Abstract
Many effectors of microtubule assembly in vitro enhance the polymerization of subunits. However, several Saccharomyces cerevisiae genes that affect cellular microtubule-dependent processes appear to act at other steps in assembly and to affect polymerization only indirectly. Here we use a mutant α-tubulin to probe cellular regulation of microtubule assembly. tub1-724 mutant cells arrest at low temperature with no assembled microtubules. The results of several assays reported here demonstrate that the heterodimer formed between Tub1-724p and β-tubulin is less stable than wild-type heterodimer. The unstable heterodimer explains several conditional phenotypes conferred by the mutation. These include the lethality of tub1-724 haploid cells when the β-tubulin–binding protein Rbl2p is either overexpressed or absent. It also explains why the TUB1/tub1-724 heterozygotes are cold sensitive for growth and why overexpression of Rbl2p rescues that conditional lethality. Both haploid and heterozygous tub1-724 cells are inviable when another microtubule effector, PAC2, is overexpressed. These effects are explained by the ability of Pac2p to bind α-tubulin, a complex we demonstrate directly. The results suggest that tubulin-binding proteins can participate in equilibria between the heterodimer and its components.
INTRODUCTION
Microtubules participate in a variety of specific functions crucial for morphological differentiation, cell growth, and cell movement. The diversity of these functions requires that microtubules assemble into quite different structures even within the same cell. Many of those structures are dynamic, allowing them to disassemble rapidly and thus provide the components necessary to form another microtubule organelle. Possible mechanisms for regulation of these processes can be envisioned at several levels: primary sequences of tubulin genes (Joshi and Cleveland, 1989; Hoyle and Raff, 1990), message stability (Pachter et al., 1987), folding and dimerization of the protein subunits (Ursic and Culbertson, 1991; Chen et al., 1994), properties of the polymer (Mitchison and Kirschner, 1984; Saxton et al., 1984), and the interaction of the polymer with non-tubulin proteins (Caceres and Kosik, 1990; Dinsmore and Solomon, 1991).
Recently, several diverse experimental approaches have identified proteins that may participate in tubulin heterodimer formation. In vitro assays for proper folding of denatured α- and β-tubulins require several protein cofactors that transiently interact with the individual polypeptide chains (Melki et al., 1996; Tian et al., 1996, 1997). These complexes of tubulin polypeptides with cofactors may be intermediates that form between release of tubulin polypeptide from the TCP1-containing ring complex and its incorporation into preexisting heterodimers by exchange. In at least some cases, those polypeptides form binary or higher-order complexes with the tubulins that are stable enough to be isolated but are still reactive (Tian et al., 1997).
Homologues of these cofactors (except cofactor C) are identified by diverse screens for mutations that affect microtubule processes in budding yeast. The processes affected include sensitivity to microtubule-depolymerizing drugs (Stearns et al., 1990), fidelity of mitotic chromosome transmission (Hoyt et al., 1990), response to overexpression of β-tubulin (Archer et al., 1995), and interactions with mitotic motors (Geiser et al., 1997). Although most of these cofactors are essential for the in vitro assay, none of their Saccharomyces cerevisiae homologues are essential for viability. Therefore, they may participate in the folding and heterodimerization of tubulin polypeptides, but there must be pathways that do not depend on them.
The genetic data alluded to above suggest that there may be multiple steps in tubulin assembly subject to cellular control. Analysis of tubulin mutants can provide access to those steps. A panel of α-tubulin mutants cold sensitive for growth arrest at their restrictive temperature with diverse microtubule phenotypes (Schatz et al., 1988). Some of the mutants arrest with no microtubules (class 1), some with a normal complement of microtubules (class 2), and the rest with an apparent excess of microtubules (class 3). This variability suggests that the conditional defects in these mutant α-tubulin proteins can affect different aspects of microtubule assembly and function. Certain of these mutations are suppressed by specific mutations in β-tubulin (Schatz et al., 1988) and others by extra copies of the mitotic check point BUB genes (Guénette et al., 1995) or by yeast homologues of the mammalian checkpoint gene RCC1 (Kirkpatrick and Solomon, 1994). However, there is too little structure–function information for tubulin to permit an understanding of the phenotype in terms of the tubulin mutation itself.
Another distinction among the tub1 mutants is uncovered when they are assayed in the presence of varying Rbl2p levels. Rbl2p binds β-tubulin to form a 1:1 complex (Melki et al., 1996; Archer et al., 1998). Rbl2p binding to β-tubulin excludes α-tubulin binding to β-tubulin. Four class 1 α-tubulin mutants are synthetically lethal with deletion of rbl2. Two of those are also synthetically lethal with overexpression of RBL2, but several other class 1, 2, or 3 mutants show no such interactions (Archer et al., 1995).
The present study analyzes and exploits the properties of one of those two mutants. The tub1-724 mutation fails to support growth at 18°C and only partially supports growth at 25°C but grows as well as wild-type cells at 30°C (Schatz et al., 1988; see Figure 5). The lethality and loss of microtubules at the nonpermissive temperature is not a consequence of degradation of α-tubulin; the steady-state α-tubulin levels in these cells is the same as that in an isogenic wild-type control (our unpublished results). Upon induction of GAL-RBL2, tub1-724 cells at permissive temperature rapidly lose assembled microtubule structures, and within 20 h <0.1% of the cells are viable (Archer et al., 1995).
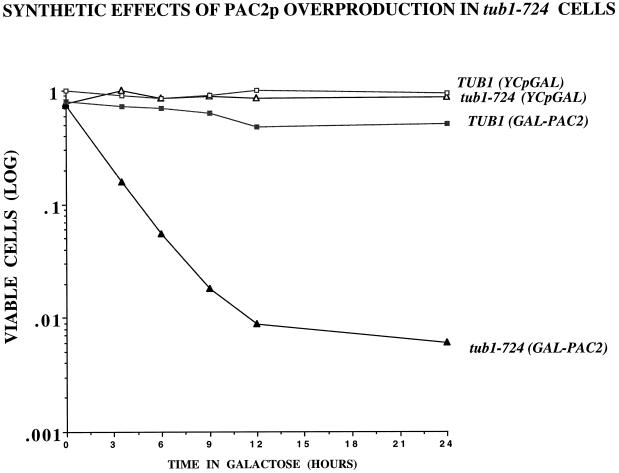
Figure 5.
Overexpressing PAC2 is lethal in tub1-724 cells. tub1-724 (triangles) and wild type (squares) containing either control plasmid (open symbols) or GAL-PAC2 (filled symbols) cells growing at 30°C were shifted to galactose-containing media at zero time, and aliquots were taken at intervals and scored for total cells and viable cells.
The data presented here demonstrate that tubulin heterodimer containing this mutant α-tubulin protein is less stable than the wild-type heterodimer. We use this property to analyze interactions between tub1-724 and altered levels of two of the cofactor homologues mentioned above. The results provide a structure–function correlation for tubulin as well as insight into the cellular activities of the β-tubulin–binding protein Rbl2p and the putative α-tubulin–binding protein Pac2p.
MATERIALS AND METHODS
Strains, Plasmids, and Media
All yeast strains are derivatives of FSY185 (Weinstein and Solomon, 1990) with the exception of the tub1 mutants (Schatz et al., 1988) (Table 1). We used standard methods for yeast manipulations (Sherman et al., 1986; Solomon et al., 1992).
Table 1.
Strains and plasmids
| Strain/plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| FSY185 | a/α; ura3-52/ura3-52, leu2-3,112/leu2-3,112, his3Δ200/his3Δ200, lys2-801/lys2-801, ade2/ADE2 | Weinstein and Solomon, 1990 |
| FSY183 | a; ura3-52; leu2-3,112; his3Δ200; lys2-801 | Weinstein and Solomon, 1990 |
| FSY157 | a; ura3-52; leu2-3,112; his3Δ200; lys2-801; Δtub1::HIS3, Δtub3::TRP1 [pRB624] | Schatz et al., 1988 |
| FSY182 | a; ura3-52; leu2-3,112; his3Δ200; lys2-801; Δtub1::HIS3, Δtub3::TRP1 [pRB539] | Schatz et al., 1988 |
| LTY8 | FSY157 plus YCpGAL | Archer et al., 1995 |
| LTY11 | FSY182 plus YCpGAL | This study |
| LTY291 | FSY157 plus pGHR | This study |
| LTY292 | FSY182 plus pGHR | This study |
| LTY374 | FSY157 plus pPA45 | This study |
| LTY376 | FSY182 plus pPA45 | This study |
| JAY47 | a/α, ura3-52/ura3-52, leu2-3,112/leu2-3,112, his3Δ200/his3Δ200, lys2-801/lys2-801, ade2/ADE2, TUB2/TUB2-LEU2-GAL-TUB2 | Archer et al., 1995 |
| LTY319 | JAY47 plus YCp50 | This study |
| LTY321 | JAY47 plus A21A | This study |
| LTY323 | JAY47 plus pLV32 | This study |
| LTY325 | JAY47 plus pA1A5106 | This study |
| LTY338 | JAY47 plus pLV30 | This study |
| LTY340 | JAY47 plus pLV38 | This study |
| LTY343 | JAY47 plus pLV36 | This study |
| LTY345 | JAY47 plus pLV37 | This study |
| LTY392 | a/α; ura3-52/ura3-52; leu2-3,112/leu2-3,112; his3Δ200/his3Δ200; lys2-801/lys2-801; TUB1/Δtub1::HIS3, TUB3/Δtub3::TRP1; [pRB539, pA5] | This study |
| LTY393 | like LTY392 but with pPA45 rather than pA5 | This study |
| LTY395 | like LTY392 but with YCpGAL rather than pA5 | This study |
| LTY396 | a/α; ura3-52/ura3-52; leu2-3,112/leu2-3,112; his3Δ200/his3Δ200; lys2-801/lys2-801; TUB1/Δtub1::HIS3,TUB3/Δtub3::TRP1, [pRB624, pPA45] | This study |
| LTY397 | like LTY396 but with pA5 rather than pA45 | This study |
| LTY440 | JAY47 plus YCpGAL | This study |
| LTY439 | JAY47 plus pLV56 | This study |
| LTY399 | like LTY396 but with YCpGAL rather than pA45 | This study |
| LTY539 | FSY183 plus pJA3 and pLV62 | This study |
| LTY541 | FSY183 plus pJA3 and pRS317 | This study |
| LTY540 | FSY183 plus YCpGAL and pLV62 | This study |
| Plasmids | ||
| YCp50 | CEN-URA3 | Kirkpatrick and Solomon, 1994 |
| pA1A5106 | TUB1-CEN-URA3 | Kirkpatrick and Solomon, 1994 |
| pA21A | RBL2-CEN-URA3 | Archer et al., 1995 |
| pA5 | GAL 1-10-RBL2-URA3 | Archer et al., 1995 |
| pGHR | GAL 1-10-HIS6-RBL2-URA3 | Archer et al., 1998 |
| pRB624 | tub1-724-CEN-LEU2 | Schatz et al., 1988 |
| pRB539 | TUB1-CEN-LEU2 | Schatz et al., 1988 |
| pPA45 | GAL 1-10-PAC2-CEN-URA3 | Alvarez et al., 1998 |
| YCpGAL | GAL1-10-CEN-URA3 | Archer et al., 1995 |
| pLV30 | tub1-704 in YCp50 | This study |
| pLV32 | tub1-724 in YCp50 | This study |
| pLV36 | tub1-737 in YCp50 | This study |
| pLV37 | tub1-747 in YCp50 | This study |
| pLV38 | tub1-714 in YCp50 | This study |
| pLV56 | GAL 1-10-PAC2-HA-HIS6-CEN-URA3 | This study |
| pLV62 | GAL1-10-PAC2-HA-HIS6-CEN-LYS2 | This study |
| pRS317 | CEN-LYS2 | Sikorski and Hieter, 1989 |
Viability Measurements and Immunofluoresence
LTY374, LTY8, LTY376, and LTY11 were grown overnight in SC-Ura-Leu raffinose media. Log phase cells were then induced with 2% galactose, and at various time points aliquots of cells were taken and counted using a haemocytometer. Known numbers of cells were then plated to SC-Ura glucose plates. Cell viability was measured as the percent of counted cells able to form colonies on the SC-Ura glucose plates. At various time points cells were fixed for immunofluorescence in 3.7% formaldehyde. Anti-β-tubulin staining was done with #206 (Bond et al., 1986) at 1:2000 in PBS containing 0.1% BSA.
Phenotypes of TUB1 or tub1-724 Heterozygous Diploids
Δtub1, Δtub3 strains containing tub1-724 or TUB1 gene on LEU2:CEN plasmids were crossed to FSY183 (wild type) containing YCpGAL, pPA45, or pA5. The diploid strains were grown to saturation overnight in SC-Ura-Leu-His glucose liquid media. The saturated cultures were serially diluted in 96-well dishes and spotted onto SC-Ura glucose and SC-Ura galactose plates.
Rescue of JAY47
JAY47 (Archer et al., 1995) was transformed with genomic CEN:URA3 plasmids containing TUB1, tub1 alleles or with CEN:URA3:RBL2. Cells were plated to SC-Leu-Ura glucose plates at 30°C and to SC-Leu-Ura galactose plates 30, 18, and 15°C. The number of colonies on galactose relative to glucose was measured.
DNA Sequencing
DNA sequencing was performed using modified T7 DNA polymerase Sequenase with the dideoxy chain termination method (United States Biochemical, Cleveland, OH).
Immune Techniques
Immunoblots.
Standard procedures were used (Solomon et al., 1992). After gel electrophoresis and transfer to nitrocellulose membranes, we blocked blots with TNT (0.025 M Tris, 0.17 M NaCl, 0.05% Tween 20, pH 7.5) for 30–120 min. Primary antibodies were incubated for >12 h at 1:3500 (#206 or #345; Weinstein and Solomon, 1990) or at 1:100 (#250; Archer et al., 1995) and then washed five times (5 min each) in TNT. Bound antibody was detected by 125I-protein A (New England Nuclear, Boston, MA) or (for 12CA5) 125I-sheep anti-mouse immunoglobulin G (New England Nuclear). Commercial preparations of anti-HA were used (Boehringer Mannheim, Indianapolis, IN).
Immunoprecipitations.
The procedure described previously (Archer et al. 1995) was used with slight modifications. The monoclonal antibodies A1BG7 (anti-α) and B1BE2 (anti-β), raised against the carboxyl-terminal 12 amino acids of Tub1p and Tub2p, respectively, were affixed to Affigel-10 beads (Bio-Rad, Hercules, CA). Yeast strains FSY157 and FSY182 were grown up at 30°C. Total protein was harvested by glass bead lysis in PME (0.1 M piperazine-N,N′-bis[2-ethanesulfonic acid], 2 mM EGTA, 1 mM magnesium chloride, pH 6.9) plus protease inhibitors and was added to antibody beads for a 1-h incubation with rotation at 4°C. We washed the beads eight times with PME plus protease inhibitors. Bound proteins were eluted by boiling in SDS sample buffer and resolved by SDS-PAGE analysis.
Purification of His6-tagged Proteins
The Ni-NTA nickel slurry and column materials were from Qiagen (Chatsworth, CA). We used protocols from the Qiagen handbook and modifications of this protocol that have been previously described (Magendantz et al., 1995).
In Vivo His6-Rbl2p-β-Tubulin Association Experiments
Yeast strains LTY291 and LTY292 are FSY157 and FSY182 transformants with a CEN pGAL-RBL2-HIS6 (pGHR). We grew LTY291 and LTY292 overnight at 30°C in selective media containing raffinose to about 2 × 109 cells per experiment. Galactose (2%) was added to induce His6-RBL2 expression. After 0, 1, and 2 h, protein was harvested by glass bead lysis in 1 ml of PME buffer plus protease inhibitors. We applied 0.85 ml of protein extract to 130 μl of Ni-NTA beads. We washed and eluted the bound proteins as previously described (Magendantz et al., 1995). Eluted proteins were subjected to SDS-PAGE analysis and probed for α-tubulin, β-tubulin, and Rbl2p and quantitated by densitometry.
In Vivo HIS6-(HA)-Pac2p-α-Tubulin Association Experiments
We grew yeast strains LTY539, LTY541, LTY439, and LTY440 overnight in selective raffinose media at 30°C. Galactose (2%) was added to induce Pac2p-(HA)-His6 and α-tubulin or β-tubulin expression. Cells (6.0 × 109) were harvested by glass bead lysis per experiment in 1.1 ml of PME buffer plus protease inhibitors. We applied 1 ml of protein extract to 25 μl of Ni-NTA beads. We washed and eluted the bound proteins as previously described (Magendantz et al., 1995). Eluted proteins were subjected to SDS-PAGE analysis and probed for α-tubulin, β-tubulin, and HA(12CA5). For Pac2p, the bead eluants represent 120 times the load of whole-cell extract. For α- and β-tubulin, the bead eluants represent 500 times the load of whole-cell extract.
RESULTS
Characterization of Cold-sensitive tub1 Mutants
The conditional loss of assembled microtubules in class 1 α-tubulin mutants could arise from cold sensitivity of any of several steps in microtubule morphogenesis. However, the synthetic lethality of Tub1-724p with both Rbl2p deletion and overexpression suggests that the mutant defect arises from a weaker heterodimer (Figure 1). If the heterodimer formed by the Tub1-724p dissociates more readily than does wild-type heterodimer, the increase in free, undimerized β-tubulin could be toxic in the absence of the β-tubulin binding capacity provided by Rbl2p. Conversely, an excess of Rbl2p, which has only minor phenotypes in a wild-type cell, could compete effectively with the mutant α-tubulin protein for β-tubulin and so diminish the level of tubulin subunits to cause loss of microtubules and cell death. The experiments described below present tests of this model.
Figure 1.
Synthetic lethal interactions between tub1-724 and altered levels of Rbl2p: a model. Cells expressing tub1-724 as their sole source of α-tubulin die when Rbl2p is either absent or overexpressed. Those relationships are explicable if the heterodimer formed by the Tub1-724p (α*β) dissociates more readily than that formed by the wild-type Tub1p (αβ). In the presence of a normal complement of RBL2, the mutant cells would have a high concentration of free β-tubulin (βfree), which may be responsible for the conditional phenotypes of the mutant (e.g., benomyl supersensitivity). In the absence of Rbl2p, the activity of βfree would increase to toxic levels. In contrast, an excess of Rbl2p could bind to β-tubulin and so enhance dissociation of the mutant heterodimer, promoting dissociation to levels below those necessary for viability.
The only difference between the primary sequences of TUB1 and tub1-724 genes predicts substitution of threonine for arginine at codon 106 (AGA becomes ACA). Arginine-106 is a highly conserved residue among α-tubulins. The possible significance of this mutation for heterodimer stability is presented in DISCUSSION.
Coimmunoprecipitation of α- and β-Tubulin from Wild-Type and tub1-724 Mutant Cells
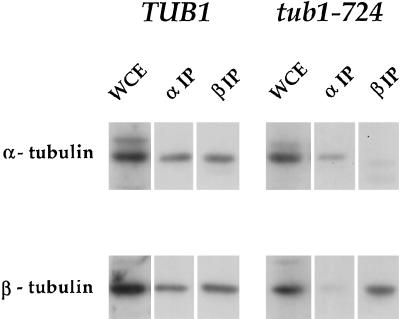
We assessed the stability of the wild-type and mutant α-β heterodimers by coimmunoprecipitation. Extracts from tub1-724 mutant cells (FSY157) and wild-type cells (FSY182) grown at 30°C were incubated with antibodies to either α-tubulin or β-tubulin coupled to Affigel beads. The beads were washed extensively to remove adventitiously adhering proteins, and specifically bound proteins were released by SDS. The tubulin chains in the immunoprecipitates were analyzed by immunoblotting (Figure 2). From extracts of wild-type cells, antibodies against each of the tubulin polypeptides coprecipitate the other chain with high efficiency; the ratio of the tubulins in the coprecipitates is comparable to the original extracts. This result suggests that under the conditions of tubulin immunoprecipitation, normal heterodimer largely remains intact. From extracts of tub1-724 cells, however, the anti-tubulin antibodies complex efficiently with the specific tubulin chain against which they are directed but precipitate the other tubulin chain only poorly.
Figure 2.
α- and β-Tubulin coimmunoprecipitate with low efficiency from tub1-724 cells. Immunoblots with anti-α-tubulin (top row) and anti-β-tubulin (bottom row) of whole-cell extracts (WCE) and the precipitates with the two antibodies (αIP and βIP) from wild-type TUB1 or mutant tub1-724 cells.
Because we recover only a small fraction of Tub1-724p heterodimer by immunoprecipitation, we cannot directly compare the stability of the mutant and wild-type heterodimers. We previously established that at least 98% of the β-tubulin in wild-type cells is in the form of α-β heterodimer (Archer et al., 1998). Because tub1-724 cells grow normally at 30°C, presumably most of the tubulin in those cells is in heterodimer in vivo. Thus, the dissociation of the heterodimer likely occurs in the course of the immunoprecipitation itself, which exposes the heterodimer to large dilutions at low temperature (4°C). Under similar conditions, the wild-type heterodimer has a half-life of ∼10 h (Archer et al., 1998).
Formation of Rbl2p-β-Tubulin Complex in Wild-Type and tub1-724 Mutant Cells
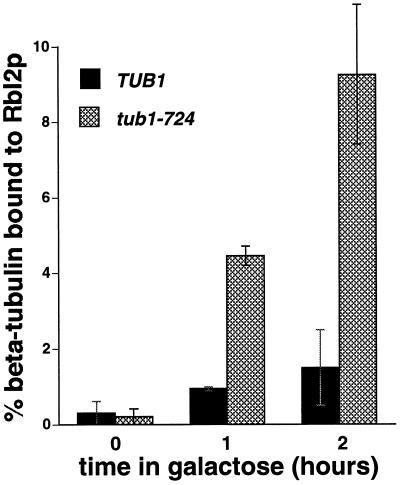
Rbl2p is complexed with β-tubulin in vivo, and the level of that complex increases as the cellular level of Rbl2p increases (Archer et al., 1995, 1998). The model presented in Figure 1 predicts that overexpressed Rbl2p will form a complex with β-tubulin more readily in tub1-724 cells than in wild-type cells. To test that possibility, we introduced a plasmid encoding His6-Rbl2p under the control of the galactose promoter into wild-type TUB1 cells or tub1-724 mutant cells. The transformants were grown at the permissive temperature for the mutant in noninducing medium and then were shifted to inducing medium containing galactose for 1 or 2 h. We used nickel-agarose beads to purify the His6-Rbl2p-β-tubulin complex. The bound proteins were eluted and analyzed by immunoblotting with antibodies against α-tubulin, β-tubulin, or Rbl2p. As expected, the levels of His6-Rbl2p-β-tubulin complex increase upon induction in both control and mutant cells, but three- to fivefold more complex forms in tub1-724 cells relative to wild type (Figure 3). In these experiments, we detect only a trace of α-tubulin bound to the nickel columns, and its level does not increase with time in galactose (Archer et al., 1998; our unpublished results). This result suggests either that Rbl2p competes more efficiently with Tub1-724p than with wild-type α-tubulin for binding to β-tubulin in vivo, or that there is a greater pool of free β-tubulin available for binding to Rbl2p in the tub1-724 mutant (see DISCUSSION). Either possibility is consistent with Tub1-724p forming a less stable heterodimer with β-tubulin than wild-type α-tubulin.
Figure 3.
The Rbl2p-β-tubulin complex in vivo is enhanced in tub1-724 cells. Cells growing in raffinose (0 h) were induced with galactose to express His6-Rbl2p for 1 or 2 h. His6-Rbl2p was isolated by affinity chromatography of the whole-cell extracts, and the levels of β-tubulin in the original extract and bound to Rbl2p were assayed by immunoblotting. The results are the averages of two independent experiments for each strain and time point with the ranges indicated by error bars. In both of these experiments, the wild-type strain produced slightly more His6-Rbl2p upon induction (our unpublished results). Solid bars, TUB1 cells; cross-hatched bars, tub1-724 cells.
Rescue of β-Tubulin Lethality by Wild-Type and Mutant α-Tubulins
An excess of either α-tubulin or Rbl2p rescues cells from β-tubulin lethality (Archer et al., 1995; Alvarez et al., 1998); the rescue likely depends on the ability of these two proteins to bind β-tubulin. Even a modest excess of α-tubulin, expressed under the control of its own promoter from a low-copy plasmid, increases the survival of cells overproducing β-tubulin by two to three orders of magnitude. If Tub1-724p binds β-tubulin with low affinity, we would expect it to rescue β-tubulin lethality poorly. To test this hypothesis, wild-type or mutant alleles of α-tubulin were introduced into JAY47, a diploid strain with a normal complement of tubulin genes plus a third, integrated copy of the β-tubulin gene TUB2 under the control of the galactose promoter. We measured the percent survivors on galactose relative to glucose at both the permissive (30°C) and the nonpermissive (18°C) temperatures (Table 2). Rescue of β-tubulin lethality by tub1-724 is substantially less efficient (0.84%) than by wild-type TUB 1 (15.4%). The efficiency of rescue is further diminished at the nonpermissive temperature for the mutant: at 18°C, tub1-724 rescues β-tubulin lethality (0.06%) to essentially the same extent as the negative control (0.03%). In contrast, four other mutant α-tubulins rescue at levels comparable to that of the wild type, and their efficiency is unaffected by the temperature of growth. In fact, the activity of those alleles persists even at 15°C (our unpublished results). These results are consistent with the conclusion that Tub1-724p binds β-tubulin with lower affinity than does wild type α-tubulin.
Table 2.
Rescue of excess β-tubulin lethality by α-tubulin alleles
| Plasmid | 30°C | 18°C |
|---|---|---|
| .04 | .03 | |
| RBL2 | 6.8 | 7.2 |
| TUB1 | 12.0 | 15.4 |
| tub1-724 | .84 | .06 |
| tub1-704 | 10.9 | 18.9 |
| −714 | 14.0 | 20.0 |
| −737 | 4.1 | 8.5 |
| −747 | 11.8 | 21.0 |
JAY47 cells, which contain an integrated GAL-TUB2 gene, carrying the indicated α-tubulin alleles on plasmids were plated to media containing either galactose or glucose and incubated at either 30 or 18°C. Rescue is reported as the percentage of cells that form colonies on galactose plates compared with glucose plates.
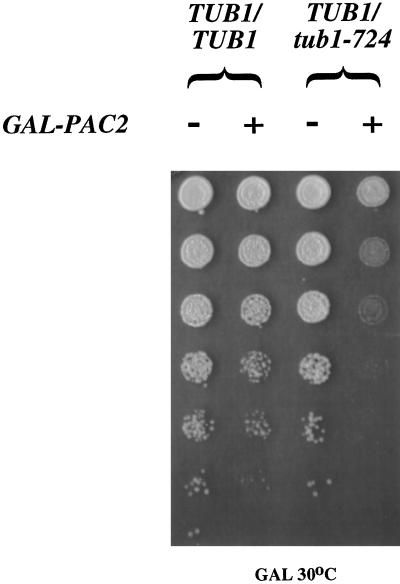
Cold Sensitivity of TUB1/tub1-724 Heterozygotes and Their Suppression by Excess Rbl2p
The tub1-724 phenotype is not completely suppressed in a heterozygote with TUB1. A diploid strain containing only single chromosomal copies of TUB1 and TUB3 plus a low-copy plasmid expressing tub1-724 is cold sensitive for growth at 18°C. In contrast, heterozygotes containing TUB1 and other tub1 mutants show the same temperature sensitivity as do wild-type cells (our unpublished results). The conditional growth of TUB1/tub1-724 heterozygotes must reflect a property of the mutant heterodimer, rather than a deficiency in tubulin levels, because diploid cells with only 50% of their wild-type complement of tubulin are wild type for growth at low temperatures (Katz et al., 1990).
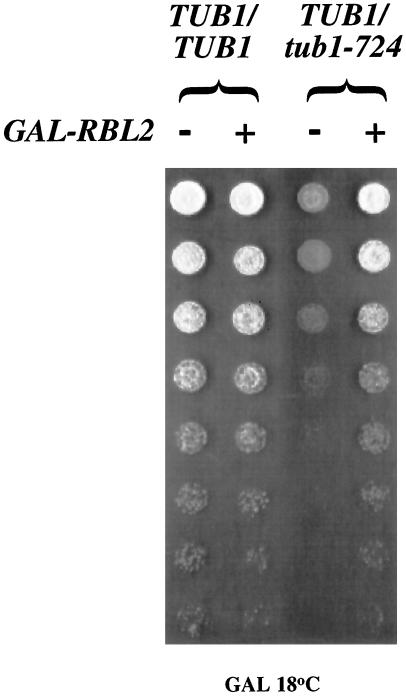
We hypothesized that the cold sensitivity of these TUB1/tub1-724 heterozygous cells is due to the free β-tubulin produced by dissociation of the mutant heterodimer. Consistent with that explanation, the cold sensitivity of the heterozygotes is substantially suppressed by overexpression of RBL2 from the galactose promoter (Figure 4). The presence of excess Rbl2p can bind the free β-tubulin and so protect the cell from its deleterious consequences. This result is in striking contrast to the lethal effect of GAL-RBL2 in cells expressing tub1-724 as their sole source of α-tubulin (see above).
Figure 4.
Overexpression of RBL2 suppresses TUB1/tub1-724 heterozygous cells. Serial (fourfold) dilutions of saturated cultures were plated to galactose-containing media and allowed to grow at 18°C. The cells were either wild-type diploids or TUB1/tub1-724 cells, carrying either YCpGAL or CEN-GAL-RBL2.
Overexpression of PAC2 in tub1-724 Cells
Pac2p is a candidate for an α-tubulin–binding protein in yeast. It is the homologue of cofactor E in the in vitro system described above. Cofactor E plays an essential role in this assay: it is believed to bind to α-tubulin after its release from the TCP1-containing ring complex (Tian et al. 1997). This binary complex is then thought to form a quaternary complex with cofactor D and β-tubulin. The cofactor E-α-tubulin complex is rather unstable and is detectable on native gels only after it is stabilized by glutaraldehyde fixation.
The Schizosaccharomyces pombe homologue of cofactor E is essential in vivo (Hirata et al., 1998). In budding yeast PAC2 is not essential, but mutations in pac2 affect microtubule functions. pac2 mutations are supersensitive to benomyl (Hoyt et al. 1997). It is required in cells deleted for cin8, which encodes a kinesin-related protein that participates in anaphase (Geiser et al., 1997), or deleted for pac10 (Alvarez et al., 1998), which affects ratios of α-tubulin to β-tubulin (Alvarez et al., 1998; Geissler et al., 1998).
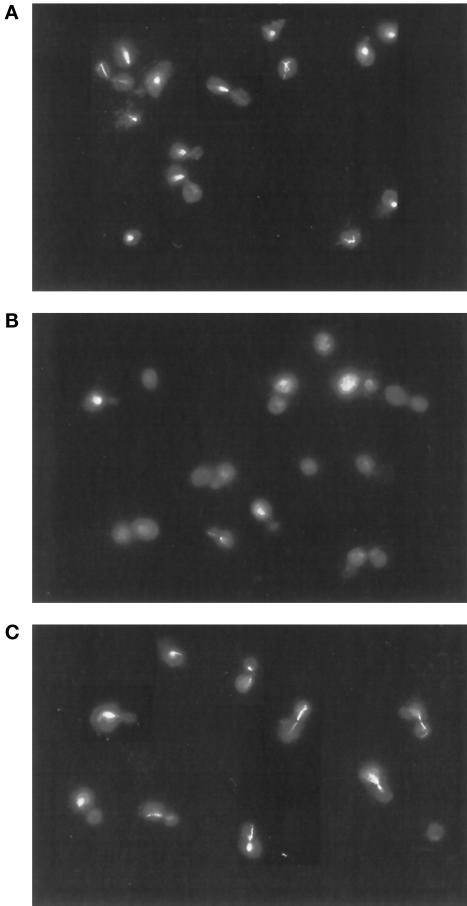
If Pac2p is an α-tubulin–binding protein, we would predict that at elevated levels it would be deleterious to cells containing the unstable tub1-724 heterodimer. As shown in Figure 5, induction of GAL-PAC2 in haploid tub1-724 cells grown at permissive temperature (30°C) causes rapid loss of viability, down 10-fold in ∼3 h. In contrast, GAL-PAC2 has only a modest effect on the viability of wild-type cells (Figure 5). In that time, the induction of GAL-PAC2 causes microtubule disassembly in the mutant but not in wild-type cells; representative micrographs are shown in Figure 6. From such fields, we find that overexpression of PAC2 increases the proportion of tub1-724 cells that have no microtubules by 10-fold (53.2 vs. 5.4%) but has no effect on wild-type cells (10.1% for both strains).
Figure 6.
Microtubule disassembly in tub1-724 cells overexpressing PAC2. Anti-tubulin immunofluorescence of tub1-724 cells containing the control plasmid YCpGAL (A) or a CEN-GAL-PAC2 (B) and wild-type cells containing a CEN-GAL-PAC2 plasmid (C). Cultures were grown in galactose for 3.5 h before fixation for immunofluorescence.
Both phenotypes of elevated Pac2p levels on tub1-724 haploid cells are the same as produced by elevated levels of Rbl2p (Archer et al., 1995). Therefore, these results could represent Pac2p binding to either β-tubulin or α-tubulin. However, the effect of GAL-PAC2 expression in TUB1/tub1-724 heterozygotes does distinguish between these two possibilities. As shown in Figure 7, overexpression of PAC2 in the heterozygotes causes a significant loss of cell viability at the permissive temperature. This result contrasts with that shown in Figure 4 above, showing that overexpression of RBL2 actually suppresses the phenotype of the TUB1/tub1-724 heterozygotes.
Figure 7.
Overexpression of PAC2 is lethal in TUB1/tub1-724 heterozygous cells. Serial (fourfold) dilutions of saturated cultures were plated to galactose-containing media and allowed to grow at 30°C. Strains were wild-type diploids or TUB1/tub1-724 cells containing either YCpGAL or CEN-GAL-PAC2.
These results are explicable if the Tub1-724p-β-tubulin heterodimer is relatively unstable (Figure 1). The increased levels of an α-tubulin–binding protein might be expected to increase free β-tubulin to toxic levels in both tub1-724 haploids and TUB1/tub1-724 heterozygotes. This outcome is in contrast to the effect noted for excess Rbl2p in the heterozygotes, where the increased capacity to bind β-tubulin would be expected to reduce its levels and so suppress the TUB1/tub1-724 phenotypes. Taken together, these results suggest that Pac2p can bind to α-tubulin in vivo and so are consistent with the conclusion of the in vitro experiments (Tian et al., 1997).
Isolation of a Pac2p-α-Tubulin Complex
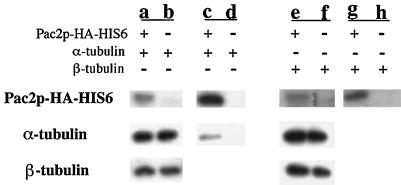
To demonstrate directly a Pac2p-α-tubulin complex, we used a form of Pac2p that contains the HA tag followed by 6 histidines at its carboxyl terminus. This modified allele is functionally indistinguishable from wild type Pac2p in both Δpac2 and tub1-724 cells (our unpublished results). We can isolate a complex containing α-tubulin and Pac2p-(HA)-His6 from extracts of cells overexpressing both proteins (Figure 8, lane c); no β-tubulin is detected in this complex. We cannot detect this complex unless both Pac2p and α-tubulin are overexpressed. In contrast, overexpression of both Pac2p and β-tubulin does not produce a complex between those two proteins (Figure 8, lane g). These results support the conclusion that Pac2p can bind α-tubulin in vivo. Overexpression of Pac2p-(HA)-His6 alone in tub1-724 cells does not produce measurable levels of the Pac2p-α-tubulin complex (our unpublished results).
Figure 8.
Binding of α-tubulin to Pac2p-(HA)-His6 in double-overexpressing cells. Whole-cell extracts (lanes a, b, e, and f) and eluants from nickel-agarose beads (lanes c, d, g, and h) were analyzed by SDS-PAGE and immunoblotting for HA-tagged Pac2p, α-tubulin, and β-tubulin. The fractions were from cells overexpressing Pac2p-(HA)-His6 and α-tubulin (a and c), Pac2p-(HA)-His6 and β-tubulin (e and g), α-tubulin alone (b and d), and β-tubulin alone (d and h). For Pac2p, the bead eluants represent 120 times the load of whole-cell extract. For α- and β-tubulin, the bead eluants represent 500 times the load of whole-cell extract.
DISCUSSION
A Tubulin Mutation That Affects Heterodimer Stability
tub1-724 is one of a set of α-tubulin mutants generated by chemical mutagenesis and selected on the basis of their conditional growth at low temperature. Because of the familiar cold lability of microtubules evident both in vivo and in vitro, a reasonable prediction might have been that mutants so selected would arrest because their microtubules were especially cold labile at temperatures permissive for wild-type cells. Instead, only a subset of the mutants arrest with no microtubules; the others have at least normal complements of assembled tubulin.
Here we have characterized the properties of the protein encoded by one of the mutants that arrest with no microtubules, tub1-724. We previously showed that cells expressing only this α-tubulin allele are dead when Rbl2p is either overexpressed or absent. Because Rbl2p is a β-tubulin–binding protein, we hypothesized that these lethal interactions could reflect an unstable heterodimer formed by Tub1-724p (Figure 1). Several of the experiments presented above demonstrate that the mutant heterodimer does act as if it were unstable relative to wild type. The mutant heterodimer does not remain intact in vitro during immunoprecipitation. Similarly, in vivo the mutant heterodimer reacts more readily with excess Rbl2p to produce Rbl2p-β-tubulin. An alternative measure of Tub1-724p binding to β-tubulin is manifest in its inability to rescue cells from β-tubulin overexpression even at permissive temperature for the mutant (Table 2); success in that assay most likely depends on the ability of the α-tubulin protein to bind β-tubulin. These results indicate that Tub1-724p has a reduced affinity for β-tubulin. However, the normal growth of the mutant cells requires that most of its tubulin be in heterodimers, rather than as free α- and β-tubulin. We previously showed that the microtubules in 50% of cells overproducing β-tubulin are completely depolymerized when β-tubulin levels are 1.4-fold greater than wild type (Weinstein and Solomon, 1990).
A weaker heterodimer could readily explain the arrest phenotype of tub1-724 cells. At the restrictive temperature, increased dissociation of the mutant heterodimer could be lethal either by decreasing the level of heterodimer below that necessary to maintain microtubules or by increasing the level of undimerized β-tubulin, which in turn causes microtubule disassembly and cell death even at modest excess (Katz et al., 1990; Weinstein and Solomon, 1990).
The single mutation in Tub1-724p predicted from the DNA sequence is loss of a positive charge at position 106. Based on the structure of tubulins reported by Nogales et al. (1998), this residue occurs in the region between the B3 and H3 loops that contact the phosphates of the nonexchangeable GTP. That site is at the postulated interface between α- and β-tubulin in the heterodimer. The wild-type arginine at this position probably contributes to phosphate binding and so may indirectly participate in α-β interactions. Further analysis to understand the physical properties of mutations in this region are under way.
This analysis of Tub1-724p provides insight into the primary molecular defect that explains the mutant phenotypes. In general, the defects of mutant tubulins are largely understood in terms of the arrest phenotype rather than their execution point. For example, mutations in yeast β-tubulin can selectively affect a subset of microtubules (Sullivan and Huffaker, 1992) or cause cells to become benomyl dependent (Huffaker et al., 1988). Similarly selective tubulin mutations have been identified in other organisms as well (Oakley and Morris, 1980). However, the precise molecular basis for the defective arrest phenotype is not yet understood. A possible exception is the disruption produced by substitution of lysine for the highly conserved glutamate at position 288 in the Drosophila β2 protein; this mutation causes an apparent packing defect, so that the protofilaments do not close to form a tubule (Fuller et al., 1987). However, the same substitution in yeast β-tubulin has no apparent effect (Praitis et al., 1991). The generalizability of the mutation found in Tub1-724p also requires further testing.
Genetic Interactions between tub1-724 and PAC2
Instability of the Tub1-724p-β-tubulin heterodimer predicts that overexpression of an α-tubulin–binding protein should be deleterious to tub1-724 cells, perhaps by producing more toxic free β-tubulin in the mutant cells. The work of Tian et al. (1997) suggests that the vertebrate homologue of the yeast protein Pac2p binds α-tubulin. As predicted, overexpression of PAC2 is lethal in tub1-724 cells and causes loss of all assembled microtubules. Consistent with this result, we can recover a complex containing Pac2p and α-tubulin from double-overexpressing cells. These results demonstrate for the first time that Pac2p can bind α-tubulin in vivo. This result does not distinguish among many possible functions for PAC2. It may act as does cofactor E in the in vitro assay, facilitating the incorporation of α-tubulin into heterodimers (Tian et al., 1997), but it is not essential for that reaction, because PAC2 is not an essential gene in vivo (Hoyt et al. 1997). Δpac2 is synthetically lethal with other microtubule mutants: Δcin8 (Geiser et al., 1997), Δpac10 (Alvarez et al., 1998), and tub1-724 (Vega, unpublished results).
Regulating Microtubule Function
The first analyses of microtubules at a molecular level focused on protein factors that could be responsible for assembly in an in vitro reaction. It is striking that so many of the genes that appear to affect microtubules in vivo almost certainly do not participate in the polymerization reaction itself. In this sense, the CIN genes (Hoyt et al., 1990; Stearns et al., 1990), the PAC genes (Geiser et al., 1997), the GIM genes (Geissler et al., 1998), and the RBL genes (Archer et al., 1995), although identified—in some cases more than once—by a wide variety of approaches, have fundamental properties in common. They are not essential for cell viability in budding yeast, and their deletion does not confer a quantitative defect in microtubule assembly. Conversely, their overexpression does not increase the level of assembly, as could be expected for a modulator of microtubule assembly. For only one of these proteins, alp1, a CIN1 homologue in fission yeast, is there evidence suggesting that it binds along the length of the microtubule (Hirata et al., 1998).
A role for these proteins arises from the in vitro system for incorporating separated tubulin chains into heterodimer. Alone among proteins that have been analyzed in such assays, the tubulin polypeptides appear to require factors that act after release from the chaperonin. Without those factors, there is no exchange of newly folded polypeptide with the exogenously added heterodimer. Some of the protein factors are homologous to gene products in S. cerevisiae and S. pombe that affect microtubule functions, and in S. pombe some of them are essential (Hirata et al., 1998). That they are not essential in S. cerevisiae, however, suggests that there must be other mechanisms for folding tubulin and forming heterodimer in those cells.
These proteins may also have alternative functions. Rbl2p levels affect how cells survive alterations in the ratios of α- to β-tubulin (Archer et al., 1995). Levels of Pac10p and the GIM genes affect those ratios (Alvarez et al., 1998; Geissler et al., 1998). It is clear that yeast cells are sensitive to those ratios. These proteins may participate in maintaining proper balance of the tubulin components, which may become an important step, especially under times of stress. Such a role could help explain why expression of RBL2 mRNA increases when cells are incubated with a microtubule-depolymerizing drug (Velculescu et al., 1997), although there is no evidence that the tubulin chains themselves are expressed in greater amounts. The results from these several approaches suggest that the early steps of microtubule morphogenesis may be crucial for cell function.
ACKNOWLEDGMENTS
We thank M. Borowski, C. Hengartner, S. Sanders, and members of our laboratory for their comments and suggestions and P. Alvarez for initiating the experiments with PAC2. This work was supported by a grant from National Institutes of Health to F.S. L.R.V. was supported in part by a predoctoral fellowship from Howard Hughes Medical Institute. J.F. was supported in part by a training grant from National Institutes of Health to the Department of Biology at Massachusetts Institute of Technology.
REFERENCES
- Alvarez P, Smith A, Fleming J, Solomon F. Modulation of tubulin polypeptide ratios by the yeast protein Pac10p. Genetics. 1998;149:857–864. doi: 10.1093/genetics/149.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J, Magendantz M, Vega L, Solomon F. Formation and function of the Rbl2p-β-tubulin complex. Mol Cell Biol. 1998;18:1757–1762. doi: 10.1128/mcb.18.3.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer JE, Vega LR, Solomon F. Rbl2p, a yeast protein that binds to β-tubulin and participates in microtubule function in vivo. Cell. 1995;82:425–434. doi: 10.1016/0092-8674(95)90431-x. [DOI] [PubMed] [Google Scholar]
- Bond JF, Fridovich-Keil JL, Pillus L, Mulligan RC, Solomon F. A chicken-yeast chimeric β-tubulin protein is incorporated into mouse microtubules in vivo. Cell. 1986;44:461–468. doi: 10.1016/0092-8674(86)90467-8. [DOI] [PubMed] [Google Scholar]
- Caceres A, Kosik K. Inhibition of neurite polarity by tau antisense oligonucleotides in primary cerebellar neurons. Nature. 1990;343:461–463. doi: 10.1038/343461a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Sullivan D, Huffaker T. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci USA. 1994;91:9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsmore J, Solomon F. Inhibition of Map2 expression affects both morphological and cell division phenotypes of neuronal differentiation. Cell. 1991;64:817–826. doi: 10.1016/0092-8674(91)90510-6. [DOI] [PubMed] [Google Scholar]
- Fuller MT, Caulton JH, Hutchens JA, Kaufman TC, Raff EC. Genetic analysis of microtubule structure: A β-tubulin mutation causes the formation of aberrant microtubules in vivo and in vitro. J Cell Biol. 1987;104:385–394. doi: 10.1083/jcb.104.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénette S, Magendantz M, Solomon F. BUB1 and BUB3 are multicopy suppressors of a Saccharomyces cerevisiae α-tubulin mutation that causes a microtubule assembly defect. J Cell Sci. 1995;108:1195–1204. doi: 10.1242/jcs.108.3.1195. [DOI] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:656–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle HD, Raff EC. Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol. 1990;111:1009–1026. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M, Macke J, Roberts B, Geiser J. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics. 1997;146:849–857. doi: 10.1093/genetics/146.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker T, Thomas J, Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi HC, Cleveland DW. Differential utilization of β-tubulin isotypes in differentiating neurites. J Cell Biol. 1989;109:663–673. doi: 10.1083/jcb.109.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number in yeast. Mol Cell Biol. 1990;10:2730–2736. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D, Solomon F. Overexpression of yeast homologs of the mammalian checkpoint gene RCC1 suppresses the class of α-tubulin mutations that arrest with excess microtubules. Genetics. 1994;137:381–392. doi: 10.1093/genetics/137.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magendantz M, Henry M, Lander A, Solomon F. Inter-domain interactions of radixin in vitro. J Biol Chem. 1995;270:25324–25327. doi: 10.1074/jbc.270.43.25324. [DOI] [PubMed] [Google Scholar]
- Melki R, Rommelaere H, Leguy R, Vandekerckhove J, Ampe C. Cofactor A is a molecular chaperone required for beta-tubulin folding: functional and structural characterization. Biochemistry. 1996;35:10422–10435. doi: 10.1021/bi960788r. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf S, Downing K. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Oakley B, Morris NR. Nuclear movement is beta tubulin-dependent in Aspergillus nidulans. Cell. 1980;19:255–262. doi: 10.1016/0092-8674(80)90407-9. [DOI] [PubMed] [Google Scholar]
- Pachter JS, Yen TJ, Cleveland DW. Autoregulation of tubulin expression is achieved through specific degradation of polysomal tubulin mRNAs. Cell. 1987;51:283–292. doi: 10.1016/0092-8674(87)90155-3. [DOI] [PubMed] [Google Scholar]
- Praitis V, Katz WS, Solomon F. A codon change in β-tubulin which drastically affects microtubule structure in Drosophila melanogaster fails to produce a significant phenotype in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4726–4731. doi: 10.1128/mcb.11.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Genetically essential and nonessential α-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986;6:3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon F, Connell L, Kirkpatrick D, Praitis V, Weinstein B. Methods for studying the yeast cytoskeleton. In: Carraway K, Carraway C, editors. The Cytoskeleton. Oxford, United Kingdom: Oxford University Press; 1992. pp. 197–222. [Google Scholar]
- Stearns T, Hoyt M, Botstein D. Yeast mutants sensitive to anti-microtubule drugs define three genes that affect microtubule function. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D, Huffaker T. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan N. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Lewis S, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan N. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Culbertson M. The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991;11:2629–2640. doi: 10.1128/mcb.11.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu V, Zhang L, Zhou W, Vogelstein J, Basrai M, Bassett D, Hieter P, Vogelstein B, Kinzler K. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Weinstein B, Solomon F. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol Cell Biol. 1990;10:5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]