Abstract
Previous studies showed that components implicated in pre-rRNA processing, including U3 small nucleolar (sno)RNA, fibrillarin, nucleolin, and proteins B23 and p52, accumulate in perichromosomal regions and in numerous mitotic cytoplasmic particles, termed nucleolus-derived foci (NDF) between early anaphase and late telophase. The latter structures were analyzed for the presence of pre-rRNA by fluorescence in situ hybridization using probes for segments of pre-rRNA with known half-lives. The NDF did not contain the short-lived 5′-external transcribed spacer (ETS) leader segment upstream from the primary processing site in 47S pre-rRNA. However, the NDF contained sequences from the 5′-ETS core, 18S, internal transcribed spacer 1 (ITS1), and 28S segments and also had detectable, but significantly reduced, levels of the 3′-ETS sequence. Northern analyses showed that in mitotic cells, the latter sequences were present predominantly in 45S-46S pre-rRNAs, indicating that high-molecular weight processing intermediates are preserved during mitosis. Two additional essential processing components were also found in the NDF: U8 snoRNA and hPop1 (a protein component of RNase MRP and RNase P). Thus, the NDF appear to be large complexes containing partially processed pre-rRNA associated with processing components in which processing has been significantly suppressed. The NDF may facilitate coordinated assembly of postmitotic nucleoli.
INTRODUCTION
The nucleolus is a prominent membraneless subnuclear compartment assembled around clusters of tandemly repeated rDNA genes from which pre-rRNA is transcribed and subsequently folded, processed, modified, and assembled into small and large ribosomal subunits (Scheer et al., 1993; Shaw and Jordan, 1995; Maden and Hughes, 1997). A remarkable aspect of the cell cycle is the disintegration of the nucleolus during mitosis and its reassembly as the daughter cells proceed toward G1 phase. A pivotal event in this process is the repression of RNA polymerase (pol) I-driven pre-rRNA synthesis between prophase and telophase (Prescott and Bender, 1962). At the same time, nucleolar components disperse to various subcellular locations as the maternal nucleoli diassemble (Goessens, 1984).
A comprehensive understanding of the locations and fates of nucleolar components during mitosis is slowly emerging (Scheer et al., 1993; Hernandez-Verdun and Gautier, 1994). For example, much of the RNA pol I transcription machinery and DNA topoisomerase I remain associated with the nucleolus organizer regions (NORs)1 of chromosomes between nucleolar disassembly in late prophase and reassembly in telophase (Scheer et al., 1993; Weisenberger and Scheer, 1995; Jordan et al., 1996; Roussel et al., 1996). In contrast, the pre-rRNA processing components appear to be concentrated in the perichromosomal regions (Hernandez-Verdun and Gautier, 1994; Weisenberger and Scheer, 1995) or in large aggregates distributed throughout the cell (Dundr et al., 1996, 1997 and references therein). Thus, the pre-rRNA processing machinery becomes physically separated from the transcriptional apparatus during mitosis. What happens to pre-rRNA transcripts that have not been assembled into ribosomes is less clear. A few studies have probed mitotic cells for the presence of pre-rRNA segments and found them to be dispersed throughout the cell with some enrichment in perichromosomal regions (Jiménez-García et al., 1994; Weisenberger and Scheer, 1995; Beven et al., 1996, Lazdins et al., 1997). However, the fate of large precursors to rRNAs during mitosis has not been investigated systematically.
In previous studies (Dundr et al., 1996, 1997) we observed that several nonribosomal nucleolar components implicated in pre-rRNA processing accumulate in numerous, relatively large cytoplasmic spherical particles termed nucleolus-derived foci (NDF) as well as in perichromosomal regions during anaphase and telophase. In contrast, specific components of the RNA pol I transcription machinery were not present in the NDF; this is in agreement with previous findings indicating the spatial separation of pre-rRNA transcription machinery and pre-rRNA processing components during mitosis. One pre-rRNA processing component found in the NDF was U3 small nucleolar (sno)RNA, which is essential for cleavage at the primary processing site of the 5′-external transcribed spacer (ETS) and subsequent processing events around the 18S rRNA region (Kass et al., 1990; Savino and Gerbi, 1990; Hughes and Ares, 1991) as well as the M phase phosphoprotein 10 component of U3 small nucleolar ribonucleoprotein (sno-RNP) implicated in 18S pre-rRNA processing in yeast (Dunbar et al., 1997; Westendorf et al., 1998). In addition, Beven et al. (1996) localized U14 snoRNA, which is involved in processing steps in the vicinity of the 18S rRNA region (Li et al., 1990; Liang and Fournier, 1995) and U3 snoRNA in numerous particles, termed prenucleolar bodies, located throughout the cytoplasm in pea root cells during anaphase. Their distribution, morphological features, appearance in anaphase, and contents suggest that they are the plant homologues of mammalian NDF. The NDF also contain several proteins believed to be involved with pre-rRNA processing including fibrillarin, a common protein component associated with numerous C/D-box snoRNAs, which are essential for nucleolytic cleavage steps and 2′-O-methylation of pre-rRNA (Maden and Hughes, 1997; Tollervey and Kiss, 1997); protein p52, a protein component of sno-RNPs (Gautier et al., 1992); and nucleolar protein B23, which is possibly involved in the endonucleolytic processing of the internal transcribed spacer 2 (ITS2) segment of pre-rRNA (Savkur and Olson, submitted). Finally, another multifunctional protein nucleolin is found in the NDF. Nucleolin is also present in a large processing complex assembled on the 5′-ETS of pre-rRNA upstream from the first processing site (Ghisolfi-Nieto et al., 1996) and functions in the first step in pre-rRNA processing (Ginisty et al., 1998). Nucleolin also possesses an RNA/RNA helicase activity (Tuteja et al., 1995), suggesting that it plays a role in modulating the secondary structure of pre-rRNA. Thus, several lines of evidence support the idea that the NDF contain essential pre-rRNA processing components.
The presence of early and late pre-rRNA processing components in the NDF raised the question of whether pre-rRNA molecules remain associated with pre-rRNA processing components after mitotic repression of pre-rRNA synthesis and nucleolar disintegration. If so, are the pre-rRNAs full length or partially processed? The relatively higher concentrations of the pre-rRNA processing components in the NDF compared with other parts of mitotic cells provide an opportunity for answering these questions. In the current study we analyzed the mitotic distribution of pre-rRNA segments using fluorescence in situ hybridization and Northern analyses with specific probes complementary to sequences distributed throughout the entire 47S transcript. We present the evidence that the NDF contain predominantly partially processed pre-rRNA transcripts lacking the 5′-ETS leader sequence and having reduced levels of the 3′-ETS segment. We also show that the NDF contain two additional pre-rRNA processing components, U8 snoRNA and hPop1, which is a protein subunit shared by RNases P and MRP.
MATERIALS AND METHODS
Cell Culture
Monkey CMT3 cells (Gerard and Gluzman, 1985) and human HeLa cells were grown on 18 × 18-mm poly-l-lysine–coated glass coverslips in DMEM (Life Technologies-BRL, Gaithersburg, MD) supplemented with 10% FCS (Life Technologies-BRL), 1% glutamine, and penicillin and streptomycin at 37°C in a 5% CO2 atmosphere. The cells were synchronized at the G1/S transition by a double-thymidine block with 2.5 mM thymidine (Bootsma et al., 1964). The cells were then released to proceed through mitosis.
Immunofluorescence
Coverslips with attached cells were washed in PBS and fixed with 3% paraformaldehyde in PBS for 20 min at room temperature, rinsed in PBS, and subsequently permeabilized with 0.2% Triton X-100 in PBS for 5 min on ice, and then were washed extensively with 1% BSA in PBS. The cells were incubated with the primary antibody diluted in PBS for 1 h, washed in PBS, and incubated with appropriate secondary antibodies conjugated with either fluorescein or Texas red (Amersham, Arlington Heights, IL) for 50 min. The cells were washed extensively with PBS, briefly in H2O and ethanol, air dried, and mounted on the slides with Mowiol (Calbiochem, La Jolla, CA) containing 1 mg/ml p-phenylenediamine. Fibrillarin was detected with human autoimmune serum S4 (kindly provided by Dr. R.L. Ochs). Protein B23 was detected using anti-B23 monoclonal antibody (mAb) (kindly provided by Dr. P.K. Chan). A 52- to 53-kDa nucleolar protein was visualized with G04 autoimmune serum (kindly provided by Dr. D. Hernandez-Verdun). Protein hPop1 was detected using rabbit polyclonal A63 antibody (a generous gift from Dr. B. Séraphin).
Hybridization Probes
The plasmid vector (pSP64) containing human U8 snoRNA (Peculis and Steitz, 1993) was kindly provided by Dr. B.A. Peculis. The hybridization antisense or sense probes were produced by in vitro transcription with biotin-16-UTP (Boehringer Mannheim, Indianapolis, IN) using SP6 RNA polymerase after linearization with EcoRI or T7 RNA polymerase after linearization with BamHI, respectively.
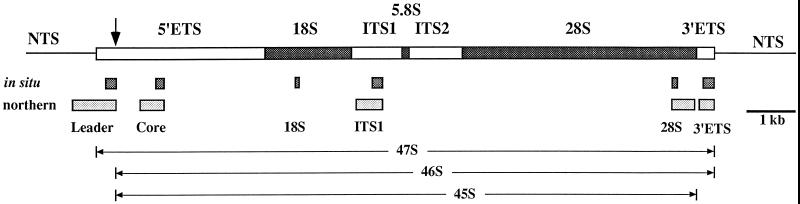
The 5′-ETS leader region of human pre-rRNA corresponding to an EcoRI–ClaI fragment of human rDNA (containing 512 nucleotides [nt] upstream from the initiation site to 422 nt downstream; nt −512/+422) derived from pBES (Wilson et al., 1982) was inserted in pBluescript SK(−) from Stratagene (La Jolla, CA). The antisense hybridization probe (+195/+422) was produced by in vitro transcription with biotin-16-UTP using T7 polymerase after linearization with BsaI. The 5′-ETS core region of human pre-rRNA, which corresponded to a SacI–KpnI fragment of human rDNA (nt +934/+1444) derived from pBSS (Wilson et al., 1982), was inserted in pBluescript SK(−). The antisense hybridization probe (+1270/+1444) was produced by in vitro transcription with biotin-16-UTP using T7 polymerase after linearization with XhoI. The linearized pTRI RNA 18S antisense template, which contains an 80-base pair (bp) insert of the human 18S rRNA gene (nt +4271/+4349) in pTRIPLEscript vector, was obtained from Ambion (Austin, TX). The hybridization antisense probe (residues −43 to +5 relative to transcription start) was produced by in vitro transcription with biotin-16-UTP using T7 RNA polymerase. The ITS1 region of human pre-rRNA corresponding to a XbaI–KpnI fragment of human rDNA (between 58 nt upstream from the 3′-end of the 18S rDNA coding region and 596 nt into ITS1; nt +5469/+6124) derived from pAXK (Erickson et al., 1981) was inserted in pBluescript SK(−). The hybridization antisense probe (+5904/+6124) was produced by in vitro transcription using T7 polymerase after linearization with BsaI. The linearized pTRI RNA 28S template containing a 115-bp cDNA fragment of the human 28S rRNA gene (nt +12,345/+12,458) inserted into the KpnI/XbaI sites of pTRIPLEscript vector was obtained from Ambion. The hybridization antisense probe (nt +37 relative to transcription start) was produced by in vitro transcription with biotin-16-UTP using T7 RNA polymerase. The 3′-ETS region of human pre-rRNA corresponding to an EcoRI–SalI fragment of human rDNA (586 nt upstream from the 3′-end of the 28S rDNA coding region to 374 nt into the 3′-ETS; +12,383/+13,343) derived from pDES (Erickson and Schmickel, 1985) was inserted in pBluescript SK(−). The antisense hybridization probe (+13,111/+13,343) was transcribed using T7 polymerase after linearization with EagI. Sense probes used in control experiments were generated using T3 polymerase after appropriate linearization of the above constructs in the pBluescript vector.
In Situ Hybridization
Cells grown on poly-l-lysine coated glass coverslips were washed with PBS and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After rinsing in PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min on ice, and then washed with PBS and finally with 2×SSC. The hybridization mixture was prepared as described by Jiménez-García et al. (1994). Briefly, 100 ng of probe and 20 μg of yeast tRNA were dried under vacuum. Ten microliters of deionized formamide were added, and the mixture was denaturated for 10 min at 70°C. The probe was immediately chilled on ice, and then the hybridization mixture was made up to final concentrations of 2×SSC, 1% BSA, and 10% dextran sulfate. Hybridization mixture (20 μl) was placed onto each coverslip and allowed to hybridize in a chamber moistened with 2×SSC/50% formamide for 16–18 h at 42°C. The coverslips were rinsed with 2×SSC/50% formamide at 37°C, 2×SSC and 1×SSC at room temperature for 30 min each. The cells were incubated with avidin-DCS-conjugated with Texas red (Vector Laboratories, Burlingame, CA) (2 μg/μl) in 4×SSC/0.25% BSA for 60–75 min, and then rinsed in 4×SSC, 4×SSC/0.1% Triton X-100, 4×SSC, and PBS. Coverslips were mounted in Mowiol (Calbiochem) containing 1 mg/ml p-phenylenediamine. When in situ hybridization was followed by immunofluorescence, after the rinsing of cells in PBS, the coverslips were incubated with anti-B23 mAb for 50 min at room temperature. After the incubation the coverslips were rinsed in PBS and incubated with sheep anti-mouse fluorescein-labeled secondary antibody (Amersham) for 45 min. The cells were washed several times with PBS, briefly with ethanol, air dried, and mounted in Mowiol (Calbiochem) containing p-phenylenediamine. In control experiments, before hybridization the cells were treated with RNase A (Sigma Chemical, St. Louis, MO) (1 μg/1 μl) in 10 mM Tris-HCl, pH 7.3, for 1 h at 37°C. In other controls sense probes were used or probes were completely omitted.
Fluorescence Microscopy
The samples were examined using laser scanning confocal microscope (Noran Instruments, Middleton, WI) with either a Nikon 60×/1.4 N.A. or 100×/1.4 N.A. planapochromat objective. The samples were subjected to an excitation wavelength of 488 nm (fluorescein) or 529 nm (Texas red) from an argon-ion laser. For double labeling, the confocal images for each fluorochrome from the same focal plane were recorded independently. Images were processed using Metamorph (Universal Imaging, West Chester, PA) and printed on a Hewlett Packard 694C printer.
Flow Cytometry
Mitotic and asynchronous CMT3 cells (5 × 106) were washed in PBS, fixed at −80°C in methanol for 15 min, pelleted, and resuspended in 500 μl of PBS. The DNA was stained by incubating each sample with 250 μl of 0.1% propidium iodide in PBS containing 10 μg/ml RNase A for 30 min at room temperature. Samples were analyzed at ∼20 cells/sec on a Becton-Dickinson (Lincoln, NJ) FACScan using CellFIT software for doublet discrimination and the RFIT (rectangular fit) model for cell cycle staging.
Northern Blot Analysis
CMT3 cells were synchronized as above by a double-thymidine block. To selectively obtain mitotic cells, the CMT3 cells were treated with nocodazole (0.04 μg/ml) for 8 h after release from the double-thymidine block and detached by mechanical shake off. The population of the cells harvested was greater than 90% mitotic cells as confirmed by flow cytometry. Asynchronous CMT3 cells were harvested with a cell scraper. Total RNA was isolated using guanidine-HCl and phenol/chloroform, and equal amounts of total RNA were loaded and separated by electrophoresis on 0.6% agarose formaldehyde gels and transferred to positively charged nylon BrightStar-Plus membranes (Ambion). The following DNA fragments were isolated from cloned human rDNA segments and used for DNA probes: the 5′-ETS leader probe was an EcoRI–SapI fragment (−512/+420); the 5′-ETS core probe was a SacI–KpnI fragment (+934/+1444); the ITS1 probe was a NarI–KpnI fragment (+5558/+6124); the 28S rRNA probe was a AccI–XbaI fragment (+12,383/+12,875 plus 30 nt from Bluescript SK(−)) derived from pDES (Erickson and Schmickel, 1985); the 3′-ETS was a BbsI–NaeI fragment (+13,029/+13,343 plus 344 nt from Bluescript SK(−)). The probes were labeled with fluorescein by random priming (Amersham) and incubated with the blots in hybridization mix for 16 h at 42°C, and then washed using the NorthernMax hybridization and wash solutions (Ambion). The hybridization signals were visualized using the Gene Images CDP-Star detection module (Amersham).
RESULTS
18S and 28S rRNA Sequences Are Present in the NDF
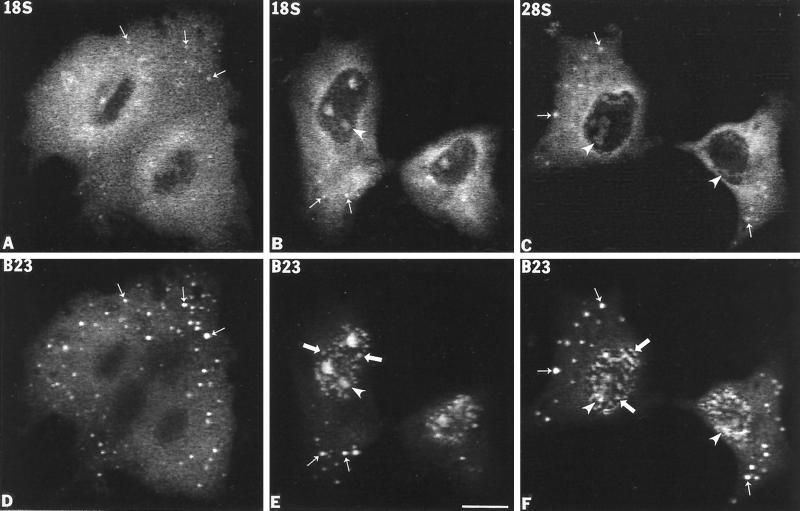
As indicated above, previous studies showed that several nucleolar components implicated in early processing events were found in the NDF and in the perichromosomal region during anaphase and telophase in various cell types. This raised the question of whether mature rRNAs or pre-rRNA transcripts are also contained in the NDF. To examine this possibility, we initially analyzed the mitotic NDF for the presence of mature 18S and 28S rRNA sequences using in situ hybridization with specific biotinylated antisense riboprobes (Figure 1). Monkey kidney CMT3 cells were used in these and most of the subsequent studies because the cells of this line consistently contained abundant and large NDF, which facilitated analyses by fluorescence in situ hybridization. During interphase the CMT3 cells labeled with antisense 18S and 28S riboprobes exhibited general nucleolar staining with a strong uniform cytoplasmic signal indicating the presence of ribosomes (our unpublished observations). From anaphase to telophase the signals of 18S (Figure 2, A and B) and 28S rRNAs (Figure 2C) uniformly covered the cytoplasm with enrichment in the perichromosomal region, whereas the chromosomes themselves were essentially negative. However, some distinct, brightly labeled foci were seen above the general cytoplasmic labeling, mostly in the peripheral regions of the cells (Figure 2, A–C, small arrows). Colocalization with protein B23 (Figure 2, D–F, small arrows) identified these foci as NDF. During late telophase the 18S (Figure 2B) and 28S rRNA (Figure 2C) signals were detected in small newly forming nucleoli (arrowheads), but not in prenucleolar bodies (PNB) that appear as distinct small spots in the nucleoplasm of postmitotic nuclei (Figure 2, E and F, large arrows). The latter structure is distinguished from the NDF in that the PNBs are only present in newly forming nuclei (Scheer et al., 1993), whereas the NDF are strictly cytoplasmic (Dundr et al., 1997).
Figure 1.
Diagram of human 47S pre-rRNA transcription region (boxed) and adjacent nontranscribed spacers (NTS; single lines). The coding regions for 18S, 5.8S, and 28S rRNAs are designated by stippled areas, and the transcribed spacer regions are designated by open areas. The arrow indicates the position of the human primary processing site (+414). The sequence locations of the biotin-labeled antisense riboprobes for in situ hybridization (dark stippled boxes) and fluorescein-labeled DNA probes for Northern analysis (gray stippled boxes) are shown below the 47S diagram. The sequence locations of the major high molecular weight pre-rRNA precursors (45S–47S) are shown at the bottom of the diagram.
Figure 2.
Sequences from 18S and 28S rRNAs are present in the NDF in CMT3 cells. 18S and 28S rRNAs were detected by fluorescence in situ hybridization using a biotin-labeled antisense riboprobe. (A) Anaphase cells labeled with an anti-18S rRNA probe. The in situ hybridization signal is diffusely distributed in the cytoplasm, but distinct bright foci predominate over the general cytoplasmic labeling. These bright foci (A, small arrows) colocalize with signal for protein B23 in the NDF (D, small arrows). (B) Late telophase cells labeled with the anti-18S rRNA probe. The signal is present in reforming postmitotic nucleoli (B, arrowheads) and in the cytoplasm with detectable labeling of NDF (B, small arrows) where it colocalizes with protein B23 (E, small arrows). (C) In early telophase cells, the signal for 28S rRNA is present predominantly over the cytoplasm with detectable NDF (C, small arrows), which colocalized with protein B23 (F, small arrows) as well as in postmitotic nucleoli (C and F, arrowheads). PNBs were also detectable with the anti-B23 antibody (E and F, large arrows). Bar, 10 μm.
The NDF Contain the Core- but Not the Leader Sequence from the 5′-ETS of pre-rRNA
The finding of sequences from 18S and 28S rRNAs in the NDF opened the possibility that the NDF also contained intact or partially processed pre-rRNAs. To address this question, we analyzed the mitotic distribution of precursor segments, again using fluorescence in situ hybridization with specific riboprobes (see Figure 1). This series of experiments began with an examination of the 5′-ETS leader pre-rRNA region. This segment of pre-rRNA is a selective marker for nascent 47S primary transcripts, which are cleaved at the primary processing site of the 5′-ETS and simultaneously degraded (Lazdins et al., 1997; Puvion-Dutilleul et al., 1997). For these experiments we employed an antisense biotinylated RNA probe complementary to the region upstream from the human primary processing site (Figure 1) combined with immunolocalization of nucleolar protein B23.
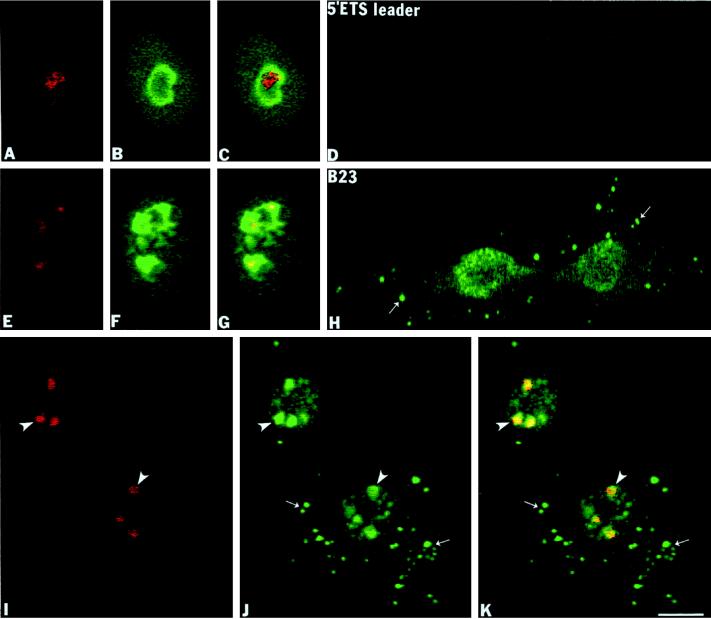
During interphase, the 5′-ETS leader pre-rRNA was present exclusively in a confined intranucleolar area, often having an elongated shape (Figure 3A) as compared with the overall nucleolar area labeled with a mAb against protein B23 (Figure 3, B and C[overlay]). No signal was visible outside of the nucleoli during interphase. Similarly, the 5′-ETS leader pre-rRNA was detected in limited areas of the partially disassembled nucleoli in early prophase (Figure 3E) relative to the more general labeling with the anti-B23 antibody (Figure 3, F and G[overlay]). This observation is in agreement with the idea that RNA pol I transcription is active in early prophase and becomes mitotically repressed in late prophase when the nuclear envelope breaks down (Prescott and Bender, 1962). During prometaphase we failed to detect any signal for the 5′-ETS leader pre-rRNA throughout the cell with only background levels of signal seen (our unpublished observations). Similarly, no signal for the 5′-ETS leader was detected in anaphase cells (Figure 3D). At the same time, protein B23 was present in the perichromosomal region and in numerous NDF (Figure 3H, small arrows). In late telophase, the 5′-ETS leader segment reappeared in small rounded postmitotic transcriptionally reactivated nucleoli (Figure 3I, arrowheads) where it colocalized with protein B23 (Figure 3, J [arrowheads] and K [yellow in overlay]). In the same cells numerous cytoplasmic NDF (Figure 3J, small arrows) and nucleoplasmic PNBs (Figure 3J, large arrows) were positively stained for protein B23, but these exhibited no signal for the 5′-ETS leader sequence (Figure 3, I and K, overlay). These data agree with the observations of Lazdins et al. (1997) who were unable to detect the 5-′ETS leader sequence in mitotic cells.
Figure 3.
The 5′-ETS leader pre-rRNA is not found in the NDF of CMT3 cells. The 5′-ETS leader pre-rRNA was detected by fluorescence in situ hybridization using an antisense biotinylated riboprobe (nucleotides +195/+424). (A–C) Interphase cells labeled with the 5′-ETS probe and anti-B23 antibody. In the nucleoli of these cells the 5′-ETS leader is localized to a restricted area (A) compared with the region labeled with anti-B23 antibody (B), which is clearly shown on the overlay (C). (E–G) Early prophase cells. In these cells the 5′-ETS leader segment is detectable in restricted areas in the disintegrating nucleoli in early (E) compared with protein B23 labeling (F) in the superimposed picture (G). (D and H) Anaphase cells colabeled with the 5′-ETS leader probe and anti-B23 antibody. In these cells the signal from the 5′-ETS leader was reduced to background levels (D) but showing the presence of protein B23 in the perichromosomal region and in NDF (H, small arrows). (I–K) Late telophase cells colabeled with the 5′-ETS leader probe and anti-B23 antibody. The 5′-ETS leader was detected only in postmitotic transcriptionally reactivated nucleoli (I, arrowheads) where it colocalized with protein B23 (J, arrowheads; K, yellow in superimposed picture). Bar, 10 μm.
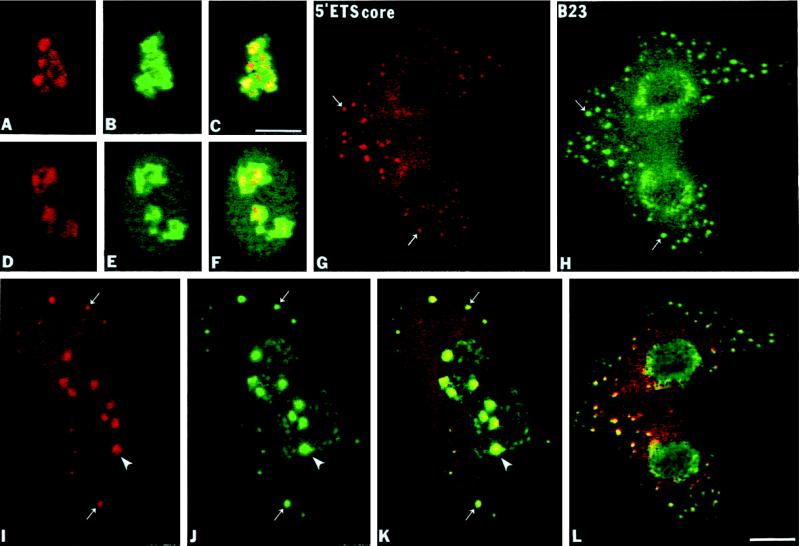
The 5′-ETS core pre-rRNA is considered to be much more stable than the 5′-ETS leader sequence in the mammalian pre-rRNA maturation pathway (Sollner-Webb et al., 1996). The 175-nt 5′-ETS core probe for the next experiments was complementary to sequences 858 nt downstream from the human primary processing site and 2214 nt upstream from 18S coding region (see Figure 1). In interphase nucleoli the 5′-ETS core pre-rRNA sequence was localized in a relatively large portion of the nucleolus with higher intensity of labeling in compact central intranucleolar regions, which probably represented dense fibrillar components (DFCs) (Figure 4A). In contrast, there was a more general staining by the anti-B23 mAb (Figure 4, B and C, overlay). The same intranucleolar labeling pattern of the 5′-ETS core sequence persisted in disintegrating nucleoli in early prophase (Figure 4D) as seen for protein B23 (Figure 4, E and F, overlay). During anaphase and telophase the 5′-ETS core segment was clearly detectable in the NDF (Figure 4, G and I, small arrows) where it colocalized with protein B23 (Figure 4, H and J, small arrows) as shown by superimposing the two signals (Figure 4, K and L). From metaphase through telophase there was also a lower level of diffuse signal from the 5′-ETS core segment throughout the cytoplasm with some enrichment in the perichromosomal region with no signal detected in the chromosomal NORs (Figure 4G). In late telophase the 5′-ETS core segment appeared in newly formed and actively synthesizing nucleoli (Figure 4I, arrowheads), again colocalizing with protein B23 (Figure 4, J[arrowheads] and K [overlay]). Thus, the 5′-ETS core segment remained stable throughout mitosis with relatively high concentrations accumulating in the NDF.
Figure 4.
The 5′-ETS core of pre-rRNA is present in the NDF of CMT3 cells. The 5′-ETS core pre-rRNA was detected by fluorescence in situ hybridization using a biotinylated antisense riboprobe (nucleotides +1272/+1446). (A–C) Interphase cells probed with the 5′-ETS core probe and with anti-B23 antibody. The 5′-ETS core segment is localized in the central regions of interphase nucleoli (A) compared with overall nucleolar labeling of protein B23 (B) and yellow areas of the superimposed picture (C). (D–F) In early prophase a similar labeling pattern of 5′-ETS signal (D) is seen when colocalized with protein B23 (E) and in the superimposed picture (F). (G and H) During anaphase the 5′-ETS core segment is enriched in the perichromosomal region and in NDF (G, small arrows) where it colocalizes with protein B23 (H, small arrows; L, yellow in overlay). Note that more NDF appeared to be labeled by immunofluorescence than by in situ hybridization because the signal faded more rapidly in the latter case. (I–K) In late telophase the 5′-ETS core segment is present mostly in reformed nucleoli (I, arrowhead) and in persisting cytoplasmic NDF (I, small arrows) where it colocalizes with protein B23 (J, arrowhead, small arrows; K, yellow in superimposed picture). Bar, 10 μm.
The ITS1 Sequence of pre-rRNA Is Present in the NDF
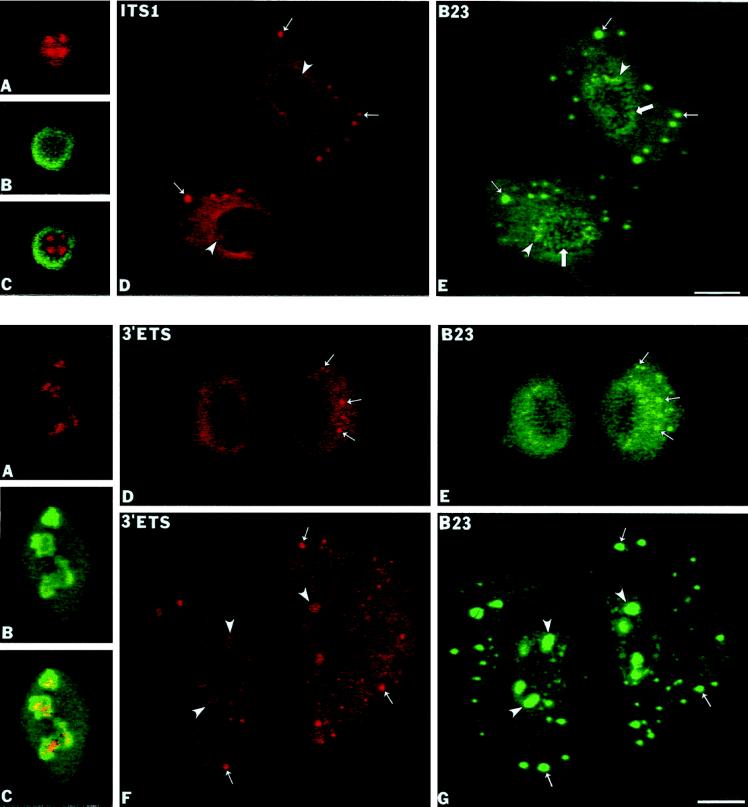
The next segment of pre-rRNA analyed was the ITS1 (Figure 1), which is also a relatively long-lived pre-rRNA sequence (Lazdins et al., 1997). During interphase the ITS1 sequence was detected throughout the nucleolus (Figure 5A) with accumulation in multiple intranucleolar regions (Figure 5, A and C, overlay) and no signal visible outside of nucleoli. A similar labeling pattern was seen in disintegrating nucleoli in early prophase (our unpublished observations). From early anaphase to late telophase the ITS1 labeling was clearly seen in numerous NDF (Figure 5D, small arrows) where it colocalized with protein B23 (Figure 5E, small arrows). As with the 5′-ETS core segment, there was also a diffuse distribution of signal throughout the cytoplasm and enrichment in the perichromosomal region, with no signal in the NORs. After reformation of the nuclear envelope in late telophase, the ITS1 signal reappeared inside daughter nuclei in newly reforming nucleoli (Figure 5D, arrowheads).
Figure 5.
The ITS1 sequence of pre-rRNA is present in the NDF of CMT3 cells. The ITS1 pre-rRNA was detected by fluorescence in situ hybridization using a biotinylated antisense riboprobe (nucleotides +5904/+6124). (A–C) Labeling of interphase nucleolus with the ITS1 probe and with the anti-B23 antibody. The ITS1 signal is concentrated in several intranucleolar regions (A) compared with protein B23, which is present mostly in nucleolar periphery as seen in panel B and the superimposed picture (C). (D and E) Late telophase cells. The ITS1 labeling appeared in reforming nucleoli (D, arrowheads) and in several cytoplasmic NDF (D, small arrows) that are also stained with anti-B23 antibody (E, small arrows). PNBs in reforming nuclei are also detected with the latter antibody (E, large arrows). Bar, 10 μm.
Distribution of the 3′-ETS Sequence of pre-rRNA
The complete removal of 3′-ETS is the second processing event occurring soon after the completion and release of the pre-rRNA transcript (Sollner-Webb et al., 1996); the 3′-ETS fragment also exhibits a very short half-life during interphase (Lazdins et al., 1997). To determine the localization of the 3′-ETS region, we used an antisense probe complementary to the last 233 nt of the 3′-ETS (Figure 1). In interphase, we detected the 3′-ETS sequence in limited intranucleolar areas (Figure 6A) relative to the overall nucleolar body visualized by anti-B23 staining (Figure 6, B and C). No signal was visible outside of the nucleoli during interphase. The distribution of the 3′-ETS sequence closely resembled the localization of the 5′-ETS leader sequence in interphase (Figure 3A). The 3′-ETS signal was also detectable during mitosis, but it was generally weaker than that of the 5′-ETS core or ITS1 signals. Virtually identical labeling patterns for the 3′-ETS and the 5′-ETS were seen in disintegrating nucleoli in early prophase (our unpublished observations). In contrast to the 5′-ETS leader segment, we were able to detect cytoplasmic labeling of the 3′-ETS from metaphase to early (Figure 6D) and late telophase (Figure 6F). The signal appeared in NDF as well as in a more general cytoplasmic labeling. The 3′-ETS signal was also seen in these areas during anaphase (our unpublished observations). The signal for the 3′-ETS in the NDF (Figure 6, D and F, small arrows) colocalized with protein B23 (Figure 6, D and E, small arrows). During late telophase the 3′-ETS segment reappeared in newly reformed nucleoli (Figure 6F, arrowheads) but PNBs identified did not show any signal for the 3′-ETS sequence (Figure 6G, large arrows). This suggests that during mitotic repression of pol I transcription at least some of the pre-rRNA transcripts are completed and properly terminated.
Figure 6.
The 3′-ETS pre-rRNA is present in the NDF of CMT3 cells. The 3′-ETS pre-rRNA segment was detected by fluorescence in situ hybridization with a biotinylated antisense riboprobe (nucleotides +13,111/+13,343). (A–C) Labeling of interphase cells with the 3′-ETS probe and the anti-B23 antibody. The 3′-ETS signal was present in restricted intranucleolar areas (A) in contrast to general nucleolar staining with anti-B23 antibody as seen in panel B and in the overlay (C). (D and E) In anaphase cells the 3′-ETS signal is detectable throughout the cytoplasm with several brighter foci (D, small arrows) that were identified as the NDF by colocalization with protein B23 (E, small arrows). (F and G) In late telophase cells the 3′-ETS signal reappeared in the reformed nucleoli (F, arrowheads), but the signal is also present in persisting cytoplasmic NDF (F, small arrows) where it colocalizes with protein B23 (G, small arrows). Bar, 10 μm.
Partially Processed pre-rRNA Is Preserved during Mitosis
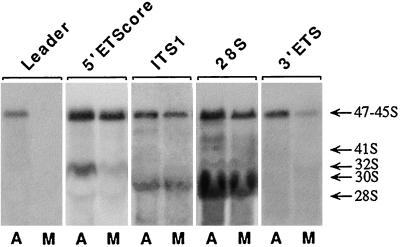
The above studies indicated that sequences from the external and internal transcribed spacers of pre-rRNA were present in the NDF, but did not rule out the possibility that some cleavage had taken place, leaving 18S, 5.8S, and 28S rRNAs plus undegraded spacer sequences in these structures during mitosis. To confirm that high molecular weight pre-rRNA is present during mitosis, total RNA was extracted from asynchronous and mitotic CMT3 cells and subjected to agarose gel electrophoresis followed by Northern hybridization using probes to various segments of the pre-rRNA transcript (Figure 1). In the asynchronous cells all probes hybridized to a prominent high molecular weight band, which appeared to be a mixture of 45S, 46S, and 47S pre-rRNAs (Figure 7). Although the 5′-ETS leader and 3′-ETS probes hybridized to the upper part of the band, it was not possible to clearly resolve these high molecular weight species of pre-rRNAs. The intensities of the bands obtained with the 5′-ETS leader and the 3′-ETS probes were lower than those with the other probes (Figure 7) under the same loads of RNA and exposure times. This is indicative of the steady-state levels of the nascent transcripts and early intermediates, i.e., there is rapid processing at the 5′- and 3′-ends resulting in reduced amounts of the full-length transcripts.
Figure 7.
Northern blot analysis of pre-rRNA transcripts. Equal amounts of total RNA from asynchronous (A) or mitotic (M) CMT3 cells were separated on multiple lanes of 0.6% agarose formaldehyde gels. The RNA was transferred to nylon membranes and probed with the fluorescein-labeled probes complementary to segments of 47S pre-rRNA as indicated above each pair of lanes (see Figure 1). The positions of major pre-rRNA species and 28S rRNA are indicated on the right.
When the relative levels of these high molecular weight pre-rRNA species were compared between asynchronous and mitotic cells using the probes in Figure 1, the patterns observed fell essentially into three categories (Figure 7). First, using the 5′-ETS leader probe, the 45–47S band was present in the asynchronous cells, but it was completely undetectable in the mitotic cells. This indicated that the primary processing event had taken place during mitosis and that virtually no 47S RNA (unprocessed transcript) remained. The second category was typified by the 5′-ETS core probe, in which there was only a slight reduction in the 45S–47S signal from the mitotic pre-rRNA. A similar pattern was observed with the ITS1 and the 28S probes, i.e., only slight reductions in 45S–47S signal were seen in the mitotic cells. Finally, with the 3′-ETS probe, the 45S–47S signal was markedly diminished in the mitotic cells, but not completely eliminated. Thus, processing had occurred at the 3′-end on most, but not all, transcripts, leaving a relatively small amount of the 3′-ETS–containing pre-rRNA. The latter point is consistent with the presence of a detectable, but reduced, signal from the 3′-ETS probe in the NDF by in situ hybridization.
In addition to the 45S–47S band, some lower molecular weight processing intermediates were observed using the 5′-ETS core, ITS1, and 28S probes (Figure 7). These bands were generally of lower intensity than the 45S–47S band. The lower molecular weight intermediates included 41S pre-rRNA (Hadjiolova et al., 1993) detected with the ITS1 and 28S probes and the 30S intermediate, which hybridized with the 5′-ETS core; these were significantly reduced in the mitotic cells. The greatest reduction was in the 30S intermediate (Figure 7), which appears to be a 18S rRNA precursor containing the core portion of the 5′-ETS (Hadjiolova et al., 1993). This suggests that processing was suppressed in mitosis, resulting in the accumulation of 45S and 46S species of pre-rRNA. High levels of 28S rRNA were seen in mitotic as well as asynchronous cells because of the overwhelming presence of ribosomes in both samples. The absence of low molecular weight species hybridizing with the spacer probes indicated that there was no accumulation of processed spacer segments. These results support the idea that the in situ hybridization signals seen in the NDF during the later stages of mitosis are derived from high molecular weight pre-rRNA species and are not coming from already-processed low molecular weight spacer sequences of pre-rRNA.
The NDF Contain U8 snoRNA
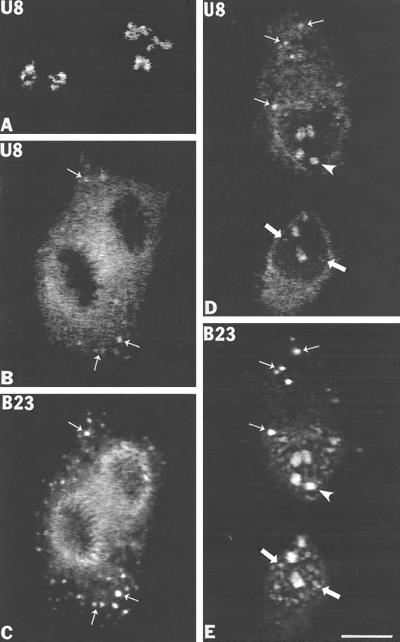
The finding of partially processed pre-rRNA in the NDF in association with processing machinery led us to search for additional processing components. The first of these was U8 snoRNA, which is involved in processing events in the ITS1, ITS2, and 3′-ETS regions (Peculis and Steitz, 1993). In situ hybridization with a biotinylated antisense riboprobe containing the entire U8 gene sequence was performed followed by immunolocalization of protein B23 using an anti-B23 mAb. During interphase, U8 snoRNA was present in individual intranucleolar subregions that were usually connected and frequently formed ring-like substructures (Figure 8A), as observed previously by Matera et al. (1994). No signal was seen outside nucleoli, and sense probe controls were negative (our unpublished observations). From prometaphase to early telophase the labeling for U8 snoRNA was dispersed in the cytoplasm with some signal in the periphery of the chromosomes (Figure 8B). In addition, from anaphase to late telophase, the U8 snoRNA signal was present in several distinct cytoplasmic foci (Figure 8, B and D, small arrows) which were positively identified as the NDF by colocalization with protein B23 (Figure 8, C and E, small arrows). In late telophase the U8 sno-RNA signal reappeared in reforming nucleoli (Figure 8D, arrowhead) as well as in several small nucleoplasmic bodies (Figure 8D, large arrows) identified as PNBs by colocalization with protein B23 (Figure 8E, large arrows). Thus, during mitosis, U8 snoRNA associates with the NDF and the perichromosomal region and accumulates in PNBs after postmitotic nuclear reformation.
Figure 8.
U8 snoRNA is present in nucleolus-derived foci (NDF) in CMT3 cells during mitosis. U8 snoRNA was detected by fluorescence in situ hybridization using a biotin-labeled antisense riboprobe. (A) Interphase cells hybridized with anti-U8 snoRNA probe. Labeling is observed exclusively in intranucleolar subdomains. (B and C). Labeling with U8 probe and with anti-B23 antibody in anaphase cells. The U8 snoRNA in situ hybridization signal is dispersed throughout the cytoplasm with some enrichment in the perichromosomal region (B) and in NDF (B, small arrows) where it colocalizes with protein B23 (C, small arrows). (D and E) During late telophase U8 snoRNA is visible inside of reforming nucleoli (D, arrowheads) but is also weakly detectable in prenucleolar bodies (D, large arrows) and in several cytoplasmic NDF (D, small arrows) that colocalize with protein B23 (E, small arrows). Bar, 10 μm.
The NDF Contain hPop1, a Protein Component Common to RNases P and MRP
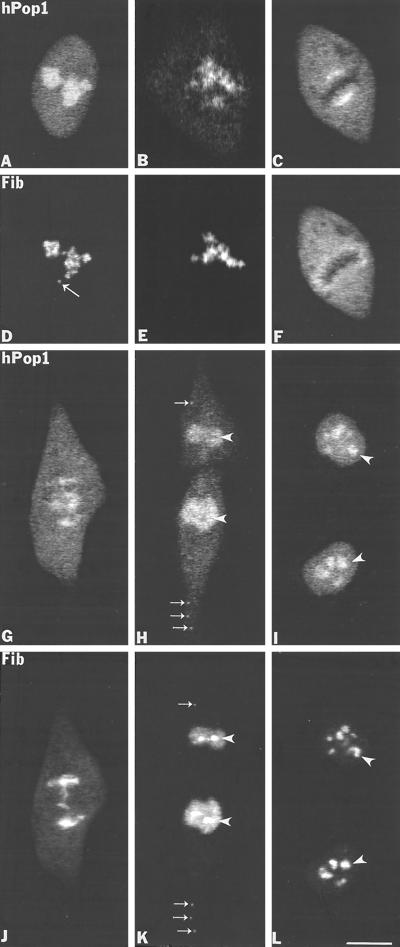
Immunolocalization studies were also performed on human protein Pop1, which is a protein component of RNase MRP and RNase P using an affinity-purified antibody (Lygerou et al., 1996b). The endoribonuclease MRP cleaves the yeast pre-rRNA at site A3 within the ITS1 region of pre-rRNA (Schmitt and Clayton, 1992; Chu et al., 1994; Lygerou et al., 1996a), and endoribonuclease RNase P plays a role in processing of the 5′-ETS, ITS1, and ITS2 regions of pre-rRNA (Chamberlain et al., 1996; Stolc and Altman, 1997). Both enzymes are structurally and functionally related and share at least three protein subunits, Pop1, Pop3p, and Pop4p (Tollervey and Kiss, 1997 and references therein). RNase MRP is present primarily in the nucleolus, but RNase P has been found in the nucleoplasm as well as the nucleolus (Jacobson et al., 1997). We found that the antibody against hPop1 (Lygerou et al. 1996b) recognized only the human Pop1 protein and did not show any labeling on monkey CMT3 cells. Therefore the immunolocalization of hPop1 was performed on HeLa cells. During interphase, hPop1 was predominantly localized in nucleoli with weak uniform nucleoplasmic labeling (Figure 9A). When hPop1 was colocalized with fibrillarin (Figure 9D), image superimposition showed that both proteins were located in the DFC, but hPop1 labeling also extended into the granular component (GC). In early prophase the hPop1 protein (Figure 9B) began to move out of disintegrating nucleoli slightly earlier than fibrillarin (Figure 9E). This reflects the presence of the hPop1 protein in the GC compared with fibrillarin, which is predominantly found in the DFC. After nucleolar disintegration in late prophase, the hPop1 protein was redistributed over the cytoplasm (our unpublished observations). In metaphase the hPop1 protein was found in the perichromosomal region near the center of the cell as highly concentrated masses on either side of the metaphase plate facing opposite poles (Figure 9C). In contrast, fibrillarin was uniformly concentrated in the perichromosomal region, completely surrounding the metaphase plate (Figure 9F). In anaphase hPop1 was associated with chromatids as they migrated to opposite poles (Figure 9G) where it colocalized with fibrillarin (Figure 9J). The hPop1 signal was also concentrated inside the mitotic spindle region (Figure 9G). As chromosomes began to decondense and the nuclear envelope reassembled, the hPop1 protein was located diffusely inside the nuclear interior but just outside of the reforming nucleoli (Figure 9H, arrowheads), which were visualized by anti-fibrillarin sera (Figure 9K, arrowheads). The hPop1 protein was clearly visible inside nucleoli only when they began to increase in size and became fully operational (Figure 9I, arrowheads). More importantly, from early anaphase to telophase, hPop1 was distinctly present in NDF (Figure 9H, small arrows) where it colocalized with fibrillarin (Figure 9K, small arrows). In summary, hPop1, which is contained in RNase MRP and/or RNase P, is present in the perichromosomal region and in the NDF during mitosis; after reassembly of the nuclear envelope it is targeted to reforming nucleoli after their reactivation. These findings further support the notion that the NDF contain components of the pre-rRNA processing machinery.
Figure 9.
The hPop1 protein subunit of human RNase P and RNase MRP is present in NDF in HeLa cells during mitosis. (A and D) Labeling of interphase cell with anti-hPop1 and the S4 autoimmune antifibrillarin serum. The hPop1 protein was predominantly localized in nucleoli with weak nucleoplasmic staining whereas fibrillarin was present in the nucleolar DFC and in a coiled body (D, long arrow). (B and E) In early prophase the hPop1 protein (B) and fibrillarin (E) disperse from disintegrating nucleolar bodies. (C and F) In metaphase the hPop1 protein is found highly concentrated at the external central portion of chromosome periphery (C), whereas fibrillarin is concentrated in the entire perichromosomal region of the metaphase plate (F). (G and J) In late anaphase the hPop1 protein was seen associated with chromatids (G) where it colocalizes with fibrillarin (J), but hPop1 was also seen inside of mitotic spindle (G). (H and K) In late telophase when the nuclear envelope is formed, hPop1 protein is concentrated in the postmitotic nuclei but it is not detectable in newly reformed nucleoli (H, arrowheads) where fibrillarin is prominantly localized (K, arrowheads). In addition, both proteins colocalized in NDF in the cytoplasm (H, K, small arrows). (I and L) In late telophase/early G1 phase the hPop1 protein reenters nucleoli (I) and is colocalized with fibrillarin (L). Bar, 10 μm.
DISCUSSION
Previous studies (Dundr et al., 1996, 1997) provided evidence that components involved in the processing of pre-rRNA become separated from the transcriptional machinery but remain associated with each other during mitosis. These components seem to be contained primarily in the perichromosomal regions and in the NDF during anaphase and telophase. The finding of high concentrations of processing components in the NDF prompted us to analyze them for pre-rRNA intermediates during mitosis. The most striking finding of this study is that partially processed pre-rRNA is present in the NDF in association with processing components. Furthermore, this pre-rRNA is predominantly a nearly intact transcript, completely lacking the extreme 5′-leader segment and partially containing the 3-′ETS. Thus, not only is pre-rRNA transcription blocked during mitosis, but pre-rRNA processing also seems to be suppressed, resulting in the partially processed transcript being carried through most of the mitotic phase.
The conclusion that partially processed pre-rRNA transcripts are present in the NDF during mitosis is based on in situ hybridization analyses with antisense probes spanning representative segments of the total length of 47S pre-rRNA. This is further supported by Northern analyses that indicate that 45S and 46S pre-rRNAs are the predominant pre-rRNAs present in mitotic cells. Thus, it is unlikely that additional cleavages within the internal portions of the pre-rRNA molecules have already taken place with the excised fragments simply protected from degradation.
The behaviors of sequences from the 5′- and 3′-ends of the 47S transcript during mitosis are important for the interpretation of the current data. The absence of any signal for the 5′-ETS leader segment during mitosis is consistent with its removal being the first processing event (Eichler and Craig, 1994; Sollner-Webb et al., 1996; Lazdins et al., 1997; Puvion-Dutilleul et al., 1997). However, Northern analyses indicated that apparently full-length transcripts contain the 5′-ETS leader sequence during interphase, suggesting that transcription and termination are completed before processing occurs. This provides evidence that a significant proportion of the 5′-ETS leader is not removed cotranscriptionally. The partial presence of the 3′-ETS segment during mitosis is also consistent with detachment of this sequence being the second processing event. The finding that a segment in the extreme 3′-end of 28S rRNA is not only strongly labeled in the NDF, but also produces a signal of intensity equal to that of the 5′-ETS core and ITS1 probes on Northern blots from mitotic cells, indicates that transcription has proceeded at least to the 3′-end of the 28S segment. In addition, a partial 3′-ETS signal indicates that a significant proportion of the pre-rRNA molecules are completed and terminated. The fact that no significant signal for any segment of pre-rRNA has been found at the NORs of mitotic chromosomes (Weisenberger and Scheer, 1995; Beven et al., 1996; Lazdins et al., 1997; this study) further suggests that the transcripts are released from the template. The presence of partially processed high molecular weight pre-rRNA in mitotic cells in these studies and in earlier work (Fan and Penman, 1971) indicates that later processing events are largely suppressed in mitosis.
The regulatory mechanism for the mitotic repression of pre-rRNA synthesis is poorly understood, although several possible explanations have been proposed. On the one hand, essential components of the rRNA transcription machinery, including the upstream binding factor (UBF), the subunits of promoter selectivity factor SL1, as well as RNA pol I, are present at the NORs of chromosomes (Weisenberger and Scheer, 1995; Jordan et al., 1996; Roussel et al., 1996; Seither et al., 1997). In addition, UBF, RNA pol I, and DNA topoisomerase I were found in the vicinity of previously active rDNA genes at NORs during metaphase and anaphase (Gébrane-Younès et al., 1997; Heliot et al., 1997; Suja et al., 1997). On the other hand, nascent pre-rRNA molecules have not been detected in the chromosomal NORs. Furthermore, Weisenberger and Scheer (1995) were not able to detect pre-rRNA processing components that are normally enriched in the 5′-ETS processing complex (Mougey et al., 1993b), including U3 sno-RNA, fibrillarin, and nucleolin, at the mitotic NORs. Weisenberger and Scheer (1995) proposed that mitotic down-regulation of pre-rRNA transcription is inhibited at the level of transcription elongation, presuming that transcriptionally engaged RNA pol I elongation complexes remain bound to the rDNA template throughout mitosis. Recently, an alternative model was proposed (Grummt, personal communication) based on the finding that mitosis-specific phosphorylation of the hTAFI110 subunit of SL1 by p34cdc2/cyclin B kinase causes repression of rDNA transcription. SL1 is essential for the assembly of preinitiation complexes at the rDNA promoter (Eberhard et al., 1993; Heix et al., 1997) by recruiting RNA pol I associated with two essential factors, TIF-IA and TIF-IC, to the template (Schnapp and Grummt, 1991). Similar mechanisms of transcription repression by mitosis-specific phosphorylation have been found for equivalent TBP–TAF complexes, TFIID of RNA pol II and TFIIIB of RNA pol III, in which assembly of preinitiation complexes is inhibited (Hartl et al., 1993; Gottesfeld et al., 1994; White et al., 1995; Leresche et al., 1996; Segil et al., 1996; reviewed by Gottesfeld and Forbes, 1997). Because we observe pre-rRNA molecules that appear to be properly terminated when transcription is mitotically repressed, our data are consistent with the latter model.
Previous studies showed that the NDF contain pre-rRNA components implicated in pre-rRNA processing events leading to release of 18S rRNA. These components include U3 (Dundr et al., 1997) and U14 sno-RNAs (Beven et al, 1996), nucleolin, and proteins associated with C/D box-containing snoRNAs, fibrillarin, and ribonucleoprotein p52 (Dundr et al., 1996, 1997). U3 and U14 are transiently associated with some 5′-ETS and 18S sequences of pre-rRNA (Beltrame and Tollervey, 1995; Liang and Fournier, 1995), and they have been suggested to assemble into a hypothetical multiple-snoRNP complex (Maxwell and Fournier, 1995; Ghisolfi-Nieto et al., 1996). In addition, electron-dense “terminal balls” at the leading ends of nascent pre-rRNA transcripts have been identified as the 5′-ETS primary processing complexes (Kass et al., 1990; Mougey et al., 1993a), which appear to contain U3 snoRNA (Kass et al., 1990), fibrillarin (Scheer and Benavente, 1990; Mougey et al., 1993b), and nucleolin (Ghisolfi-Nieto et al., 1996; Ginisty et al., 1998). Thus, the NDF may be a means of preserving the integrity of these processing complexes until the reformed postmitotic nucleoli become fully functional.
In this study we show that mitotic NDF during anaphase and telophase also contain components involved in later stages of pre-rRNA processing. One of these is U8 snoRNA, which is essential for production of 5.8S and 28S rRNAs (Peculis and Steitz, 1993). Localization studies on U8 snoRNA reflect involvement in later pre-rRNA processing events showing labeling in the periphery of the DFC compared with the early processing components, U3 and U14 snoRNAs, which are closer to the FC (Matera et al., 1994; Beven et al., 1996). The presence of early and late processing components in the NDF suggests that they are associated with pre-rRNA transcripts as a unit even though they are topologically separated during interphase.
We have also shown in this study that NDF during anaphase and telophase contain hPop1, a protein component common to RNase MRP and RNase P (Lygerou et al., 1996b). RNase MRP is implicated in the cleavage of pre-rRNA at site A3 within ITS1 (Lygerou et al., 1994). RNase P participates in cleavages in the 5′-ETS, ITS1, and ITS2 of the primary 35S pre-rRNA transcript in yeast (Chamberlain et al., 1996; Stolc and Altman, 1997). Thus, it seems likely that nucleolar endonucleases remain associated with nascent pre-rRNA, which further supports the idea that multiple processing components are contained in complexes whose overall organization is preserved during mitosis.
What is the advantage to the cell in packaging partially processed pre-rRNA with associated processing components into the NDF and carrying these through mitosis? One possibility is that, during mitosis, the supply line that provides proteins and RNAs for continued assembly of ribosomes has been shut down. In this case, processing may be suppressed and the partially processed transcripts preserved until these components are again made available. This would imply that partially processed pre-rRNA transcripts reenter the daughter nuclei and the assembly process resumes in the reformed nucleoli. An immediate supply of preribosomal particles ready to be made into ribosomes may provide efficiency to the daughter cells for rapid recovery from the dimunition of the synthetic machinery after mitosis. This could be especially important since the rate of pre-rRNA transcription appears to be at a low level during early G1 phase and does not reach substantial rates until late G1 phase (Voit et al., 1997). Work in this laboratory suggests that most of the nucleolar proteins and snoRNAs in the NDF reenter the reforming nucleoli at the end of mitosis (Dundr et al., 1996, 1997; this work), although we have not determined the fate of the pre-RNA molecules. Recent work by Zatsepina et al. (1997) also showed that hypotonically induced nucleolar bodies structually similar to the NDF can reassemble into nucleoli after cells are returned to isotonic conditions during interphase. This supports the idea that nucleoli may be reassembled from previously existing nucleolar components as suggested by Phillips and Phillips (1973). A study by Medina et al. (1995) indicates that RNAs radiolabeled in G2 phase accumulate in nucleoli in telophase and early G1 phase. However, the labeled RNAs found in the newly formed daughter nucleoli were not identified.
A second possibility is that the pre-rRNP particles containing partially processed pre-rRNAs do not need to reenter nucleoli for the final part of assembly. In this scenario, the assembly and processing are completed in the cytoplasm. As the ribosomal subunits are released, the processing components may be recycled by migration back into the nucleoli. Finally, the partially processed pre-rRNA may be frozen in place and serve as a scaffold for maintaining the organization of the processing machinery during mitosis. In this scenario, as the processing components are released and reenter the newly forming nuclei and nucleoli in telophase, the persisting pre-rRNA transcripts may be degraded as the processing system is reactivated. Currently, there is insufficient information about the fate and role of partially processed pre-rRNA during mitosis to favor or reject any of these hypotheses.
ACKNOWLEDGMENTS
The authors thank Drs. Bertrand Séraphin, Danièle Hernandez-Verdun, Pui K. Chan, and Robert L. Ochs for providing antibodies; Brenda A. Peculis for the plasmid expressing U8 snoRNA; and James E. Sylvester for rDNA-containing plasmids. This work was supported by National Institute of Health grants GM-28349 and AI-34277.
Footnotes
Abbrevations used: DFC, dense fibrillar component; ETS, external transcribed spacer; GC, granular component; ITS, internal transcribed spacer; NDF, nucleolus-derived foci; NOR, nucleolus organizer region; PNB, prenucleolar body; pol, polymerase; snoRNA, small nucleolar RNA; sno-RNP, small nucleolar ribonucleoprotein.
REFERENCES
- Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven AF, Lee R, Razaz M, Leader DJ, Brown JWS, Shaw PJ. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J Cell Sci. 1996;109:1241–1251. doi: 10.1242/jcs.109.6.1241. [DOI] [PubMed] [Google Scholar]
- Bootsma D, Budke L, Vos O. Studies on synchronous division of tissue culture cells initiated by excess thymidine. Exp Cell Res. 1964;33:301–309. doi: 10.1016/s0014-4827(64)81035-1. [DOI] [PubMed] [Google Scholar]
- Chamberlain J, Pagan-Ramos E, Kindelberger DW, Engelke D. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar DA, Wormsley S, Agentis TM, Baserga SJ. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Leno GH, Lewis N, Rekosh D, Hammarskjöld M-L, Olson MOJ. Location of the HIV-1 Rev protein during mitosis: inactivation of the nuclear export signal alters the pathway for postmitotic reentry into nucleoli. J Cell Sci. 1996;109:2239–2251. doi: 10.1242/jcs.109.9.2239. [DOI] [PubMed] [Google Scholar]
- Dundr M, Meier UT, Lewis N, Rekosh D, Hammarskjöld M-L, Olson MOJ. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–417. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- Eberhard D, Tora L, Egly J-M, Grummt I. A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res. 1993;21:4180–4186. doi: 10.1093/nar/21.18.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- Erickson JM, Rushford C, Dorney C, Wilson G, Schmickel R. Structure and variation of human ribosomal DNA: molecular analysis of cloned fragments. Gene. 1981;16:1–9. doi: 10.1016/0378-1119(81)90055-x. [DOI] [PubMed] [Google Scholar]
- Erickson JM, Schmickel RD. A molecular basis for discrete size variation in human ribosomal DNA. Am J Hum Genet. 1985;37:311–325. [PMC free article] [PubMed] [Google Scholar]
- Fan H, Penman S. Regulation of synthesis and processing of nucleolar components in metaphase-arrested cells. J Mol Biol. 1971;59:27–42. doi: 10.1016/0022-2836(71)90411-6. [DOI] [PubMed] [Google Scholar]
- Gautier T, Dauphin-Villemant C, André C, Masson C, Arnoult J, Hernandez-Verdun D. Identification and characterization of a new set of nucleolar ribonucleoproteins which line the chromosomes during mitosis. Exp Cell Res. 1992;200:5–15. doi: 10.1016/s0014-4827(05)80065-5. [DOI] [PubMed] [Google Scholar]
- Gébrane-Younès J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- Gerard RD, Gluzman Y. New host cell system for regulated simian virus 40 DNA replication. Mol Cell Biol. 1985;5:3231–3240. doi: 10.1128/mcb.5.11.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessens G. Nucleolar structure. Int Rev Cytol. 1984;87:107–158. doi: 10.1016/s0074-7696(08)62441-9. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Wolf VJ, Dang T, Forbes DJ, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie J-P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- Hartl P, Gottesfeld J, Forbes DJ. Mitotic repression of transcription in vitro. J Cell Biol. 1993;120:613–624. doi: 10.1083/jcb.120.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heix J, Zomerdijk JC, Ravanpay A, Tjian R, Grummt I. Cloning of murine RNA polymerase I-specific TAFs: conserved interactions between the four subunits of the species-specific transcription factor TIF-IB/SL1. Proc Natl Acad Sci USA. 1997;94:1733–1738. doi: 10.1073/pnas.94.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliot L, Kaplan H, Lucas L, Klein C, Beorchia A, Doco-Fenzy M, Menager M, Thiry M, O’Donohue M-F, Ploton D. Electron tomography of metaphase nucleolar organizer regions: evidence for a twisted-loop organization. Mol Biol Cell. 1997;8:2199–2216. doi: 10.1091/mbc.8.11.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Gautier T. The chromosome periphery during mitosis. Bioessays. 1994;16:179–185. doi: 10.1002/bies.950160308. [DOI] [PubMed] [Google Scholar]
- Hughes JMX, Ares M. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao L-G, Taneja K, Singer RH, Wang Y, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jiménez-García LF, Segura-Valdez ML, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Lazdins IB, Delannoy M, Sollner-Webb B. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997;105:481–495. doi: 10.1007/BF02510485. [DOI] [PubMed] [Google Scholar]
- Leresche A, Wolf VJ, Gottesfeld JM. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- Li HD, Zagorski JJ, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W-Q, Fournier MJ. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic pre-rRNA by RNase MRP in vitro. Science. 1996a;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Séraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996b;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Maden BEH, Hughes JMX. Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma. 1997;105:391–400. doi: 10.1007/BF02510475. [DOI] [PubMed] [Google Scholar]
- Matera AG, Tycowski KT, Steitz JA, Ward DC. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–1299. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Medina FJ, Cerdido A, Fernández-Gómez ME. Components of the nucleolar processing complex (pre-rRNA, Fibrillarin, and Nucleolin) colocalize during mitosis and are incorporated to daughter cell nucleoli. Exp Cell Res. 1995;221:111–125. doi: 10.1006/excr.1995.1358. [DOI] [PubMed] [Google Scholar]
- Mougey EB, O’Reilly M, Osheim Y, Miller OL, Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993a;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Mougey EB, Pape LK, Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing in the 5′ external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993b;13:5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Phillips DM, Phillips SG. Repopulation of postmitotic nucleoli by preformed RNA. J Cell Biol. 1973;58:54–663. doi: 10.1083/jcb.58.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Puvion E, Bachellerie J-P. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5′ETS leader probe. Chromosoma. 1997;105:496–505. doi: 10.1007/BF02510486. [DOI] [PubMed] [Google Scholar]
- Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Benavente R. Functional and dynamic aspects of the mammalian nucleolus. Bioessays. 1990;12:14–21. doi: 10.1002/bies.950120104. [DOI] [PubMed] [Google Scholar]
- Scheer U, Thiry M, Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP and essential for cell viability. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Schnapp A, Grummt I. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem. 1991;266:24588–24595. [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Seither P, Zatsepina O, Hoffmann M, Grummt I. Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma. 1997;106:216–225. doi: 10.1007/s004120050242. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B, Tyc K, Steitz JA. Ribosomal RNA processing in eukaryotes. In: Zimmerman R, Dahlberg A, editors. Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Synthesis. Boca Raton, FL: CRC Press; 1996. pp. 469–490. [Google Scholar]
- Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11:2926–2937. doi: 10.1101/gad.11.21.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suja JA, Gébrane-Younès J, Géraud G, Hernandez-Verdun D. Relative distribution of rDNA and proteins of the RNA polymerase I transcription machinery at chromosomal NORs. Chromosoma. 1997;105:459–469. doi: 10.1007/BF02510483. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Tuteja N, et al. Human DNA helicase IV is nucleolin, an RNA helicase modulated by phosphorylation. Gene. 1995;160:143–148. doi: 10.1016/0378-1119(95)00207-m. [DOI] [PubMed] [Google Scholar]
- Voit R, Schäfer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D, Scheer U. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J Cell Biol. 1995;129:561–575. doi: 10.1083/jcb.129.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf JM, Konstantinov KN, Wormsley S, Shu MD, Matsumoto-Taniura N, Pirollet F, Klier FG, Gerace L, Baserga SJ. M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol Biol Cell. 1998;9:437–449. doi: 10.1091/mbc.9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Gottlieb TM, Downes CS, Jackson SP. Cell cycle regulation of RNA polymerase III transcription. Mol Cell Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GN, Szura LL, Rushford C, Jackson D, Erickson J. Structure and variation of human ribosomal DNA: the external transcribed spacer and adjacent regions. Am J Hum Genet. 1982;34:32–49. [PMC free article] [PubMed] [Google Scholar]
- Zatsepina OV, Dudnic OA, Chentsov YS, Thiry M, Spring H, Tredelenburg MF. Reassembly of functional nucleoli following in situ unraveling by low-ionic-strength of cultured mammalian cells. Exp Cell Res. 1997;233:155–168. doi: 10.1006/excr.1997.3556. [DOI] [PubMed] [Google Scholar]