Abstract
In the yeast Saccharomyces cerevisiae, Sic1, an inhibitor of Clb-Cdc28 kinases, must be phosphorylated and degraded in G1 for cells to initiate DNA replication, and Cln-Cdc28 kinase appears to be primarily responsible for phosphorylation of Sic1. The Pho85 kinase is a yeast cyclin-dependent kinase (Cdk), which is not essential for cell growth unless both CLN1 and CLN2 are absent. We demonstrate that Pho85, when complexed with Pcl1, a G1 cyclin homologue, can phosphorylate Sic1 in vitro, and that Sic1 appears to be more stable in pho85Δ cells. Three consensus Cdk phosphorylation sites present in Sic1 are phosphorylated in vivo, and two of them are required for prompt degradation of the inhibitor. Pho85 and other G1 Cdks appear to phosphorylate Sic1 at different sites in vivo. Thus at least two distinct Cdks can participate in phosphorylation of Sic1 and may therefore regulate progression through G1.
INTRODUCTION
The yeast Saccharomyces cerevisiae becomes committed to a new cycle of cell division when it reaches a critical size in the presence of sufficient nutrient. This point occurs late in G1 phase and is known as Start (Hartwell et al., 1974). After passing through Start, haploid cells are no longer sensitive to mating pheromone (Hereford, 1974) and initiate DNA replication, spindle pole body duplication, and bud growth, which are critical for further cell cycle events, namely mitosis and cytokinesis. In most eukaryotic cells, the onset of S phase is prevented until cells grow to a critical size (Killander and Zetterberg, 1965), and this control is mainly exerted late in G1 phase.
Genetic analysis indicated that a unique cyclin-dependent kinase (Cdk) encoded by CDC28 is essential not only for Start but also for S phase and mitosis (Nasmyth, 1993). The Cdc28 kinase is a member of the highly conserved Cdk family found among eukaryotic cells and is activated by binding of cyclin (Nasmyth, 1993). Cyclin is now known to form a family whose members have a conserved domain called the cyclin box and to bind to Cdk, generating an active complex (Morgan, 1995). In budding yeast, distinct cyclin-Cdc28 complexes are required at different stages of the cell cycle. At Start, Cdc28 is activated by binding of three G1 cyclins, Cln1, Cln2, and Cln3 (Nasmyth, 1993). Individually, none of the genes for these three G1 cyclins is essential, but mutants in which all three CLN genes are deleted arrest in G1 (Richardson et al., 1989). A recent report showed that CLN3 is involved in activation of the transcription factors SCB binding factor (SBF) and MluI binding factor (MBF), whereas CLN1 and CLN2 have overlapping function in promotion of Start-related events, including budding, DNA replication, and cessation of Clb degradation (Dirick et al., 1995; Stuart and Wittenberg, 1995). After Start, six B-type cyclins (Clb) associate with Cdc28 to promote DNA replication (Clb5 and Clb6) (Epstein and Cross, 1992; Schwob and Nasmyth, 1993) and mitosis (Clb1 to Clb4) (Richardson et al., 1992; Surana et al., 1991). In contrast to yeast cells, in which unique Cdk functions in progression of cell cycle, vertebrate cells have various Cdks (Cdk1 to Cdk8) and cyclins (cyclins A–H), and different combinations of Cdk and cyclin are used at different stages of the cell cycle. For instance, Cdk4 and Cdk6 complexed with cyclin D are required for G1 progression, whereas Cdc2-cyclin B is required for mitosis (Nigg, 1995).
In a cln1 cln2 double mutant, Start-related events, including budding, DNA replication, spindle pole body duplication, and termination of Clb degradation, are all delayed until the cell reaches a size that is much larger than a wild-type cell at Start (Dirick et al., 1995). However, transcription of SBF- and MBF-regulated genes is activated with normal timing (Dirick et al., 1995; Stuart and Wittenberg, 1995), because activation of SBF and MBF transcription factors are carried out by Cln3-Cdc28 (Tyers et al., 1993; Dirick et al., 1995). The delay in DNA replication in a cln1 cln2 mutant is suppressed by deletion of SIC1, which encodes a Cdk inhibitor (CKI) of Clb5, 6-Cdc28 kinases (Mendenhall, 1993; Dirick et al., 1995). Cln2-Cdc28 was shown to phosphorylate Sic1 in vitro (Schwob et al., 1994). Recently, in vitro experiments demonstrated that ubiquitination of Sic1 requires phosphorylation of the inhibitor by Cln-Cdc28 (Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997a). These findings and other biochemical and genetic evidence (Schneider et al., 1996; Tyers, 1996) suggest that Cln1, 2-Cdc28 kinases are likely to target Sic1 for degradation by phosphorylating the CKI and thus enabling the activation of Clb5, 6-Cdc28 kinases. Then, a cln1 cln2 mutant must phosphorylate Sic1 to tag it for degradation in the absence of Cln1, 2-Cdc28 kinase activity. What kinase carries out this function? Can Cln3-Cdc28 kinase activate another kinase to carry out that function or can it directly phosphorylate Sic1?
In budding yeast, although a unique Cdk (Cdc28) functions in progression of the cell cycle, there exists a Cdk family whose members function in various cellular events: Ccl1 cyclin-Kin28 kinase (Valey et al., 1993, 1995; Cismowski et al., 1995) and Srb11 cyclin-Srb10 kinase (Liao et al., 1995) are involved in phosphorylation of the C-terminal domain of the largest subunit of RNA polymerase II, and Pho80 cyclin-Pho85 kinase phosphorylates the transcriptional activator Pho4 to repress transcription of PHO genes (Kaffman et al., 1994). Although whether these Cdks are involved in cell cycle regulation is not clear yet, Pho85 kinase was shown to associate with G1 cyclin homologues, including Pcl1, Pcl2, and Pcl9 (Espinoza et al., 1994; Measday et al., 1994, 1997). Expression of the genes coding for these Pcl proteins is periodical: that of PCL1 is peaked in G1 and is regulated by SBF as CLN1 and CLN2 are (Nasmyth and Dirick, 1991; Ogas et al., 1991); that of PCL9 peaks at the end of M and is regulated by Swi5 (Aerne et al., 1998); and that of PCL2 reaches the maximum between those of PCL1 and PCL9, being regulated by SBF and Swi5 (Measday et al., 1994; Aerne et al., 1998). PHO85 is not essential for cell growth (Uesono et al., 1987), but it becomes indispensable when both CLN1 and CLN2 are absent (Espinoza et al., 1994; Measday et al., 1994). Similarly, either PCL1 or PCL2 is required for G1 progression when both CLN1 and CLN2 are deleted (Measday et al., 1994). These genetic results suggest that Pho85 kinase activity is required in the absence of Cln1, 2-Cdc28 kinase activity.
To clarify the role of PHO85 in the regulation of G1 progression, we studied whether Pho85 kinase can function as a Sic1 kinase. We find that the Pcl1-Pho85 complex can phosphorylate Sic1 in vitro, and that PHO85 affects the stability of Sic1 in vivo. We also show that three consensus sites for phosphorylation by Cdk in the Sic1 molecule are important for prompt degradation of the CKI, and that the Pho85 kinase is involved in phosphorylation of one of the three sites in normal cell cycle progression.
MATERIALS AND METHODS
Strains and Media
Escherichia coli DH5α and BL21 strains (Sambrook et al., 1989; Studier et al., 1990) were used for plasmid construction and production of fusion proteins, respectively. Yeast strains used were MFY115 (MATα leu2 ura3 trp1 ade1 his GAL+) and MFY116 (MFY115 pho85Δ) (Nishizawa, Suzuki, Fujino, Oguchi, and Toh-e, unpublished data). MFY151 (MATa ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3 GAL cln1::hisG cln2 METp-CLN2[TRP1] pho85::LEU2) and MFY152 (MFY151 sic1::URA3) were derived from K3652 and K4900 (Dirick et al., 1995), respectively, by disrupting the PHO85 locus with a LEU2 fragment. Yeast cells were grown in synthetic dextrose (SD) medium containing 0.67% Difco (Detroit, MI) yeast nitrogen base, 2% glucose, and appropriate nutritional supplements or SGal medium in which galactose replaces glucose in SD (Rose et al., 1990).
PCR Cloning and Mutagenesis
DNA fragments encoding the open reading frames of SIC1, CLB2, CLB5, PCL1, and PCL2 were cloned by PCR using primers listed in Table 1. The primers were synthesized to incorporate an NcoI site at the start codon and a BglII or XhoI site at the 3′ end of the ORF. PCR reaction was carried out in a 50-μl reaction mixture containing 100 ng of yeast chromosomal DNA, a 20 pM concentration of each primer, a 200 μM concentration of each dNTP, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, and 5 U of Taq DNA polymerase (Pharmacia, Piscataway, NJ). The mixture was incubated at 95°C for 30 s, at 55°C for 1 min, and at 72°C for 2 min, a cycle that was repeated 30 times. After the PCR reaction, excess primers were removed with a Microspin S-200 HR column (Pharmacia), and the DNA was cleaved with restriction enzymes appropriate for cloning the fragments into plasmid pSP73.
Table 1.
Primers used for PCR cloning and mutagenesis
| SIC1 | MN107 | GACTATTACACGACCATGGCTCCTTCCACC |
| MN108 | TTTCAGATCTTGAATGCTCTTGATCCCTAG | |
| SIC1 mutant | MN113 | ACACGACCATGGCTCCTTCCGCCCCACCAAGGTCC |
| MN114 | TGCAAGGTCAAAAGGTCCCCCAAAAGCCTT | |
| MN115 | TTAATGGGCTTACGGCCCCTCAACGCTCGCC | |
| CLB2 | MN103 | TGATCTTATCCATGGCCAACCCAA |
| MN104 | GCCCCTCTTCTCGAGCATGCAAGG | |
| CLB5 | MN105 | CACCTTTACTGAACCATGGGAGAGAACCAC |
| MN106 | CTAATAGATCTAAGATTAAATAGATTTTGA | |
| PCL1 | MN109 | GTAAAGTAATACCATGGGTGAATACAGC |
| MN110 | CCACATTAAAACTCGAGTTGACTCATGA | |
| PCL2 | MN118 | TTACTACAAACCATGGCAAACTACGAAGCC |
| MN121 | CCCAGTTTTCAAGATCTCAGGGCGCGC |
DNA encoding Sic1 variants with specific amino acid substitutions within three phosphorylation regions were also constructed by PCR as described above using cloned SIC1 as template and primers listed in Table 1. DNA fragment encoding a T5A point mutation was cloned by PCR using MN113 and MN108 primers. A truncated DNA fragment bearing the T33V or S76A mutation at its 5′ end was mixed with the fragment encoding the full length of the wild-type SIC1 ORF, followed by denaturation and annealing to form a heteroduplex fragment. The gaps were filled with Taq DNA polymerase at 72°C for 3 min, and the resulting fragment was subjected to PCR using MN107 and MN108 primers to obtain the full-length DNA fragment encoding Sic1 T33V or S76A variant. To construct DNA fragments bearing a double mutation, a pair of fragments each bearing a single mutation (T5A and S76A or T5A and T33V) was subjected to heteroduplex formation, filling the gaps, and PCR as described above. The T5A S76A fragment was then mixed with a truncated fragment bearing T33V mutation, followed by heteroduplex formation, filling the gaps and PCR amplification to generate a fragment containing the T5A T33V S76A point mutations. The triple mutant fragment was then subjected to PCR using MN107 and MN108 primers to generate a fragment containing the T33V S76A double mutation. All constructions were confirmed by DNA sequencing.
Construction of Plasmids
Plasmid pAT484 carrying the PHO85 gene without the intron sequence was cleaved with PstI and BamHI, and the ends were converted to the blunt end and SalI site, respectively. The blunt end of the PHO85 fragment was converted to BamHI, and the resulting BamHI–SalI fragment was cloned into pGEX-4T-2 (Pharmacia) for production of glutathione S-transferase (GST)-Pho85 fusion protein in bacteria. To produce GST-tagged proteins in yeast, pKT10 plasmid was modified by inserting a fragment encoding GST downstream of the TDH3 promoter (Tanaka et al., 1990). A BamHI–SalI PHO85 fragment was then inserted downstream of the tag sequence to generate a plasmid expressing GST-Pho85. A pSP73-based plasmid, pMF906, was constructed by inserting a GAL10 promoter fragment derived from pBM252 (Johnston and Davis, 1984) and the 3 × hemagglutinin (HA) epitope fragment into the vector to contain NcoI, BglII, and XhoI sites between the two fragments. An NcoI–BglII fragment encoding SIC1 and a TRP1–ARS1–CEN4 fragment derived from pMF557 (Nishizawa et al., 1994) were incorporated into pMF906 to generate plasmids that direct overproduction of HA-tagged protein under the control of the GAL10 promoter. To regulate expression of PHO85 by methionine, plasmid pMF925 containing the MET3 promoter derived from pHAM8 plasmid (Korch et al., 1991) and an ARS1–CEN3–URA3 fragment was digested with BamHI and SalI, and a BamHI–SalI PHO85 fragment was inserted to generate plasmid pMF927.
Purification of GST-tagged Protein
E. coli BL21 cells (Studier et al., 1990) harboring the pGEX-PHO85 plasmid was grown to midlog phase and production of GST-Pho85 protein was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 1 mM and incubation at 37°C for 3 h. The cells were then collected by centrifugation at 5000 × g for 5 min and washed once with PBS (20 mM phosphate buffer, pH 7.2, 150 mM NaCl), followed by suspension in PBS containing 10 mM EDTA. After addition of lysozyme to a concentration of 0.5 mg/ml, the suspension was placed on ice for 20 min and subjected to sonication for 15 s, which was repeated three times. The bacterial lysate was centrifuged at 15,000 × g and 4°C for 15 min, and the supernatant was saved for purification of the GST-Pho85 protein. The fusion protein was absorbed to a glutathione-Sepharose 4B column (Pharmacia) equilibrated with PBS containing 0.1% Triton X-100, and the column was washed with the same buffer. GST-Pho85 protein was eluted with 5 mM glutathione in 20 mM Tris-HCl, pH 7.5, and the solution containing the GST-tagged protein was dialyzed against buffer A (20 mM phosphate buffer, pH 7.2, 150 mM NaCl, 10 mM KCl, and 10% glycerol).
Immunoblot Analysis of Sic1 Stability
To analyze the stability of an HA-tagged mutant and the wild-type Sic1 proteins, yeast cells harboring appropriate plasmid were grown to midlog phase in 50 ml of SD medium lacking tryptophan, harvested by centrifugation, and washed twice with distilled water and once with SGal medium. After being resuspended in 50 ml of SGal medium lacking tryptophan, the cells were incubated at 30°C for 3 h. Glucose was then added to a final concentration of 2% to shut off the production of the tagged Sic1 protein while incubation at 30°C was continued. At 1-h intervals from 0 (at the time of addition of glucose) to 3 h, 10 ml of culture were removed, and the cells were collected, washed, and resuspended in buffer A containing 1 mM PMSF and a proteinase inhibitor mixture (2.5 μg/ml aprotinin, leupeptin, pepstatin A, and antipain, 50 μg/ml l-1-tosylamide-2-phenylethylchloromethyl ketone and Nα-p-tosyl-l-lysine chloromethyl ketone). The cells were disrupted by vortexing with glass beads (0.45 mm diameter), and the extracts were cleared by centrifugation at 12,000 × g for 5 min at 4°C.
To analyze the effect of PHO85 on Sic1 stability, pho85Δ cells harboring two plasmids (MET3p-PHO85 and GAL10p-SIC1-HA) were grown as described above, except that the media were supplemented with 10 mM methionine to repress PHO85 expression during the induction of HA-Sic1. At the end of the induction period, cells were harvested and washed twice with distilled water and once with SD medium, followed by resuspension in 100 ml of SD medium lacking methionine. The suspension was divided into two equal portions, and one of them was supplemented with 10 mM methionine whereas the other was not. Both were incubated at 30°C, and cell extracts were prepared periodically as described above.
Plasmid pMT290 (Willems et al., 1996) was cleaved with PvuII to obtain a fragment containing GAL1p-CLN2-HA, which was used to transform MFY115 and MFY116. Production of Cln2-HA was induced by incubating the transformant cells in 50 ml of SGal medium lacking leucine at 30°C for 3 h and was shut off by addition of glucose. At 15-min intervals from 0 to 45 min, 5 ml of culture were removed, and cell extracts were prepared as described above.
Proteins (40–50 μg) were separated on an SDS-10% polyacrylamide gel and electrotransferred onto a nitrocellulose membrane, and the blot was soaked in TTBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.03% Tween 20) containing 5% nonfat milk for 1 h, followed by incubation with affinity-purified anti-GST antibody (GST Ab, 1:5000 dilution) or anti-HA monoclonal antibody (HA Ab, 1:100 dilution, clone 12CA5; Boehringer Mannheim, Indianapolis, IN) in TTBS containing 1% nonfat milk for 30 min at room temperature. The membrane was rinsed with TTBS three times for 10 min each, and the GST or HA fusion proteins were then probed by incubating with horseradish peroxidase–conjugated goat antibodies to rabbit or mouse immunoglobulin G in TTBS containing 1% nonfat milk for 30 min at room temperature, followed by washing of the blot with TTBS as described above. The proteins were visualized with a Renaissance chemiluminescence system (New England Nuclear, Boston, MA) and by exposing to a hyperfilm ECL (Amersham, Arlington Heights, IL). The amount of proteins loaded onto each lane was quantitated by probing the blot with anti-actin monoclonal antibody (clone C4, Boehringer Mannheim). Immunoblot images were captured using a ScanJet IIcx/T scanner (Hewlett-Packard, Palo Alto, CA) and DeskScan II software and were quantitated with NIH Image software, version 1.61 (National Institutes of Health, Bethesda, MD).
Immunoprecipitation and Kinase Assay
Yeast cells harboring a plasmid encoding Pcl1-HA, Clb2-HA, or the wild-type or E53A mutant Pho85 proteins tagged to GST, were grown in 10 ml of SD medium lacking appropriate nutrient to midlog phase. Production of HA-tagged proteins was then induced by transferring the cells to 10 ml of SGal medium after washing with distilled water and incubating at 30°C for 3 h. The cells were harvested and resuspended in 0.5 ml of lysis buffer (50 mM Tris-HCl buffer, pH 7.5, 2 mM sodium pyrophosphate, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 1 mM PMSF, and the proteinase inhibitor mixture). The cells were disrupted by vortexing with glass beads (0.45 mm diameter), and the extracts were cleared by centrifugation at 12,000 × g for 5 min at 4°C. To 50 μl of the extract containing ∼200 μg of protein, 1 μl of affinity-purified GST Ab or 4 μl of HA Ab (clone 12CA5) were added, and the mixture was incubated on ice for 1 h. Thirty microliters of protein A-Sepharose 6B (Pharmacia) suspended in the lysis buffer were then added, and the incubation was continued for 1 h at 4°C. The immunoprecipitates were recovered by centrifugation at 800 × g for 1 min at 4°C and washed three times with radioimmunoprecipitation assay buffer (lysis buffer supplemented with 150 mM NaCl) and twice with kinase assay buffer (10 mM HEPES, pH 7.2, 10 mM MgCl2, 50 mM NaCl, 2 mM EDTA, 1 mM DTT, and 0.02% Triton X-100).
To activate bacterial GST-Pho85, yeast extracts were prepared from cdc28-4 mutant cells grown at 24°C essentially as described (Deshaies and Kirschner, 1995), except that spheroplasts were prepared by digestion with Zymolyase 100T (1 mg/ml; Seikagaku, Tokyo, Japan) at 30°C for 20 min, and washed spheroplasts were resuspended in YEB buffer (125 mM potassium acetate, 30 mM HEPES-KOH, pH 7.2, 3 mM EDTA, 3 mM EGTA, 2 mM DTT, 1 mM PMSF, and the proteinase inhibitor mixture) containing 0.1% Nonidet P-40 and disrupted with several strokes of a Teflon pestle homogenizer. The lysate was cleared by centrifugations at 16,000 × g for 10 min and then at 45,000 rpm for 30 min (55.2 Ti rotor; Beckman Instruments, Palo Alto, CA), followed by spin column gel filtration on Sephadex G-25 (Pharmacia) to desalt the cleared lysate. Activation reaction mixtures (10 μl) contained 5 μl of the yeast extracts, 2 μl of purified GST-Pho85, GST-Pho85E53A, or GST (∼50 ng), 1 μl each of YEB, 10× ATP mixture (10 mM ATP, 350 mM creatine phosphate, 20 mM HEPES buffer, pH 7.2, 10 mM magnesium acetate, 500 μg/ml creatine kinase), and 10× reaction buffer (50 mM magnesium acetate, 10 mM DTT, 10 mM PMSF, and the protease inhibitor mixture). Reactions were incubated at 30°C for 30 min and terminated by adding 180 μl of ice-cold immunoprecipitation buffer (IPB; 50 mM glycerol 2-phosphate, 100 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 1 mM PMSF, and the protease inhibitor mixture) and 10 μl of bacterial lysate containing cyclin. The GST-Pho85-cyclin complex was recovered by immunoprecipitation with 2 μl of affinity-purified GST Ab at 4°C for 1 h, followed by mixing with 50 μl of protein A-Sepharose beads in IPB at 4°C for 1 h. Immunoprecipitates were washed three times with IPB and twice with the kinase assay buffer.
The protein A beads were then resuspended in 40 μl of the reaction buffer containing 2.5 μM ATP, 0.5 μCi of [γ-32P]ATP, 2 μg/ml purified GST-Sic1 fusion protein and incubated for 30 min at 30 or 36°C to inactivate contaminating Cdc28 kinase activity when using bacterially produced kinases (Wittenberg and Reed, 1988). The reaction was stopped by adding 15 μl of 4× SDS loading buffer (0.2 M Tris-HCl, pH 6.8, 0.2 M DTT, 8% SDS, 0.4% bromophenol blue, 40% glycerol), and the proteins were denatured by incubating at 95°C for 3 min before electrophoresis on an SDS-polyacrylamide gel and were analyzed by autoradiography.
Incorporation of 32P into the Wild-Type or Mutant Sic1 In Vivo
To label the HA-tagged mutant or wild-type Sic1 with 32PO4, yeast cells harboring appropriate plasmid were grown in 2 ml of SD medium lacking tryptophan and inoculated into 20 ml of low-phosphate medium (Toh-e et al., 1973) containing 50 μM potassium phosphate and 2% glucose but lacking tryptophan, which was incubated at 30°C for 15 h. The cells were collected by centrifugation and washed twice with distilled water and once with low-phosphate medium containing 50 μM potassium phosphate and 2% galactose but lacking tryptophan. After being resuspended in the same medium, 125 μCi of 32PO4 (8500–9120 Ci/mmol, New England Nuclear) were added to the culture, which was incubated at 30°C for 3 h. The cells were harvested, washed, and resuspended in buffer A containing 0.1% Triton X-100, the proteinase inhibitor mixture, and phosphatase inhibitors (5 mM NaF, 5 mM NaVO3, 2.5 mM β-glycerophosphate, 100 nM okadaic acid). The cell extracts were prepared as described above, and Sic1-HA was immunoprecipitated from 40 μl of extract using 40 ng/μl HA Ab and 20 μl of protein A-Sepharose suspended in buffer A containing 0.1% Triton X-100, the protease inhibitor mixture, and the phosphatase inhibitors. After being washed with the same buffer, the beads were suspended in 40 μl of water and 15 μl of 4× SDS loading buffer without DTT. Proteins were denatured by incubating at 95°C for 3 min, separated on an SDS polyacrylamide gel, and subjected to autoradiography or immunoblotting.
Growth and Viability of cln1 cln2 pho85 and cln1 cln2 pho85 sic1 Mutants
K3652, K4900, MFY151, and MFY152 cells were grown in SD medium lacking methionine at 30°C to sustain their growth with CLN2. When cell concentration reached 0.6–0.7 × 107 cells/ml, the culture was divided into two equal portions, and one was supplemented with 10 mM methionine to shut off CLN2 expression. The culture was then incubated at 24°C, and at 1.5-hr intervals from 0 (at the time of addition of methionine) to 9 h, aliquots were removed to count cell number with a hemocytometer, and at 3-hr intervals, aliquots removed from the methionine-supplemented medium were diluted and plated onto SD medium lacking methionine to determine the number of viable cells.
RESULTS
Pho85 Kinase Can Phosphorylate Sic1 In Vitro
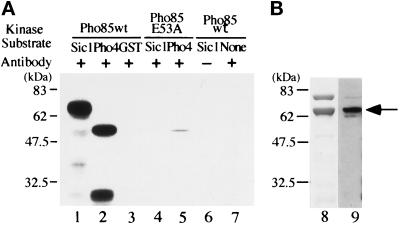
To clarify the role of Pho85 kinase in progression of G1, we first tested whether Pho85 kinase can phosphorylate Sic1 in vitro. GST-Pho85 fusion protein was immunoprecipitated from yeast cell extracts and subjected to kinase assay using GST-Sic1 or T7 gene10-Pho4 (Ogawa et al., 1995) as substrate. As shown in Figure 1, Sic1 and Pho4 (lanes 1 and 2) were phosphorylated, whereas GST (lane 3) was not, indicating that phosphorylation did not occur in the GST portion of the fusion protein. When the Pho85 E53A mutant that lacks kinase activity (Fujino et al., 1994; Nishizawa, Suzuki, Fujino, Oguchi, and Toh-e, unpublished data) was immunoprecipitated and used for kinase assay, almost no phosphorylation was detected in Sic1 or Pho4 (Figure 1, lanes 4 and 5), indicating that Pho85 kinase activity was responsible for the phosphorylation of Sic1. In the absence of GST-Sic1, phosphorylated proteins were not detected (Figure 1, lane 7). The mobility of the phosphorylated protein (Figure 1, lane 1) corresponds to those of GST-Sic detected by staining the gel with Coomassie brilliant blue (Figure 1, lane 8) and by immunoblotting with GST Ab (Figure 1, lane 9), indicating that the phosphoprotein in Figure 1, lane 1, was GST-Sic1. Thus Pho85 kinase could phosphorylate Sic1 as well as Pho4 in vitro.
Figure 1.
(A) Immunoprecipitated GST-Pho85 can phosphorylate Sic1 in vitro. GST-Pho85 (the wild type) or GST-Pho85E53A mutant proteins were immunoprecipitated from yeast cell extracts with affinity-purified anti-GST antibody (GST Ab) and subjected to kinase assay using GST-Sic1 (lanes 1, 4, and 6), T7 gene 10-Pho4 (lanes 2 and 5), or GST (lane 3) as substrate. Control experiments were carried out without antibody (lane 6) and without external substrate (lane 7). The band seen at the bottom of lane 2 is a degradation product of Pho4. Molecular sizes are indicated to the left. (B) Profile of GST-Sic1 preparation. A GST-Sic1 protein preparation used as substrate for kinase assay was separated on an SDS polyacrylamide gel, followed by staining the gel with Coomassie brilliant blue (lane 8) and by immunoblotting with GST Ab (lane 9), respectively. The position of GST-Sic1 is designated by an arrow. Note that no significant difference in the mobilities of phosphorylated and unphosphorylated GST-Sic1 was observed with the gel system used in this study.
Pcl1 is a Cyclin Partner of Pho85 to Phosphorylate Sic1
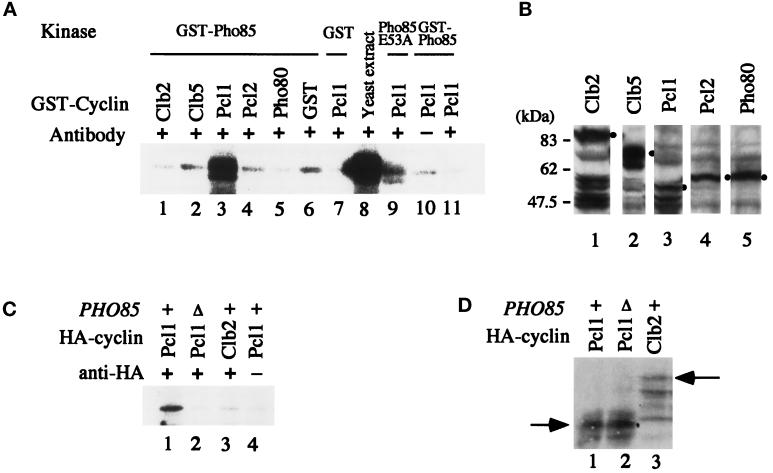
Because Pho85 kinase is a Cdk, it should require cyclin to phosphorylate Sic1. To identify the cyclin partner, we reconstituted the cyclin-Pho85 complex in vitro using bacterially produced proteins and analyzed its ability to phosphorylate Sic1. When we simply mixed GST-Pho85 purified from E. coli with bacterial cell extract containing various cyclin, we failed to detect any Sic1 kinase activity (our unpublished results). We, therefore, tried to activate bacterial Pho85 kinase using yeast extracts (Deshaies and Kirschner, 1995). To circumvent contamination by Cdc28 kinase activity when assaying Sic1 phosphorylation, we used yeast extracts prepared from temperature-sensitive cdc28 mutant cells (cdc28-4) and assayed Sic1 phosphorylation at 36°C to inactivate Cdc28 kinase (Wittenberg and Reed, 1988). After activation, bacterial Pho85 kinase was mixed with bacterial extracts containing Clb2, Clb5, Pcl1, Pcl2, Pho80, or GST alone and was immunoprecipitated with anti-GST Ab. When the immunocomplex was subjected to Sic1 kinase assay, Pcl1-Pho85 combination strongly phosphorylated Sic1 (Figure 2A, lane 3). The observations that the mobility of the phosphorylated band in Figure 2A, lane 3, corresponded to that obtained by GST-Pho85 immunoprecipitate from yeast extracts (lane 8) in the experiment depicted in Figure 1, and that almost no phosphorylated band was detected when the Sic1 substrate was not included in the reaction mixture (lane 11), indicate that the proteins phosphorylated by Pcl1-Pho85 was GST-Sic1. When GST alone (lane 7) or the nonfunctional Pho85 E53A mutant fused to GST (lane 9) were combined with Pcl1, little Sic1 kinase activity was observed. These results indicate that Sic1 phosphorylation was dependent on functional Pho85 kinase and not on other kinases present in the yeast extracts used to activate bacterial Pho85. Although the Pho85 E53A mutant failed to bind Pho80 and to phosphorylate Pho4 in vitro (Fujino et al., 1994; Nishizawa, Suzuki, Fujino, Oguchi, and Toh-e, unpublished data), it might interact with Pcl1 to some extent to give weak phosphorylation of Sic1. Because Clb2, Clb5, Pcl1, Pcl2, and Pho80 cyclins were all present in E. coli extracts, although with different degrees of degradation (Figure 2B), mere absence of cyclin was not the cause of failure to detect phosphorylation of Sic1 in the case of Clb2, Clb5, Pcl2, and Pho80. With respect to functioning of GST-cyclin fusions, GST-Pho80, when complexed with bacterial Pho85, could phosphorylate Pho4 in vitro (Nishizawa, Suzuki, Fujino, Oguchi, and Toh-e, unpublished data).
Figure 2.
The Pcl1-Pho85 complex can phosphorylate Sic1 in vitro. (A) GST-Pho85 (the wild type), GST-Pho85E53A mutant protein, or GST purified from E. coli was incubated with the yeast extracts prepared from cdc28-4 cells at 30°C for 30 min and then mixed with bacterial cell extracts containing various GST-tagged cyclin as designated. The cyclin-Pho85 complexes were immunoprecipitated with GST Ab and subjected to kinase assay carried out at 36°C for 30 min with GST-Sic1 as substrate. As control experiments, bacterial extracts containing GST alone were used as a cyclin source (lane 6), purified GST was used as a kinase source (lane 7), GST Ab was not added to the reaction mixture (lane 10), and no external substrate was added to the kinase assay reaction (lane 11). For positive control, GST-Pho85 was immunoprecipitated from yeast cell extracts as described in the legend to Figure 1 and subjected to the kinase assay (lane 8). Note that the bands observed in lanes 1, 2, 4–6, 10, and 11 are probably nonspecific phosphorylation of a protein present in bacterial extracts, because it is observed even in the absence of the Sic1 substrate (lane 11). (B) Cell extracts were prepared from E. coli BL21 producing GST-cyclin as indicated and analyzed by immunoblot using affinity-purified GST Ab. Although degradation of the fusion proteins was observed in the case of Clb2, Clb5, and Pcl1 (lanes 1–3), the band showing the slowest mobility corresponds to the intact form of each GST-cyclin, as indicated by a dot. The positions of the molecular weight markers are shown at the left. (C) Extracts were prepared from the wild-type (+) or pho85Δ cells producing Pcl1-HA or Clb2-HA as indicated, and HA Ab (clone 12CA5) was added (+) to precipitate cyclin and associated kinase. As a negative control, the antibody was not added (−) to the reaction (lane 4). The immunocomplex was then subjected to kinase assay using purified GST-Sic1 as substrate. (D) The yeast cell extracts used in C were analyzed for the presence of HA-cyclin by immunoblot using HA Ab. The lane numbers correspond to those of C, and the positions of Pcl1-HA and Clb2-HA are indicated by arrows.
To further confirm that Pcl1-Pho85 complex can phosphorylate Sic1, we overproduced HA-tagged Pcl1 in yeast and analyzed whether an immunoprecipitate with HA Ab can phosphorylate Sic1 in vitro. As shown in Figure 2C, the immunoprecipitate obtained from PHO85+ cells but not from pho85Δ cells showed Sic1 kinase activity (lanes 1 and 2). Phosphorylation of Sic1 was not detected when immunoprecipitated Clb2 was used as a kinase source (Figure 2C, lane 3). Pcl1-HA and Clb2-HA were produced in pho85Δ and PHO85+ cells, respectively, as shown in Figure 2D (lanes 2 and 3).
Sic1 Becomes Stable in pho85Δ Mutant Cells
To analyze whether PHO85 is involved in phosphorylation and subsequent degradation of Sic1 in vivo, we tested whether PHO85 affects the stability of Sic1 by comparing its amount in PHO85+ and pho85Δ cells. When Sic1 was overproduced under the direction of the GAL10 promoter, it appeared to accumulate to a greater degree in the mutant cells than in the wild-type cells (Figure 3B, Sic1 wild-type). Although the two strains are isogenic (Nishizawa, Suzuki, Fujino, Oguchi, and Toh-e, unpublished data), it is more conclusive to compare the stability in the same strain. For that purpose, we expressed PHO85 under the MET3 promoter (pMF927) in pho85Δ cells harboring GAL10p-SIC1-HA plasmid, so that the production of Pho85 kinase is regulated by methionine (Korch et al., 1991). In the absence of methionine, pho85Δ cells transformed with pMF927 could repress expression of acid phosphatase under the high-phosphate-concentration condition, confirming the production of functional Pho85 (Nishizawa, unpublished results).
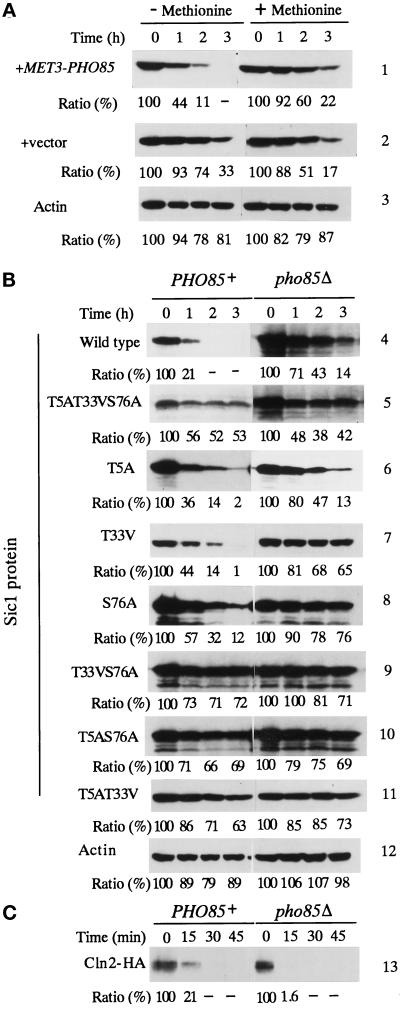
Figure 3.
Stability of the wild-type and mutant Sic1 in the presence and absence of PHO85. (A) pho85Δ cells harboring MET3-PHO85 (column 1) or vector (column 2) and GAL10-SIC1-HA plasmids were grown in the medium containing galactose and methionine to accumulate Sic1-HA. The cultures were divided into two portions and transferred to SD medium, and one was supplemented with methionine to repress production of Pho85, whereas the other was not. Portions of cells were removed periodically, and the cell extracts were prepared to assay the amount of remaining Sic1-HA in the cell by immunoblotting. Proteins loaded onto each lane were quantitated by immunoblotting of actin with anti-actin antibody (column 3). (B) HA-tagged wild-type or mutant Sic1 was produced in PHO85 (left panel) or pho85Δ (right panel) cells by growing them in SGal medium. Glucose was added to turn off the production of Sic1-HA proteins and portions of the cells were removed periodically to assay the amount of remaining HA-tagged proteins in the cell by immunoblotting. Proteins loaded onto each lane were quantitated by probing the blot with anti-actin antibody (column 12). Quantitation of Sic1 protein is expressed as a ratio, taking the value at 0 h as 100. (C) Stability of Cln2-HA in PHO85 (left panel) or pho85Δ (right panel) cells was studied by a periodical assay of the amount of the tagged Cln2 remaining in the cell by immunoblotting.
The transformants were grown in galactose medium supplemented with methionine to accumulate Sic1-HA in the absence of Pho85 kinase. The cells were then transferred to glucose medium to halt further Sic1-HA production, and the cultures were divided into two equal portions: one kept methionine free and the other supplemented with methionine. As shown in Figure 3A, in the presence of Pho85 kinase (medium lacking methionine), most of the accumulated Sic1 was degraded after 2 h (11% of the initial amount was remaining), whereas a significant amount of Sic1 remained (60% of the initial amount) in the absence of the kinase (methionine-containing medium) after the same period (Figure 3A, column 1). In pho85Δ transformed with vector and GAL10p-SIC1-HA plasmid, the stability of Sic1 was similar in the presence and absence of methionine (Figure 3A, column 2), indicating that addition of methionine did not cause stabilization of Sic1. These results indicated that Sic1 was more stable in vivo in the absence of PHO85.
Different Roles of Cdc28 And Pho85 Kinases in Phosphorylation of Sic1
It is believed that phosphorylation of Sic1 by Cln-Cdc28 and subsequent degradation of Sic1 is required for the initiation of DNA replication (Dirick et al., 1995; Schneider et al., 1996). We now demonstrated that Pho85 kinase can phosphorylate Sic1 in vitro and that PHO85 affects Sic1 stability in vivo. If Pho85 kinase is really involved in phosphorylation of Sic1 in vivo, do two different Cdk-cyclin complexes participate in one reaction, or does Sic1 phosphorylation by Pho85 have physiological relevance only in the absence of CLN1 and CLN2? To answer these questions, we focused on three consensus sites for phosphorylation by Cdk present in the Sic1 molecule, T5, T33, and S76 (consensus sequences are TPPR, TPQK, and SPQR, respectively). On the other hand, the Pho85 kinase was shown to phosphorylate the serine residues in the SPXI/L of Pho4 (O’Neill et al., 1996) and in the SPXDL sequence of Gsy2 (Huang et al., 1996) (X stands for any amino acid residue). Therefore, it is possible that Cdc28 and Pho85 kinases phosphorylate different sites in Sic1. To test this idea, we first constructed a Sic1 variant bearing amino acid substitutions within these phosphorylation sites and analyzed whether the mutant molecule is phosphorylated by Pho85 or Cdc28 in vitro. If it is the case, the two kinases are likely to phosphorylate different sites of Sic1. However, Cdc28 and Pho85 could phosphorylate the triple Sic1 mutant efficiently in vitro (our unpublished results). This raises a possibility that Cln-Cdc28 kinase phosphorylates Sic1 at sites other than the three consensus ones. Therefore, we next analyzed whether the three consensus sites in Sic1 are functionally important in vivo by studying Sic1 stability and the overproduction effect of the triple mutant protein. As shown in Figure 3B, the triple mutant protein was more stable than the wild-type Sic1, regardless of the presence of PHO85, suggesting that the three consensus phosphorylation sites were important for proper degradation of Sic1. In accord with this, overproduction of the triply altered Sic1 protein inhibited the growth of both PHO85+ and pho85Δ cells (Figure 4A, 8), and cells were arrested with elongated buds (Figure 4C). On the other hand, overproduction of the wild-type Sic1 protein was tolerable to both PHO85+ and pho85Δ cells (Figure 4A, 1). Taken together, the three consensus phosphorylation sites in Sic1 were functionally important in vivo; that is, phosphorylation of these sites was required for degradation of Sic1.
Figure 4.
Growth inhibition of PHO85 or pho85Δ cells by overproduction of various Sic1 mutant proteins. (A) PHO85 (left panel) or pho85Δ (right panel) cells harboring plasmids expressing HA-tagged wild-type or various mutant Sic1 under the direction of GAL10 promoter were streaked on SD (top panel) or SGal medium (bottom panel) and photographed after 2 d (SD medium) or 5 d (SGal medium) at 30°C. Sic1 proteins are: 1, the wild type; 2, S76A; 3, T5A; 4, T33V; 5, T33VS76A; 6, T5AS76A; 7, T5AT33V; 8, T5AT33VS76A; and 9, vector alone. (B) Production of the wild-type and mutant Sic1-HA was analyzed by immunoblotting. The numbers of the slots correspond to those described in A. (C) Morphology of PHO85 (top panel) or pho85Δ (bottom panel) overproducing HA-tagged wild-type (left panel) or T5AT33VS76A mutant (middle panel) Sic1. Cells were photographed after grown in SGal or SD (right panel) medium for 4.5 h.
We next asked whether Pho85 kinase is involved in phosphorylation of any of the three sites by studying the effect on cell growth of overproduction of Sic1 mutant proteins that have only one or two of the consensus sites altered. When two of the three sites were mutated, such mutant proteins inhibited the growth of both PHO85 and pho85Δ cells (Figure 4A, 5–7). When mutant proteins with only one altered site were overproduced, PHO85 cells could grow regardless of the mutation site, although with different efficiency, whereas pho85Δ cells could tolerate only overproduction of the T5A mutant protein that had intact T33 and S76 residues (Figure 4A, 2–4). The presence of Sic1 protein was confirmed by Western blotting analysis (Figure 4B). The growth inhibition was shown to correlate with degradation of Sic1p mutants: in PHO85 cells, T5A, T33V, and S76A mutant proteins were degraded promptly (Figure 3B, columns 6–8), whereas T33VS76A, T5AS76A, and T5AT33V were stable (Figure 3B, columns 9–11). In the absence of PHO85, all Sic1 mutant proteins tested were stable (Figure 3B, columns 5, 7–11), except that the T5A mutant was degraded as efficiently as the wild-type Sic1 (Figure 3B, columns 4 and 6). Stability of Cln2-HA did not differ significantly in PHO85+ and pho85Δ cells (Figure 2C, column 13), indicating that the pho85Δ mutation did not cause general stabilization of cellular proteins. These results indicate that at least two consensus phosphorylation sites were required for prompt degradation of Sic1 and imply that Pho85 kinase is required when the T5 residue is one of the two consensus phosphorylation sites remaining in the Sic1 molecule.
Pho85 Kinase and In Vivo Phosphorylation of Sic1
We next analyzed whether the three consensus phosphorylation sites of Sic1 are really phosphorylated in vivo. Labeling of Sic1-HA with radioactive Pi demonstrated that T5A and T5AS76A mutant proteins as well as the wild-type Sic1 were labeled both in PHO85 and pho85Δ cells (Figure 5A, lanes 1, 2, 4, 6, 7, and 9), although labeling was more efficient (approximately threefold) in the presence of PHO85 (Figure 5A, lanes 4 and 9, when normarized to the content of Sic1-HA). The reason for this difference is unknown. On the other hand, a T33VS76A mutant was labeled in PHO85 (20% of the phosphorylation level of the wild-type Sic1) but not to a detectable level in pho85Δ cells (Figure 5A, lanes 3 and 8), suggesting that PHO85 was involved in phosphorylation of T5 residue. Incorporation of radioactivity into the triple Sic1 mutant was not detected in either type of cells (Figure 5A, lanes 5 and 10). Western analysis shown in Figure 5B indicates that the amounts of the wild-type and mutant Sic1 proteins were not significantly altered.
Figure 5.
In vivo phosphorylation of the wild-type and mutant Sic1 proteins. PHO85 or pho85Δ cells harboring plasmids expressing HA-tagged wild-type or various mutant Sic1 as indicated were grown in low-phosphate medium, and 32PO4 was added upon induction of Sic1-HA. Sic1 proteins were immunoprecipitated from cell extracts with HA Ab and subjected to SDS-PAGE, followed by autoradiography (A) or Western blot analysis (B). The levels of phosphorylation (A) and the amounts of Sic1 protein (B) were quantitated by densitmetory and expressed as ratio against those of the wild-type Sic1 protein in PHO85+ cells. In A, the levels of phosphorylation were also indicated as normalized values based on the amounts of Sic1 proteins.
Effect of a Deletion of SIC1 from cln1 cln2 pho85 Strain
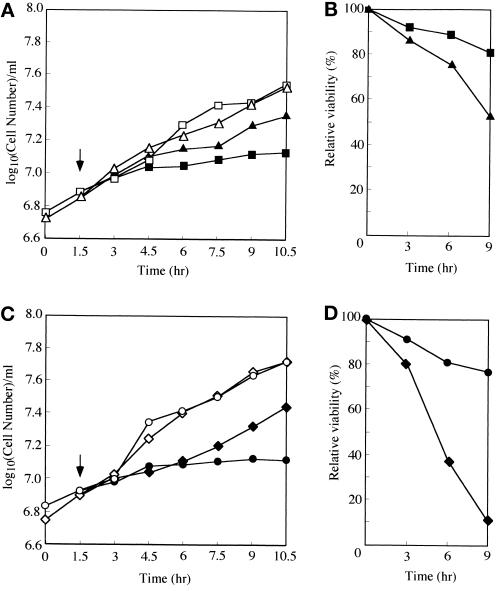
Our biochemical and genetic evidence indicates that Pho85 kinase is involved in turnover of Sic1 through its phosphorylation. According to this model, the growth defect of the cln1 cln2 pho85 mutant should be caused by loss of a means to phosphorylate Sic1, tagging it to its destruction pathway, and therefore could be suppressed by a sic1Δ mutation. To test this, we constructed cln1 cln2 pho85 triple (MFY151) and cln1 cln2 pho85 sic1 quadruple (MFY152) mutants whose growth was supported by MET3-CLN2 (Dirick et al., 1995) and analyzed whether these strains could grow in the presence of methionine, in which CLN2 expression is shut off. At 24°C, although the triple mutant cells stopped dividing within 3 h after cessation of CLN2 expression, the quadruple mutant continued to grow up to 9 h (Figure 6C). At 30°C, growth of the triple and quadruple mutants continued for a shorter period, that is, up to 1.5 and 3 h, respectively (Nishizawa, unpublished results). Thus it appears that inactivation of SIC1 rescued the growth defect of the triple mutant, suggesting that Pho85 kinase is required for degradation of Sic1. However, when the cells were plated onto medium lacking methionine to determine their viability, that of the quadruple mutant dropped rapidly (<10% after 9 h in the methionine-supplemented medium), whereas a majority of the triple mutant (∼80%) were viable after the same period (Figure 6D). A deletion of SIC1 in the cln1 cln2 strain also suppressed the growth defect of the double mutant (Dirick et al., 1995; Figure 6A) and caused an increase in the loss of viability (Figure 6B), although the rate of the viability loss is much slower in the cln1 cln2 sic1 than in the cln1 cln2 pho85 sic1 mutant (Figure 6, B and D), indicating that a sic1 deletion itself was not responsible for the rapid loss of the viability of the quadruple mutant. A pho85Δ sic1Δ double mutant is viable at 24°C (Nishizawa, unpublished results; Aerne et al., 1998). These results suggest a possibility that, in the absence of Cln1, 2-Cdc28 activities, PHO85 is required for cell viability in addition to phosphorylation of Sic1.
Figure 6.
Effect of a deletion of SIC1 on growth (A and C) and viability (B and D) of cln1 cln2 or cln1 cln2 pho85 mutants. Growth of the mutants was sustained by CLN2 in the absence of methionine (open symbols), and at the time indicated by an arrow, the cultures were divided into two equal portions, and one was supplemented with methionine to shut off CLN2 expression (closed symbols). While continuing incubation of cultures at 24°C, cell number was counted periodically with a hemocytometer (A and C), and appropriate dilutions of the cultures were plated periodically onto SD medium lacking methionine to determine the number of cells that could form a colony (B and D). ▪, cln1 cln2; ▴, cln1 cln2 sic1; •, cln1 cln2 pho85; ♦, cln1 cln2 pho85 sic1.
DISCUSSION
cln1 cln2 double mutant cells become able to initiate replication of DNA after attaining a large cell size (Dirick et al., 1995). For such cells to initiate DNA replication, Sic1 must be degraded, and subsequent activation of Clb5, 6-Cdc28 kinase promotes DNA replication and probably budding (Dirick et al., 1995). CLN3 or either CLB5 or CLB6 is required for the viability of the cln1 cln2 mutant, because cln1 cln2 cln3 or cln1 cln2 clb5 clb6 mutant cells are arrested completely at G1 (Richardson et al., 1989; Schwob and Nasmyth, 1993). The delay in the onset of S phase in the cln1 cln2 double mutant is suppressed by introduction of a sic1 null mutation (Dirick et al., 1995), which also suppresses cln1 cln2 cln3 lethality, although the resulting quadruple mutant is quite unhealthy (Schneider et al., 1996). Therefore, persistence of Sic1 is largely responsible for the growth defect, namely the delay in the initiation of DNA replication. Then what induces Sic1 degradation in the absence of CLN1 and CLN2? Genetic evidence indicates that PHO85 function is required at G1 when both of CLN1 and CLN2 are absent (Espinoza et al., 1994; Measday et al., 1994). In this paper, we demonstrated that Pho85 kinase could phosphorylate Sic1 in vitro by the observations that 1) the combination of Pho85 and Pcl1, both produced in E. coli, could phosphorylate Sic1 (Figure 2A), and 2) immunocomplex containing GST-Pho85 and Pcl1-HA obtained from yeast extracts could phosphorylate Sic1 depending on the activity of Pho85 and functional PHO85, respectively (Figures 1 and 2C). We also demonstrated that Pho85 was involved in phosphorylation of Sic1 in vivo (Figure 5), that PHO85 affected the stability of cellular Sic1 (Figure 3), and that a deletion of SIC1 appeared to rescue growth arrest of the cln1 cln2 pho85 triple mutant (Figure 6). From these biochemical and genetic evidence, we suggest that the Pho85 kinase fulfills the role.
Verma et al. (1997a) reported that Sic1 is phosphorylated in vivo in cln1 cln2 cells overproducing Cln3. Cln3-Cdc28 may directly phosphorylate Sic1, or activation of PCL1 expression by CLN3 (Tyers et al., 1993; Dirick et al., 1995; Stuart and Wittenberg, 1995) consequently activates Pcl1-Pho85 kinase to phosphorylate Sic1. These two possibilities are not mutually exclusive, and both may function in vivo. Overexpression of CLB5 can suppress the lethality of a cln1 cln2 cln3 triple mutation (Schwob and Nasmyth, 1993). In this case, overproduced Clb5 should somehow activate the Sic1 degradation pathway that requires phosphorylation of Sic1 (Figures 3 and 4; Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997a), because Sic1 is largely responsible for the lethality of the triple mutant (Schneider et al., 1996). A recent report that Sic1 phosphorylated by Clb5-Cdc28 can be ubiquitinated in vitro may explain the suppression effect (Skowyra et al., 1997). However, because overproduced Clb5 protein can activate PCL1 expression (Schwob and Nasmyth, 1993), it is possible that, again in this case, Pcl1-Pho85 becomes active to phosphorylate Sic1. Alternatively, overproduced Clb5 can overcome the inhibition by Sic1 simply through the dosage effect.
The lethality of the cln1 cln2 cln3 triple mutant is suppressed by a deletion of SIC1, although the resulting quadruple mutant is very unhealthy (Schneider et al., 1996). In the case of the cln1 cln2 pho85 mutant, its growth arrest appeared to be rescued by inactivation of SIC1 (Figure 6C), but the resulting quadruple mutant lost its viability very rapidly (Figure 6D), suggesting that entering a new cell cycle in the absence of both Cln1, 2-Cdc28 and Pho85 kinase activities was detrimental to yeast cells, and that, in the absence of Cln1, 2-Cdc28, Pho85 kinase may have a specific role, in addition to phosphorylation of Sic1, in cell growth, which cannot be substituted by Cln3-Cdc28.
PCL1 may have a specialized role(s) in G1 to S progression in diploid cells, because a pcl1 null mutation causes severe growth defect in diploid cells but no prominent phenotypes in haploid cells (Espinoza et al., 1994), and a cln1 cln2 pcl1 triple mutation results in slow growth phenotype in haploid but is lethal in diploid (Espinoza et al., 1994). However, we do not know at present whether Pho85 is a Cdk partner of Pcl1 to fulfill the role or whether Pcl1 associates with yet unknown Cdk. Because, in haploid cells, cln1 cln2 pho85 is arrested at G1, whereas cln1 cln2 pcl1 is not (Espinoza et al., 1994), it is possible that Pho85 uses another cyclin, in addition to Pcl1, to phosphorylate Sic1. Pcl9 is a likely candidate whose expression is also periodical with a maximum in the boundary of M and G1 (Aerne et al., 1998). Although we could not detect phosphorylation of Sic1 by Pcl2-Pho85 in vitro, it is still possible that Pcl2-Pho85 phosphorylates Sic1 in vivo, because the cln1 cln2 pcl1 pcl2 mutant is arrested at G1 (Measday et al., 1994).
Ubiquitination of Sic1 through CDC34-SKP1-CDC53-CDC4 requires phosphorylated Sic1 (Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997b), and Cln-Cdc28 appears to play a primary role in the modification of the CKI (Feldman et al., 1997; Skowyra et al., 1997). Here we demonstrated that at least two consensus Cdk phosphorylation sites were required for efficient degradation of Sic1 in the wild-type cells (Figures 3B and 4A), that Pho85 became necessary for the degradation of the CKI when the T5 residue was one of the two remaining sites (Figure 3B), and that Pho85 was involved in phosphorylation of the T5 residue in vivo (Figure 5). We, therefore, speculate that Pho85 may preferentially phosphorylate T5, whereas other G1 Cdks act on T33 and S76. When mutant Sic1 protein lacking either T33 or S76 is overproduced in pho85Δ cells, Cdc28 kinase could phosphorylate the remaining T33 or S76, but much less efficiently T5, thus leaving a large portion of the mutant protein uniquely phosphorylated, and therefore, the mutant protein is resistant to degradation. In the absence of Pho85 kinase, the wild-type and T5A mutant Sic1 proteins should be phosphorylated to a similar extent if T5 is phosphorylated solely by Pho85. However, it was not the case (Figure 5, lanes 6 and 7), probably because of phosphorylation of T5 to a certain extent by a Cdk other than Pho85. This phosphorylation may be stimulated by phosphorylation of the other two sites, because phosphorylation, if any, was not detectable in the T33VS76A mutant (lane 8).
Why should two kinases work on Sic1 at different sites? One explanation is a ticketing to prompt progression through G1. Because Sic1 is more stable in the absence of PHO85, its degradation is more efficient if it is phosphorylated at three sites than if it is phosphorylated at two sites. In this regard, it should be noted that the involvement of Pho85 in connecting nutritional conditions to cell cycle is widely considered. When nutrient including Pi is sufficient, Pho85 is kept active to phosphorylate Pho4 to repress PHO5 expression (Kaffman et al., 1994; O’Neill et al., 1996) and Gsy2 to prevent unnecessary accumulation of glycogen (Huang et al., 1996). Pho85 activity may also be targeted to Sic1, ensuring its degradation and thus prompt passage through G1. When nutrient is limited, cells are arrested in G1, and Sic1 should not be degraded to prevent cells from entering S phase without a sufficient supply of nutrients. Under this circumstance, Pho85 becomes inactive as Sic1 kinase to reduce the level of Sic1 phosphorylation.
While we were preparing this manuscript, we noticed a paper by Verma et al. (1997a) reporting that four sites of Sic1, T5, T33, T45, and S76, are mainly phosphorylated in vivo. Their results suggest that three sites in any combination should be intact for phosphorylation and subsequent ubiquitination of Sic1 in vitro (Verma et al., 1997a). Although we could not detect phosphorylation at T45 in vivo (Figure 5, lanes 5, 8, and 10), we think it unlikely that Pho85 is involved in T45 phosphorylation. If Pho85 acted on T45, the T5A mutant would be phosphorylated at only two sites and should become as stable as the double mutants that have two remaining phosphorylation sites (Verma et al., 1997a). However, the T5A mutant was degraded as efficiently as the wild-type Sic1 in pho85Δ (Figure 3B), suggesting that the remaining three sites were phosphorylated in the absence of PHO85.
Our demonstration that at least two distinct Cdks can work on phosphorylation of Sic1 implies the presence of a regulatory network of yeast cell cycle by a Cdk family, as in higher eukaryotes. Future work, including whether regulation of Pcl1-Pho85 kinase by nutrient conditions is carried out through CLN3 that regulates PCL1 expression or through a yet unknown CKI that is specific to Pcl1-Pho85, will reveal a mechanism that shows how the regulatory network is coordinated by the Cdk family members.
ACKNOWLEDGMENTS
We thank E. Amano for technical assistance, N. Ogawa for Pho4 protein, B. Futcher for CLN2-HA plasmid, K. Nasmyth for K3652 and K4900 strains, L. Johnston for communicating results before publication, and J. Tkacz for reading of the manuscript. This work was supported by grants-in-aid for scientific research from the Monbu-sho of Japan (to M.N. and A.T.).
REFERENCES
- Aerne BL, Johnson AL, Toyn JH, Johnston LH. Swi5 controls a novel wave of cyclin synthesis in late mitosis. Mol Biol Cell. 1998;9:945–956. doi: 10.1091/mbc.9.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismowski MJ, Laff GM, Solomon MJ, Reed SI. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Kirschner M. G1 cyclin-dependent activation of p34CDC28 (Cdc28p) in vitro. Proc Natl Acad Sci USA. 1995;92:1182–1186. doi: 10.1073/pnas.92.4.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Espinoza FH, Ogas J, Herskowitz I, Morgan DO. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fujino M, Nishizawa M, Yoon S-J, Oguchi T, Toh-e A. In: Characterization of Pho85 kinase of Saccharomyces cerevisiae, In: Phosphate in Microorganisms: Cellular and Molecular Biology. Torriani-Gorini A, Silver S, Yagil E, editors. Washington, DC: American Society for Microbiology; 1994. pp. 70–75. [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hereford LM. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- Huang D, Farkas I, Roach PJ. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O’Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Killander G, Zetterberg A. A quantitative cytochemical investigation of the relationship between cell mass and initiation of DNA synthesis in mouse fibroblasts in vitro. Exp Cell Res. 1965;40:12–20. doi: 10.1016/0014-4827(65)90285-5. [DOI] [PubMed] [Google Scholar]
- Korch C, Mountain HA, Bystrom AS. Cloning, nucleotide sequence, and regulation of MET14, the gene encoding the APS kinase of Saccharomyces cerevisiae. Mol Gen Genet. 1991;229:96–108. doi: 10.1007/BF00264218. [DOI] [PubMed] [Google Scholar]
- Liao S-M, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, Vuuren HJJv, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Control of the yeast cell cycle. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Taga S, Matsubara A. Positive and negative transcriptional regulation by the yeast GAL11 protein depends on the structure of the promoter and a combination of cis elements. Mol Gen Genet. 1994;245:301–312. doi: 10.1007/BF00290110. [DOI] [PubMed] [Google Scholar]
- Ogas J, Andrews BJ, Herskowitz I. Transcriptional activation of CLN1, CLN2 and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- Ogawa N, Noguchi K, Sawai H, Yamashita Y, Yompakdee C, Oshima Y. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transcription pathway of Pi signals in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Richardson HE, Wittenberg C, Cross F, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Harbor Spring Laboratory; 1990. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorf JW. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher B, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Tamanoi F, Kajiro Y, Matsumoto K, Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990;10:4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A, Ueda Y, Kakimoto S, Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973;113:727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Tanaka K, Toh-e A. Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and structural characterization of PHO85. Nucleic Acids Res. 1987;15:10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rbp1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Valey JG, Simon M, Faye G. The Kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997a;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Verma R, Feldman RMR, Deshaies RJ. Sic1 is ubiquitinated in vitro by a pathway that requires Cdc4, Cdc34, and cyclin/Cdk activities. Mol Biol Cell. 1997b;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI. Control of the yeast cell cycle is associated with assembly/disassembly of the Cdc28 protein kinase complex. Cell. 1988;54:1061–1072. doi: 10.1016/0092-8674(88)90121-3. [DOI] [PubMed] [Google Scholar]