or as complexes, and what limits assembly and thus determines flagellar length. In this paper we explore assembly mechanisms of a multisubunit flagellar ATPase, the outer dynein arm.
Recent observations suggest that dynein arms may be preassembled in the cytoplasm and transported to the flagellum as a complex. In Paramecium, a dynein complex has been isolated from the cytoplasm that has the same sedimentation rate (22S) as extracted axonemal outer dynein arms and contains a heavy chain (HC)1 with a similar vanadate cleavage pattern and antibody cross- reactivity to an axonemal HC present in extracted 22S axonemal dynein (Fok et al., 1994). In Chlamydomonas, an inner-row dynein subunit (p28), extracted from the flagellar cytoplasmic matrix with nonionic detergent, sediments on sucrose gradients as part of a larger complex, suggesting that at least some inner-row dynein subunits assemble before their association with doublet microtubules (Piperno and Mead, 1997).
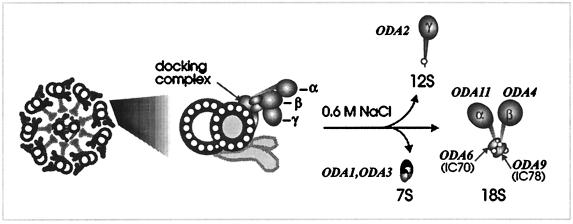
Indirect evidence suggests that some Chlamydomonas dynein mutants with assembly defects accumulate partially assembled complexes in their cytoplasm. When Chlamydomonas gametes of opposite mating type are mixed, fusion results in temporary dikaryons with four flagella and a common cytoplasm. Fusion between gametes containing mutations at different loci can lead to cytoplasmic complementation with assembly of functional components into the flagella and restoration of normal or near-normal motility (Luck et al., 1977; Johnson and Rosenbaum, 1993). However, certain combinations of the 14 known outer-dynein arm assembly (oda) loci fail to complement in dikaryons (Huang et al., 1979; Kamiya, 1988; Luck and Piperno, 1989; Koutoulis et al., 1997; for a general discussion of nonallelic noncomplementation in Chlamydomonas, see Dutcher and Lux, 1989). Kamiya (1988) assayed cytoplasmic complementation by measuring restoration of beat frequency in temporary dikaryons that formed between pairwise combinations of oda mutant gametes. Based on his data, oda mutants fall into one of three groups defined by their inability to complement either oda1 (oda1 and oda3), oda2 (oda2, 4, 6, 7, and 9), or oda5 (oda5, 8, and 10); these data are summarized in Table 1. One explanation for this lack of cytoplasmic complementation is that each gamete preassembles nonmutant subunits into partial dynein complexes that are incapable of dissociation and reassociation into wild-type complexes when gametes of opposite mating type fuse. Alternatively, absence of one protein may cause instability of a binding partner/partners or sequestration of the remaining proteins in a compartment that is unavailable to the assembly mechanism. When outer dynein arms are extracted from Chlamydomonas flagellar axonemes, they typically dissociate into three smaller subcomplexes that can be separated by sucrose gradient fractionation into 18S, 12S, and 7S components (Piperno and Luck, 1979; Takada and Kamiya, 1994) as illustrated diagrammatically in Figure 1. Our data show that complexes do exist in the cytoplasm before their attachment onto axonemal microtubules, but these complexes are not identical to the complexes produced by extraction from axonemes.
Table 1.
Complementation in temporary dikaryons between oda strainsa
| oda1 | oda2 | oda3 | oda4 | oda5 | oda6 | oda7 | oda8 | oda9 | oda10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| oda1 | − | |||||||||
| oda2 | + | − | ||||||||
| oda3 | − | + | − | |||||||
| oda4 | + | − | + | − | ||||||
| oda5 | + | + | + | + | − | |||||
| oda6 | + | − | + | − | + | − | ||||
| oda7 | + | − | + | − | + | − | − | |||
| oda8 | + | + | + | + | − | + | + | − | ||
| oda9 | + | − | + | + | + | − | + | + | − | |
| oda10 | + | + | + | + | − | + | + | − | + | − |
+ Indicates complementation, defined as dikaryon beat frequency greater than 35 Hz, and is based on the data reported in Table 3 of Kamiya (1988) in which the mean frequency of all dikaryons indicated above as noncomplementing (−) was 26 ± 4 Hz (n = 22), while that of all complementing (+) dikaryons was 42 ± 3 Hz (n = 33).
Figure 1.
Diagram illustrating the relationship between outer-row dynein arms in situ and after dissociation by 0.6 M NaCl into 7S, 12S, and 18S particles. The loci (ODA1, ODA2, etc.) that are known to encode subunits of each particle are also indicated.
Outer dynein arms in Chlamydomonas contain 3 HCs of ∼500 kDa each (HCα, HCβ, and HCγ), 2 intermediate chains (IC78, and IC70), about 10 light chains (LCs) ranging from 22 to 8 kDa, and a 7S factor of three proteins that form an outer dynein arm attachment site or docking complex (DC105, DC62.5, and DC25) (Piperno and Luck, 1979; Pfister et al., 1982; King and Witman, 1990; Takada and Kamiya, 1994; Koutoulis et al., 1997). For this report we tested outer-dynein arm subunit stability in the cytoplasm of wild-type and 11 assembly mutant strains by probing Western blots of cytoplasmic extracts with several subunit-specific antibodies. To test the state of dynein assembly in these extracts, we used the same antibodies to probe Western blots of immunoprecipitates prepared with a single subunit-specific antibody. The 11 loci marked by oda mutations include those known to encode five of the enzyme subunits and two proteins of the dynein attachment complex (summarized in Table 2 and Figure 1). Gene products of the remaining 6 loci are unknown. Two additional mutations, pf13 and pf22, also reduce outer-row dynein assembly, but because cells carrying these mutations have short paralyzed flagella (Huang et al., 1979), they represent a different class of flagellar assembly defect than the oda mutants and were not included in this study. Our results show that dynein subunits preassemble in the cytoplasm and that both preassembly of dynein complexes and protein instability contribute to dikaryon cytoplasmic noncomplementation. These results provide new information on subunit interactions, dynein mutant phenotypes, and the process of flagellar assembly.
Table 2.
Characteristics of outer-dynein arm assembly mutants
| Mutation | Gene product | Beat frequency | Assembly defect | Reference |
|---|---|---|---|---|
| oda1 | DC63 | 25 Hza | Docking complex + outer arm | bc |
| oda2 (pf28) | HCγ | 25 Hza | Outer arm | bd |
| oda3 | DC105 | 25 Hza | Docking complex + outer arm | be |
| oda4 | HCβ | 25 Hza | Outer arm | bf |
| oda5 | ? | 25 Hza | Outer arm | b |
| oda6 | IC70 | 25 Hza | Outer arm | bg |
| oda7 | ? | 25 Hza | Outer arm | b |
| oda8 | ? | 25 Hza | Outer arm | b |
| oda9 | IC78 | 25 Hza | Outer arm | bh |
| oda10 | ? | 25 Hza | Outer arm | b |
| oda11 | HCα | 55 Hz | HCα + 16 kD LC | bi |
Wild-type beat frequency = 60 Hz.
MATERIALS AND METHODS
Mutant Strains
All of the mutant strains of Chlamydomonas reinhardtii used in this study have been described previously (see Table 2). The oda2 allele used was pf28 (Mitchell and Rosenbaum, 1985). Cells were maintained on minimal medium using standard procedures (Harris, 1989).
Cell Cytoplasmic Extract
Chlamydomonas cells were grown in 500 ml of liquid M medium (Sager and Granick, 1953) with aeration in continuous light to a density of 106 cells/ml, harvested by centrifugation (550 × g for 6 min at 22°C), and resuspended in ice-cold HMDEK (10 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.1 mM EDTA, 25 mM potassium chloride, pH 7.4) to a total of 500 μl. The suspension was transferred to a 1.5-ml microfuge tube that contained an equal volume of acid-washed glass beads (1 mm) and vortexed at setting 6.5 on a Genie II vortexer for 1 min. Cell suspensions were then centrifuged using a Beckman L8 centrifuge at 48,000 × g, at 4°C for 2 h. Supernatants were either used for immunoprecipitation as described below or mixed with 0.25 volume of 4× sample buffer (8% SDS, 40% glycerol, 125 mM Tris-HCl, pH 6.8, with Pyronin Y added as tracking dye) and β-mercaptoethanol, to a final concentration of 0.7 M, and stored at −20°C for SDS-PAGE. Pellets were resuspended in 500 μl HMDEK, mixed with 0.25 volume 4× sample buffer, and stored at −20°C for SDS-PAGE. Protein concentration was determined by the Bradford dye binding method using BSA as a standard (Bradford, 1976).
Axonemal Preparation
Axonemes were isolated by the method of Witman et al. (1978). Cells were grown in 500 ml of liquid M medium (Sager and Granick, 1953) with aeration in continuous light to a density of 106 cells/ml, harvested by centrifugation at 550 × g for 6 min at 22°C, washed with 10 mM HEPES, pH 7.4, centrifuged again, and resuspended in 10 ml HMDS (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM DTT, and 4% sucrose). Resuspended cells were deflagellated with 400 μl 50 mM dibucaine (CIBA Pharmaceutical, CIBA-GEIGY, Summit, NJ) and diluted with 10 ml ice-cold HMDS containing 2 mM EGTA and 2 mM phenylmethylsulfonyl fluoride, and cell bodies were removed by centrifugation at 4°C for 7 min at 1,550 × g. The supernatant was collected and recentrifuged as above. Cell-free supernatant was then centrifuged at 31,000 × g to pellet axonemes, which were resuspended in HMDEK and an equal volume of 2× sample buffer. β-Mercaptoethanol was added to a final concentration of 0.7 M, and samples were stored at −20°C.
SDS-PAGE and Western Blotting
Samples were prepared and run with Tris-glycine-buffer (Laemmli, 1970) in 5% stacking gels and 5, 7, or 12% separating gels (designated in text) prepared from stocks that contained 30% acrylamide and 0.4% bis-acrylamide. Broad Range protein standards (New England Biolabs, Beverly, MA) of 212, 158, 116, 97.2, 66.4, 55.6, and 42.7 kDa were used, and gels were either stained with Coomassie Blue to show total protein or transferred to immobilon membrane (Millipore, Bedford, MA) for Western blotting following the recommendations of Burnette (1981). Gels were soaked in transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol) for 10 min and transferred either at 200 mA for 12 h (Figures 2, 3, 4A, and 5) or at 300 mA for 6 h (Figures 4B and 6). Protein standard lanes were separated from sample lanes and stained with amido black. Antibody binding and detection were performed as directed in the POD chemiluminescence kit (Boehringer Mannheim, Indianapolis, IN). Briefly, transferred blots were blocked with 1% POD blocking solution for 1 h at room temperature, incubated with the primary antibody in 0.5% POD blocking solution for 3 h at room temperature, washed with TBST (50 mM Tris base, 150 mM NaCl, pH 7.5, with 0.1% Tween-20 [vol/vol]) 2 × 10 min, washed with 0.5% POD blocking solution 2 × 10 min, incubated with secondary antibody in 0.5% POD blocking solution for 1 h at room temperature, washed with TBST four times for 10 min, and then incubated with developing solution for 1 min and exposed to Biomax film (Eastman Kodak, Rochester, NY). Antigen quantitation was estimated by comparison with a blot of an antigen dilution series (2, 1, 0.8, 0.6, 0.4, 0.2, 0.1 x WT control) processed in parallel.
Figure 2.
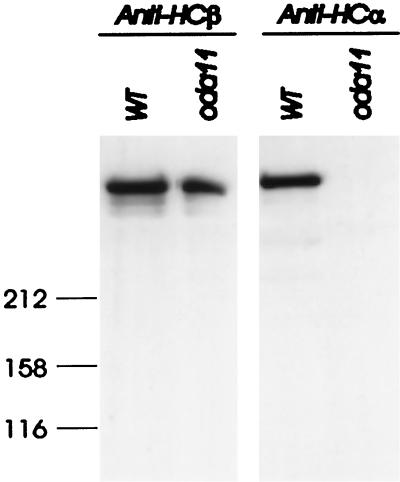
Specificity of antibody B3B. WT and oda11 flagella, run on 5% gels and blotted to PVDF membrane, were probed with C11.6 (left panel), which detected equal levels of HCβ in both samples. A parallel blot probed with antibody B3B (right panel) detected a single band in WT flagella that is missing in flagella of HCα assembly mutant oda11. Antibodies were detected with a peroxidase-linked secondary antibody and chemiluminescent detection.
Figure 3.
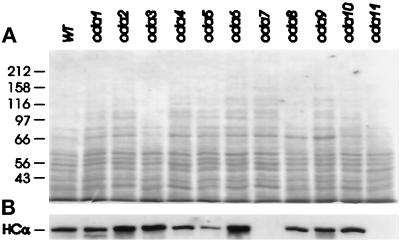
Western blots of HCα in WT and oda cytoplasmic extracts. Samples of cytoplasmic extracts from wild-type and oda1-oda11 cells were separated on 7% gels and blotted to PVDF membrane. Each lane was loaded with 20 μg of total protein. (A) Gel stained with Coomassie blue. (B) Western blot probed with B3B (anti-HCα). No HCα was detectable in either oda7 or oda11; levels in oda5 are 50% of WT.
Figure 4.
Western blots of WT and oda cytoplasmic extracts test subunit stability. Cytoplasmic extracts of WT and oda1-oda11 prepared as in Figure 2 were probed with mAbs C11.6 (anti-HCβ), 25–8 (anti-HCγ), 1878A (anti-IC78), and 1869A (anti-IC70). (A) Outer dynein arm proteins (indicated along the right margin) present in cytoplasmic extracts of WT and oda1–11. Most oda mutant samples display antigen levels similar to WT, but there are several exceptions as discussed in text. Multiple bands below HCβ in WT, oda1, and oda3 are due to the breakdown of HCβ. No detectable HCβ was found in oda4, no HCγ was found in oda2, no IC78 was found in oda9, and no IC70 was found in oda6 (at this exposure; see text and Figure 3C). 1878A (anti-IC70) cross-reacted with cytoplasm proteins migrating faster than 78 kDa (asterisk) that are apparently unrelated to flagellar dynein. (B) Western blots of a second set of independently prepared cytoplasmic extracts from oda5-oda10 probed with 1878A (anti-IC78) and 1869A (anti-IC70) that illustrate the variability in the reductions of IC70 in oda9 and IC78 in oda6 (compare with bottom two panels in panel A). (C) Upper and lower panels show the results of a 10-fold increase in exposure time for lanes oda5–oda7 from the bottom panels of Figure 3A and Figure 3B, respectively, and reveal an IC70 band in oda6 cytoplasmic extracts.
Figure 5.
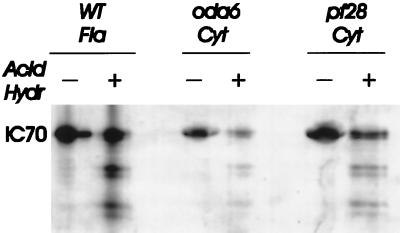
Fragmentation of 70-kDa antigens by acid hydrolysis. Proteins of ∼70 kDa were purified by SDS-PAGE from WT axonemes, oda6 cytoplasmic extract, and pf28 cytoplasmic extract, digested by partial hydrolysis with formic acid, separated on a 12% gel, blotted to PVDF membrane, and probed with 1869A (anti-IC70). All three samples generate similar fragmentation patterns.
Figure 6.
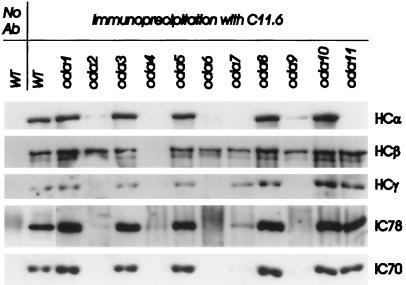
Immunoprecipitation of preassembled complexes. Immunoprecipitates with C11.6 (anti-HCβ) or no antibody (control) from WT and oda1–oda11 cytoplasmic extracts were separated on 7% gels, blotted to PVDF membrane, and probed with B3B (anti-HCα), C11.6 (anti-HCβ), 25–8 (anti-HCγ), 1878A (anti-IC78), or 1869A (anti-IC70). Immunoprecipitates of oda1 extracts showed the presence of all five major outer-dynein arm proteins at equivalent levels to WT, as did immunoprecipitates of oda3, 5, 8, and 10. The immunoprecipitate of oda11 lacked only HCα; the immunoprecipitate of oda7 lacked HCα and had greatly reduced levels of IC70 and IC78, whereas no proteins coprecipitated with HCβ in oda2, oda6, or oda9. The oda4 lanes serve as a no-antigen control.
Antibodies
Anti-HCβ mAb C11.6 was concentrated by ammonium sulfate precipitation from hybridoma culture supernatants and used at a 1:100 dilution. It was generated from the same fusion as C11.13 (Mitchell and Rosenbaum, 1986). Anti-IC70 mAb 1869A ascites was used at 1:2000 dilution. Anti-IC78 mAb 1878A hybridoma culture supernatant was kindly donated by Dr. G. Witman (King et al., 1985) and was used at a 1:2 dilution. Anti-HCα polyclonal antibody B3B was produced in a rabbit by immunization with a purified bacterial fusion protein. A 1-kilobase (kb) PmlI/HpaI fragment of HCα cDNA pBcA6 (Mitchell and Brown, 1997), which encodes amino acids 512–838 of HCα (a region unrelated to other DHC sequence and that contains the HCα EPAA repeat element), was cloned into vector pGEX-4T-2 (Pharmacia Biotech, Piscataway, NJ) at a SmaI site and was transformed into DH5αF′ E. coli. Fusion protein expression was induced for 3 h with 0.1 mM isopropyl-β-thiogalactopyranoside at 37°C. Fusion protein was solubilized and purified by the method of Frangioni and Neel (1993), run on an SDS-PAGE gel, transferred to nitrocellulose, and visualized with Ponceau S. Nitrocellulose strips containing the protein of interest were dissolved in dimethyl sulfoxide, mixed with adjuvant, and used for immunization. Specific antibodies were affinity purified from whole sera using Western blots of the fusion protein (Frangioni and Neel, 1993) and were used at a dilution of 1:50. Anti-HCγ mAb 25–8, kindly donated by Dr. G. Piperno (King and Witman, 1988), was used at a 1:10 dilution. Peroxidase-labeled goat anti-mouse or goat anti-rabbit secondary antibodies (Bio-Rad Laboratories, Hercules, CA) were used at a 1:6000 dilution.
Immunoprecipitation
Cell extracts (0.5 ml) were mixed in a 15-ml conical tube with an equal volume of ice-cold immunoprecipitation (IP) buffer (HMDEK, 75 mM NaCl, 0.01% thimersol, 0.5 mM PMSF, 3% BSA, 0.1% Triton X-100, pH 7.5) and preabsorbed with 25 μl of 50/50 (vol/vol) protein A agarose in IP buffer for 30 min on ice. mAb anti-HCβ antibody C11.6 was added to preabsorbed extracts and incubated for 3–4 h at 4°C. A 100-μl volume of 50/50 (vol/vol) protein A agarose (Sigma Chemical, St. Louis, MO) in IP buffer was added, and the tubes were mixed gently for 1 h. Agarose beads were washed three times with IP buffer containing 0.05% Triton X-100. Immune complexes were eluted by addition of 2× sample buffer containing 0.7 M β-mercaptoethanol and incubation for 2 min in boiling water. The quantity of HCβ immunoprecipitated with antibody C11.6 from wild-type and oda mutant cytoplasmic extracts was determined from preliminary Western blots. Subsequent loads of immunoprecipitate samples were adjusted to include equal amounts of HCβ.
Partial Acid Hydrolysis
Western blots of proteins subjected to partial acid hydrolysis were generated by a modification of the method described by Cleveland et al. (1977). Briefly, cytoplasmic extracts or whole axonemes were run on an SDS-PAGE gel along with molecular weight markers. The gel was stained in 0.1% Coomassie blue, 50% methanol, and 10% acetic acid for 30 min and destained in 5% methanol and 10% acetic acid for 45 min. Bands of ∼70 kDa molecular mass were cut from the gel and soaked 30 min in 0.125 M Tris/HCl, pH 6.8, 0.1% SDS, 1 mM EDTA. Slices were then lyophilized and either stored frozen at −20°C (controls) or incubated with 70% formic acid for 16 h at 37°C, washed with 50% methanol, and lyophilized. Gel slices were then rehydrated with buffer (1% SDS, 10 mM Tris/HCl, pH 8, 0.1% β-mercaptoethanol, and 10% glycerol) for 6 h, pushed into the bottom of an SDS-PAGE well, and overlayed with 10 μl of Tris/HCl buffer containing 20% glycerol for electrophoresis and transfer to PVDF membranes.
RESULTS
Western Blots of Cytoplasmic Extracts
Based on previous studies of flagellar regeneration, Chlamydomonas cells maintain a cytoplasmic pool of flagellar subunits sufficient to assemble two half-length flagella (reviewed by Johnson and Rosenbaum, 1993). Our first goal was to determine, for several outer-dynein arm subunits, whether similar levels of protein were present in cytoplasmic extracts of wild-type (WT) cells and of outer-dynein arm assembly mutants. Preliminary tests showed that our previously described HCα antibody C2.14 (Mitchell and Rosenbaum, 1986) lacked sufficient sensitivity for these studies. Therefore, a polyclonal rabbit serum (B3B) was produced by immunization with a bacterial GST fusion protein encoding a portion of HCα (see MATERIALS AND METHODS). The specificity of this antibody is demonstrated in Figure 2, in which Western blots of flagellar proteins from WT cells and from cells harboring the oda11 mutation (which blocks HCα assembly; see Table 2) were probed with C11.6 (anti-HCβ) and with B3B. Antibody C11.6 detects HCβ in both WT and oda11 flagella (left panel), whereas B3B detects HCα in WT but not oda11 flagella (right panel). When cytoplasmic extracts of WT and 11 oda mutants were probed with this antibody, it was found that similar signals were present in all but three strains. The amount of HCα was less than half the normal levels in oda5 and was completely missing in oda7 and oda11 (Figure 3). In three independent cytoplasmic extracts, the level of HCα in oda5 varied between 40% and 60% of WT, whereas no antigen was detectable in either oda7 or oda11. Since the HCα gene has been shown to be closely linked to oda11, but not oda7 (Sakakibara et al., 1991), our data support the assignment of oda11 as the HCα locus and suggests that HCα is synthesized normally in oda7 but is unstable in this mutant background. Identical results were obtained with extracts from an oda7 strain generated by two rounds of backcrosses to wild-type strain 137c, supporting linkage between oda7 and this apparent defect in HCα stability.
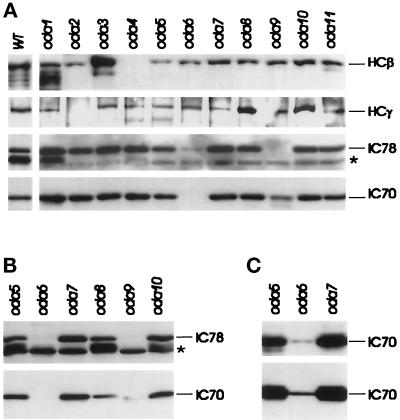
HCβ, like HCα, was present at reduced levels in oda5 cytoplasm and was undetectable in oda4 cytoplasm (Figure 4A). Absence of HCβ in oda4 is expected based on previous identification of oda4 as the HCβ locus, whereas its reduction in oda5 suggests that HCs may have reduced stability in the absence of the oda5 gene product. In addition, proteolytic fragments of HCβ were always more prominent in oda1 and oda3 than in any of the other strains. These two loci encode gene products that form part of the DC (Table 2), and lack of HCβ stability therefore indicates that the DC likely interacts with HCβ in the cytoplasm, altering its susceptibility to proteolysis. Similar analysis of HCγ revealed the presence of this HC in the cytoplasm of all oda mutants except oda2 (Figure 4A), which has previously been identified as a mutation in the HCγ gene. No quantitative conclusions could be drawn regarding HCγ levels in the remaining strains using this antibody.
When these same blots were probed with anti-IC78 and anti-IC70 (Figure 4A, bottom two panels) two unanticipated results were found. The anti-IC78 antibody, whose specificity for a single flagellar dynein IC has already been documented (King et al., 1985, 1986), cross-reacted with cytoplasmic protein bands of slightly lower apparent molecular weight than IC78 (asterisk in Figure 4). These bands showed variable levels of signal intensity unrelated to the strain used, but their presence in oda9 cytoplasm suggests that they are not due to breakdown of the IC78 protein since oda9 is a mutation in the IC78 gene. Since flagellar and cytoplasmic dynein ICs are homologous (Wilkerson et al., 1995), these bands might be due to antibody cross-reactivity to a cytoplasmic dynein IC. IC78 was not only absent in oda9 cytoplasm, but was also reduced in oda5 (40% of WT) and oda6 (10% of wt) as well. Similarly, IC70 was not only missing in oda6, but was also reduced to 60% in oda5 and 30% in oda9. Both the level of IC70 in oda9 cytoplasm and the level of IC78 in oda6 cytoplasm varied among independent sample preparations, from the levels seen in the bottom two panels of Figure 4A to those illustrated in Figure 4B. We were unable to determine the cause of this variation, but the reduction of IC78 in oda6 was consistently greater than the reduction of IC70 in oda9.
When blots of oda6 cytoplasmic extract probed with anti-IC70 were exposed for extended periods, small amounts (2% of WT) of antigenic protein that migrated identically to flagellar outer dynein arm IC70 were detectable (Figure 4C). Sequence analysis has shown that oda6–95, the allele used in this study, is a frameshift that encodes a truncated protein 10% the size of full-length IC70, and Western blots with an anti-IC70 mAb confirmed that oda6–95 flagella lack detectable IC70 (Mitchell and Kang, 1993). To further test the nature of this antigen, patterns generated by partial acid hydrolysis of the oda6–95 cytoplasmic 70-kDa antigen were compared with patterns generated from WT flagellar IC70 and from 70-kDa antigens present in the cytoplasmic extract of an oda2 strain (pf28 allele). All three samples consisted of SDS-PAGE–purified 70-kDa proteins, which were then subjected to hydrolysis by formic acid, separation of hydrolysis products by SDS-PAGE, and Western blotting with mAb 1869A. All three samples showed identical fragment patterns (Figure 5). From this we concluded that the 70-kDa antigen in oda6 cytoplasm represents a bona fide IC70 protein present at very low levels. In spite of its presence, no IC70 has been seen in oda6 flagella (Mitchell and Kang, 1993) or in coprecipitates with HCβ from oda6 cytoplasm (detailed below). We do not believe the IC70 seen in Figure 4C results from the appearance of spontaneous revertants in our cultures since the spontaneous reversion frequency of oda6–95 is low (Mitchell and Kang, 1991), and since we have never observed phenotypic revertants in these cultures. However, residual levels of gene expression in frameshift mutations such as oda6–95 can result from +1, +2, −1, or −2 shifts during ribosomal translation (Weiss et al., 1987), and a string of four cytosines upstream of the oda6–95 mutation site could allow a −1 translational shift that would result in synthesis of a full-length protein with only eight amino acids different from WT. We previously analyzed an intragenic pseudo-revertant of oda6–95 (oda6-r88), in which a second frameshift mutation had occurred 23 codons upstream of the oda6–95 mutation. Since this revertant allele generates an IC70 protein that assembles into flagellar outer dynein arms (Mitchell and Kang, 1993), the hypothesized gene product generated by a translational frameshift of oda6–95 mRNA should also support dynein assembly, but may be synthesized at a rate too low to allow coassembly with other subunits.
Westerns of Immunoprecipitates from Cytoplasmic Extracts
Our second goal was to determine whether immunoprecipitation of one dynein subunit from a native cytoplasmic extract would result in coprecipitation of a preassembled complex. For these experiments we chose an anti-HCβ mAb, since it has already been demonstrated that an outer-dynein arm complex containing two HCs, two ICs, and several LCs can be immunoprecipitated from crude flagellar extracts using anti-HCβ mAb C11.13 (Mitchell and Rosenbaum, 1986). mAb C11.6, which was prepared from the same hybridoma fusion as C11.13 and recognizes the same HCβ epitope (see MATERIALS AND METHODS), was used because of instability of the C11.13 hybridoma cell line. When C11.6 immunoprecipitates of WT cytoplasmic extracts were transferred and probed with anti-HCα, anti-HCβ, anti-HCγ, anti-IC78, and anti-IC70 antibodies, all five subunits were found to be present (Figure 6, WT lanes).
We then repeated this procedure with oda mutant cytoplasmic extracts. Since HCβ is present in all oda mutants (except oda4), this experiment should reveal proteins associated with HCβ in each mutant. Immunoprecipitates from cytoplasmic extracts of oda1 contained all five tested subunits (HCα, HCβ, HCγ, IC78, and IC70) as did immunoprecipitates of oda3, oda5, oda8, and oda10 extracts, demonstrating that none of these five mutations prevent association of HCs and ICs. The immunoprecipitate from oda4 cytoplasm showed no antigenic protein in any blot, which was expected since the oda4 mutation disrupts the antigen target used for these immunoprecipitations. Immunoprecipitates from oda11 contain all the major outer dynein arm proteins except HCα, but the oda2, oda6, oda7, and oda9 mutations had more drastic effects. Those from extracts of oda2, oda6, and oda9 showed the presence of only HCβ (Figure 6). Absence of HCγ (oda2) or the IC dimer (oda6 and oda9) thus prevent preassembly of HCβ with the remaining tested subunits, even though those subunits are present in the cytoplasm at approximately WT levels. Although faint HCα bands are detectable in oda2 and oda9 immunoprecipitates (Figure 6, top panel), with longer exposures such bands became visible in all lanes except those already shown to completely lack this antigen (see Figure 3) and are due to nonspecific binding to the protein A agarose used in the immunoprecipitation. Immunoprecipitates of oda7 revealed a complex between HCβ, HCγ, and faint traces of IC78 and IC70, but did not contain HCα. Both IC70 and IC78 are present at normal levels in oda7 cytoplasm (oda7 lanes in Figure 4); therefore, their reduction in oda7 immunoprecipitates indicates an effect on IC–HC interactions. The IC dimer appears essential for joining HCs together since neither HCα nor HCγ coprecipitate with HCβ when the IC dimer is missing (oda6 and oda9 lanes in Figure 6). The oda7 defect thus appears to allow an association between the ICs and HCs that is strong enough to support assembly between HCβ and HCγ, but not strong enough to withstand immunoprecipitation.
DISCUSSION
In this report we have examined the stability and assembly state of several outer-dynein arm subunits that reside in a cytoplasmic pool of flagellar precursor proteins. Although the existence of a Chlamydomonas flagellar precursor pool has been established, the assembly state of proteins within this precursor pool has not been previously examined. We show, by coprecipitation of five flagellar outer-dynein arm subunits with a mAb against one subunit, that large preassembled flagellar complexes exist within this cytoplasmic pool (Figure 6). A preassembled complex of flagellar radial spoke proteins has been observed in Chlamydomonas cytoplasmic extracts (Diener et al., 1996), and inner-row dyneins may also preassemble (Piperno and Mead, 1997), which suggests that many flagellar components assemble to varying degrees in a cytoplasmic pool before flagellar assembly.
We then examined 11 outer-dynein arm assembly mutants, to determine whether lack of flagellar assembly reflected loss of subunit stability, a block to cytoplasmic preassembly, or potential defects in transport and binding of preassembled complexes onto flagellar doublet microtubules. Defects in each of these three categories were observed. We confirmed the previous identification of gene products for oda2 (HCγ), oda4 (HCβ), oda6 (IC70), and oda9 (IC78) loci, since each antigen is missing in the corresponding mutant strain (Figures 3 and 4). The absence of detectable HCα antigen in either oda11 flagella (Figure 2 and Sakakibara et al., 1991) or cytoplasm (Figure 3) also supports the identification of oda11 as the HCα locus (although the oda11 and HCα loci are genetically linked, direct evidence of allelism is lacking [Sakakibara et al., 1991]). At least four oda mutations affect the abundance of a subunit that is not the mutant gene product, a result that we interpret as an alteration in subunit stability. There was no detectable HCα antigen in oda7 cytoplasmic extracts even though all other tested outer-dynein arm proteins were present at approximately WT levels (Figure 3), which may indicate that the WT oda7 gene product interacts with and stabilizes HCα. The only subunit known to directly interact with HCα is its tightly associated 16-kDa LC (Mitchell and Rosenbaum, 1986), which has recently been cloned and identified as a thioredoxin homologue (Patel-King et al., 1996). The oda7 locus probably does not encode the 16-kDa LC, since absence of this LC (along with HCα) from outer-dynein arms of oda11 flagella does not prevent assembly of the remaining outer-dynein arm subunits (Sakakibara et al., 1991), whereas the oda7 mutation clearly does (Kamiya, 1988). In addition to its affect on HCα stability, oda7 also prevents preassembly of the remaining subunits into a stable complex; the two remaining HCs remain associated during immunoprecipitation, but the ICs are largely lost. Could oda7 be essential for a posttranslational modification that is needed both for stability of HCα and for a strong HC–IC interaction? Two possibilities include regulation of the redox states of the 16-kDa (HCα-associated) and 19-kDa (HCβ-associated) thioredoxin homologous LCs (Patel-King et al., 1996), and regulation of the phosphorylation state of HCα, which is the only outer-dynein arm subunit known to be phosphorylated in vivo (Piperno and Luck, 1981; King and Witman, 1994). The significance of thioredoxin homology in these LCs has not yet been determined, and the physiological role of HCα phosphorylation is also presently unknown.
The two other mutations that alter stability have reciprocal effects, a mutation in IC70 (oda6), reducing IC78 abundance, and a mutation in IC78 (oda9), affecting IC70 levels (Figure 4). These two proteins can be purified from partially dissociated flagellar extracts as a stable heterodimer (Mitchell and Rosenbaum, 1986), and although full-length in vitro translation products are apparently stable as individual proteins (King et al., 1995), dimerization may be required for stability in vivo. Absence of this dimer, in turn, prevents cytoplasmic preassembly of HCα, HCβ, and HCγ (Figure 6).
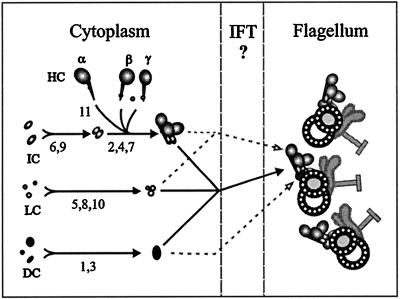
In five strains (oda1, 3, 5, 8, 10) all five HC and IC subunits tested were preassembled in the cytoplasm, but this preassembled complex could not be transported from the cytoplasm and bound onto flagellar doublets. The data of Kamiya (1988) suggest that these loci encode subunits of two additional preassembled complexes (Table 1). We hypothesize that loci in each group encode proteins that form preassembled complexes, and that the HC–IC complex described here is the complex disrupted by oda2, 4, 6, 7, or 9. This is the first direct demonstration that lack of complementation among oda mutants results from the prevention of complex preassembly. The oda1 and oda3 gene products copurify from flagella as subunits of a 7S DC that can assemble into flagella independently of other outer-dynein arm subunits (Takada and Kamiya, 1994; Koutoulis et al., 1997); therefore, lack of cytoplasmic complementation between oda1 and oda3 (Kamiya, 1988) suggests that the gene products of these loci also preassemble into a separate complex in the cytoplasm. The gene products of oda5, 8, and 10 are not known, but since a mixture of 12S and 18S dynein complexes purified from flagella can rescue either oda2 or oda5 axonemes in vitro (Takada and Kamiya, 1994), 12S and 18S dynein may contain all of the components of both the preassembled HC–IC complex (disrupted in oda2) and the hypothesized oda5, 8, 10 complex. The combined 12S and 18S fractions consist of only three HCs (encoded by oda2, 4, and 11), two ICs (encoded by oda6 and 9), and eight LCs (Piperno and Luck, 1979; Pfister et al., 1982), which suggests that the oda5, 8, and 10 complex consists of LCs. A model of outer-arm dynein assembly based on the data and these assumptions is presented in Figure 7. In wild-type cells, all three complexes come together in the cytoplasm to form a complete dynein arm, which then moves into the flagellar compartment for attachment to doublet microtubules (heavy arrows). Mutations block this process at the steps indicated by the numbers, each of which refers to one of the oda mutations. Dashed lines in the model show assembly steps that still occur even when other pathways are blocked (e.g., the DC can assemble onto doublets in the absence of other components, and the remainder of the outer arm can then add onto the docking site in the flagellum, as occurs after the fusion of oda1 and oda2 gametes). Evidence that the entire arm preassembles in wild-type cells includes the observations that oda5 affects the level of HCα and HCβ in cytoplasmic extracts (Figures 2 and 3), and that HCβ is more susceptible to proteolysis in oda1 and oda3 extracts (Figure 4), as well as the data of Fok et al. (1994) on Paramecium extracts. As an alternative to this model, the oda5, 8, and 10 complex could be required only in the cytoplasm, where it could act to modify the HC–IC complex into an assembly-competent form.
Figure 7.
Model of the dynein assembly process. Solid arrows indicate steps thought to occur in wild-type cells, where all of the subunits are hypothesized to preassemble in the cytoplasm (left side) before moving into the flagellum (right side). An intermediate compartment (IFT) may be required for transport between the cytoplasm and flagellum. Assembly mutants oda1–oda11 block this process at the steps indicated by each number in the diagram and affect the preassembly of one of three complexes, an IC–HC complex (top), a LC complex (middle), or a docking complex (bottom). Dashed arrows show assembly pathways occurring during cytoplasmic complementation of assembly mutants.
In conclusion, our data reveal that outer-dynein arm HC and IC subunits exist as a preassembled complex in a cytoplasmic precursor pool, and that at least three oda mutations that disrupt this complex have major effects on the abundance of another subunit in addition to the mutant gene product. It is unknown whether there is a sequential order in the wild-type assembly process, although temporary dikaryon complementation studies suggest that many paths can lead to assembly of a complete complex. Only the identification of gene products of the remaining oda loci will reveal whether they all encode enzyme subunits or whether some encode cytoplasmic assembly factors. When flagella and axonemes of several oda mutants were tested for the presence of outer-dynein arm subunits by Western blot, trace amounts of some subunits were seen, but in every case these subunits remained tightly associated with axonemes after detergent extraction (Mitchell, unpublished observation). It thus appears that dynein subunits that are not bound to doublet microtubules do not accumulate in the flagellar compartment in these mutants, and that the flagellar pool is very small relative to the total cytoplasmic pool. We do not know whether this flagellar pool size increases during flagellar assembly or dikaryon complementation. Having established that dynein subunits undergo extensive cytoplasmic assembly before their movement into the flagellar compartment, it remains to be determined whether specific transport or targeting mechanisms such as the recently identified intraflagellar transport (IFT) particles (Cole et al., 1998; Pazour et al., 1998) are involved in bringing these complexes to their ultimate destinations.
ACKNOWLEDGMENTS
We acknowledge George Witman and Gianni Piperno for providing antibodies, Joel Rosenbaum and Steven King for communicating results before publication, and Margaret Maimone, Beth Mitchell, and Win Sale for critical comments on the manuscript. This work was supported by grant 44228 from the National Institute of General Medical Sciences to D.R.M.
Footnotes
Abbreviations: DC, docking complex; HC, heavy chain; IC, intermediate chain; IP, immunoprecipitation; LC, light chain.
The two thioredoxin-related outer-arm LCs were initially identified as 14-kDa IC-associated and 16-kDa HCα-associated proteins (Patel-King et al., 1996), and a Tctex2 homologous LC was identified as a 19-kDa HCβ-associated protein (Patel-King et al., 1997). More recently, this assignment has been modified to assign the two thioredoxin homologues as the 16-kDa (HCα) and 19-kDa (HCβ) LCs, and the Tctex2 homologue as the 20- kDa IC-associated LC (S. King, personal communication).
REFERENCES
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burnette WN. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child FM, Apter MN. Experimental inhibition of ciliogenesis and ciliary regeneration in Arbacia embryo. Biol Bull. 1969;137:394–395. [Google Scholar]
- Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- Dentler WL. Microtubule-membrane interactions in cilia and flagella. Int Rev Cytol. 1981;72:1–47. doi: 10.1016/s0074-7696(08)61193-6. [DOI] [PubMed] [Google Scholar]
- Diener DR, Cole DG, Rosenbaum JL. Cytoplasmic precursors of flagellar radial spokes exist as large complexes. Mol Biol Cell Suppl. 1996;7:47a. (Abstract). [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Lux FG., III Genetic interactions of mutations affecting flagella and basal bodies in Chlamydomonas. Cell Motil Cytoskeleton. 1989;14:104–117. doi: 10.1002/cm.970140120. [DOI] [PubMed] [Google Scholar]
- Fok AK, Wang H, Katayama A, Aihara MS, Allen RD. 22S axonemal dynein is preassembled and functional prior to being transported to and attached on the axonemes. Cell Motil Cytoskeleton. 1994;29:215–224. doi: 10.1002/cm.970290304. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook. San Diego: Academic Press; 1989. [Google Scholar]
- Huang B, Piperno G, Luck DJL. Paralyzed flagella mutants of Chlamydomonas reinhardtii. J Biol Chem. 1979;254:3091–3099. [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJL. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Suhan JP. The role of the flagellar transition region: inferences from the analysis of a Chlamydomonas mutant with defective transition region structures. J Cell Sci. 1991;99:731–740. [Google Scholar]
- Johnson KA, Rosenbaum JL. The basal bodies of Chlamydomonas reinhardtii do not contain immunologically detectable DNA. Cell. 1990;62:615–619. doi: 10.1016/0092-8674(90)90105-n. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. Flagellar regeneration in Chlamydomonas: a model system for studying organelle assembly. Trends Cell Biol. 1993;3:156–161. doi: 10.1016/0962-8924(93)90136-o. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc Natl Acad Sci USA. 1985;82:4717–4721. doi: 10.1073/pnas.82.14.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 1986;134:291–306. doi: 10.1016/0076-6879(86)34097-7. [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS, Wilkerson CG, Witman GB. The 78,000-Mr intermediate chain of Chlamydomonas outer dynein is a microtubule-binding protein. J Cell Biol. 1995;131:399–409. doi: 10.1083/jcb.131.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Witman GB. Structure of the gamma heavy chain of the outer arm dynein from Chlamydomonas flagella. J Cell Biol. 1988;107:1799–1808. doi: 10.1083/jcb.107.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Witman GB. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J Biol Chem. 1990;265:19807–19811. [PubMed] [Google Scholar]
- King SM, Witman GB. Multiple sites of phosphorylation within the alpha heavy chain of Chlamydomonas outer arm dynein. J Biol Chem. 1994;269:5452–5457. [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng H, Takada S, Witman GB. The Chlamydomonas reinhardtii oda3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luck DJL, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck DJL, Piperno G. Dynein arm mutants of Chlamydomonas. In: Warner FD, Satir P, editors. Cell Movement: The Dynein ATPases. Vol. 1. I.R. Gibbons, New York: Alan R. Liss; 1989. pp. 49–60. [Google Scholar]
- Mitchell DR, Brown KS. Sequence analysis of the Chlamydomonas alpha and beta dynein heavy chain genes. J Cell Sci. 1994;107:635–644. doi: 10.1242/jcs.107.3.635. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Brown KS. Sequence analysis of the Chlamydomonas reinhardtii flagella α dynein gene. Cell Motil Cytoskeleton. 1997;37:120–126. doi: 10.1002/(SICI)1097-0169(1997)37:2<120::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J Cell Biol. 1991;113:835–842. doi: 10.1083/jcb.113.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Reversion analysis of dynein intermediate chain function. J Cell Sci. 1993;105:1069–1078. doi: 10.1242/jcs.105.4.1069. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. Protein-protein interactions in the 18S ATPase of Chlamydomonas outer dynein arms. Cell Motil Cytoskeleton. 1986;6:510–520. doi: 10.1002/cm.970060510. [DOI] [PubMed] [Google Scholar]
- Musgrave A, De Wildt P, Van Etten I, Pijst H, Scholma C, Kooyman R, Homan W, Van den Ende H. Evidence for a functional membrane barrier in the transition zone between the flagellum and cell body of Chlamydomonas eugametos gametes. Planta. 1986;167:544–553. doi: 10.1007/BF00391231. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, Benashski SE, Harrison A, King SM. Two functional thioredoxins containing redox-sensitive vicinal dithiols from the Chlamydomonas outer dynein arm. J Biol Chem. 1996;271:6283–6291. doi: 10.1074/jbc.271.11.6283. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, Benashski SE, Harrison A, King SM. A Chlamydomonas homolgue of the putative murine t complex distorter Tctex-2 is an outer arm dynein light chain. J Cell Biol. 1997;137:1081–1090. doi: 10.1083/jcb.137.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for retrograde particle movement (“IFT”) in the flagellum. J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Fay RB, Witman GB. Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. 1982;2:525–547. doi: 10.1002/cm.970020604. [DOI] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJL. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Luck DJL. Axonemal adenosine triphosphatases from flagella of Chlamydomonas reinhardtii. J Biol Chem. 1979;254:3084–3090. [PubMed] [Google Scholar]
- Piperno G, Luck DJL. Inner arm dyneins from flagella of Chlamydomonas reinhardtii. Cell. 1981;27:331–340. doi: 10.1016/0092-8674(81)90416-5. [DOI] [PubMed] [Google Scholar]
- Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Gardner LC, Mitchell DR, Dutcher SK. Mutations in the SUP-PF-1 locus of Chlamydomonas reinhardtii identify a regulatory domain in the β-dynein heavy chain. J Cell Biol. 1994;126:1495–1507. doi: 10.1083/jcb.126.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall SJ, Cavalier-Smith T, McVittie A, Warr JR, Hopkins JM. Developmental and control processes in the basal bodies and flagella of Chlamydomonas reinhardtii. Dev Biol Suppl. 1967;1:43–83. [Google Scholar]
- Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Child FM. Flagellar regeneration in protozoan flagellates. J Cell Biol. 1967;34:345–364. doi: 10.1083/jcb.34.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardtii. Ann NY Acad Sci. 1953;466:18–30. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Mitchell DR, Kamiya R. A Chlamydomonas outer arm dynein mutant missing the α heavy chain. J Cell Biol. 1991;113:615–622. doi: 10.1083/jcb.113.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RE. Differential protein synthesis and utilization during cilia formation in sea urchin embryos. Dev Biol. 1977;61:311–329. doi: 10.1016/0012-1606(77)90301-3. [DOI] [PubMed] [Google Scholar]
- Takada S, Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J Cell Biol. 1994;126:737–745. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Wilkerson C, Kamiya R, Witman G. The oda1 gene encodes a 62-kDa component of a complex necessary for outer dynein docking on the doublet microtubules of Chlamydomonas flagella. Mol Biol Cell Suppl. 1996;7:568a. (Abstract). [Google Scholar]
- Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harbor Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Koutoulis A, Pazour GJ, Witman GB. The 78,000 Mr intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Witman GB. Molecular analysis of the γ heavy chain of Chlamydomonas flagellar outer-arm dynein. J Cell Sci. 1994;107:497–506. doi: 10.1242/jcs.107.3.497. [DOI] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. J Cell Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]