Abstract
The fission yeast pob1 gene encodes a protein of 871 amino acids carrying an SH3 domain, a SAM domain, and a PH domain. Gene disruption and construction of a temperature-sensitive pob1 mutant indicated that pob1 is essential for cell growth. Loss of its function leads to quick cessation of cellular elongation. Pob1p is homologous to two functionally redundant Saccharomyces cerevisiae proteins, Boi1p and Boi2p, which are necessary for cell growth and relevant to bud formation. Overexpression of pob1 inhibits cell growth, causing the host cells to become round and swollen. In growing cells, Pob1p locates at cell tips during interphase and translocates near the division plane at cytokinesis. Thus, this protein exhibits intracellular dynamics similar to F-actin patches. However, Pob1p constitutes a layer, rather than patches, at growing cell tips. It generates two split discs flanking the septum at cytokinesis. The pob1-defective cells no longer elongate but swell gradually at the middle, eventually assuming a lemon-like morphology. Analysis using the pob1-ts allele revealed that Pob1p is also essential for cell separation. We speculate that Pob1p is located on growing plasma membrane, possibly through the function of actin patches, and may recruit proteins required for the synthesis of cell wall.
INTRODUCTION
Coordination of cell growth with maintenance of proper cell morphology is an intriguing problem in cell biology. It has been well documented that actin cytoskeleton plays important roles for both cell growth and the integrity of cell morphology, and a cascade involving small GTP-binding proteins is implicated in the regulation of actin organization (Van Aelst and D’Souza-Schorey, 1997; Hall, 1998). However, much remains to be studied in order to understand how such coordination is substantiated throughout the cell cycle.
In the budding yeast Saccharomyces cerevisiae, formation of buds has been studied cytologically and genetically, and many gene products engaged in this process have been identified. For example, bud emergence is known to require Cdc42p, a small GTP-binding protein of the Rho subfamily, Cdc24p, a GDP-GTP exchange factor for Cdc42p, and Bem1p, a protein that contains SH3 domains and interacts with Cdc24p (Peterson et al., 1994, and citations therein). Other small GTP-binding proteins, Rho3p and Rho4p, are required for bud growth (Matsui and Toh-e, 1992). Cortical F-actin patches are observed at the budding site and the tip of the growing bud. The actin cytoskeleton, with the aid of type I myosin, Myo2p, appears to transport secretory vesicles to the budding site, and this process is likely to be under the control of the aforementioned molecules (Johnston et al., 1991).
Unlike S. cerevisiae, the fission yeast Schizosaccharomyces pombe has cylindrical cell shape and grows by elongation at cell tips. During interphase, F-actin is localized at growing ends in patches (Marks and Hyams, 1985; Balasubramanian et al., 1997). In M phase, an actin ring is generated at the midline of the cell (Marks and Hyams, 1985; Balasubramanian et al., 1997) while the patches translocate from the tips to the central region (Balasubramanian et al., 1998). In a similar manner to animal cells, the ring constricts with the aid of type II myosin to achieve cytokinesis (Balasubramanian et al., 1997; Kitayama et al., 1997; May et al., 1997; Motegi et al., 1997). F-actin patches remain beside the division plane until the completion of a septum, most likely aiding its formation (Balasubramanian et al., 1998).
Mutations in a variety of genes are known to affect polarized growth and/or cylindrical morphology of S. pombe cells. The function of cdc42 is required for polarized cell growth. Loss of its functional product, Cdc42p, results in growth arrest and in small- and round-cell morphology. Expression of an activated form of Cdc42p also causes rounding of cells (Miller and Johnson, 1994). The ral1/scd1 gene is homologous to S. cerevisiae CDC24 and encodes a putative guanine-nucleotide exchange factor for Cdc42p (Chang et al., 1994). Cells defective in ral1/scd1 are viable but have a round shape (Fukui and Yamamoto, 1988). The orb mutants also show round morphology at the restrictive temperature (Verde et al., 1995). In contrast to these round cells, which appear to result from loss of polarity, the tea mutants often display branched cells, suggesting mislocalization of their growing tips (Mata and Nurse, 1997).
The conventional small GTP-binding protein, Ras1p, also affects cell morphology in fission yeast. Cells of the ras1 deletion mutant are short and fat, in addition to being sterile, suggesting that they are defective in the maintenance of cell morphology (Fukui et al., 1986). Two pathways have been established to function downstream of Ras1p, although evidence suggesting a third possible pathway exists (Hakuno et al., 1996). One of the two is a MAP kinase cascade that transmits the mating pheromone signal and regulates nuclear gene expression, which is essential for mating but not for the maintenance of cell morphology (Wang et al., 1991; Nielsen et al., 1992; Xu et al., 1994). The other is a cascade involving Cdc42p, which is essential for both mating and normal cell morphology. Cells defective in ral1/scd1, encoding a homologue of S. cerevisiae Cdc24p, and cells defective in ral3/scd2, encoding a homologue of S. cerevisiae Bem1p, are both reminiscent of the ras1 mutant cells in their morphology and sterility (Fukui and Yamamoto, 1988; Chang et al., 1994). These proteins constitute analogous cascades in S. pombe and S. cerevisiae, together with respective Cdc42 proteins. The cascade in S. pombe is regulated by Ras1p, whereas the counterpart in S. cerevisiae is regulated by Rsrp (Bud1p). Rsrp is a small GTP-binding protein similar to, but distinct from, Ras proteins. Simultaneous overexpression of Cdc42p and Ral1/Scd1p restores wild-type cell morphology to the ras1 mutant of S. pombe (Chang et al., 1994).
In this article, we describe isolation and characterization of a new S. pombe gene, pob1, which is required for cell elongation. Localization of Pob1p during the cell cycle mimics that of F-actin patches, although they do not strictly coincide. Pob1p is structurally and functionally related to Boi1p and Boi2p, which are implicated in budding in S. cerevisiae (Bender et al., 1996; Matsui et al., 1996). However, Pob1p turned out to play an essential role also in cell separation, a job not so far ascribed to S. cerevisiae Boi proteins. Possible molecular function of the Boi family proteins will be discussed.
MATERIALS AND METHODS
Yeast Strains, Genetic Procedures, and Media
S. pombe strains used in this study are listed in Table 1. General genetic procedures for S. pombe were according to Gutz et al. (1974). Complete medium YE, minimal medium SD (Sherman et al., 1986), and minimal medium MM (Moreno et al., 1990) were used. S. pombe cells were transformed by either electroporation (Prentice, 1992) or a lithium acetate method (Okazaki et al., 1990).
Table 1.
S. pombe strains used in this study
| Strain | Genotype |
|---|---|
| JW100 | h− ade6-M216 leu1 pob1+-3HA (+kanR) |
| JX268 | h90 ade6-M216 leu1 ura4-D18 ras1::ura4+ ste8-D1 |
| JX584 | h− ade6-M216 leu1 pob1-664 |
| JX645 | h90/h90 ade6-M216/ade6-M210 leu1/leu1 ura4-D18/ura4-D18 |
| pob1::ura4+/pob1+ | |
| JX646 | h90 wee1::ura4+ leu1 ura4-D18 pob1-664 |
| JX1001 | h+ ade6-704 pob1::(nmt1-GFP-pob1++sup3.5) |
| JY1 | h− wild type |
| JY333 | h− ade6-M216 leu1 |
| JY878 | h90 ade6-M216 leu1 ura4-D18 |
| JZ489 | h90/h90 ade6-M216/ade6-M210 leu1/leu1 ura4-D18/ura4-D18 |
| JZ1005 | h− wee1::ura4+ leu1 ura4-D18 |
Cloning and Sequencing of the pob1 Gene
To identify a new fission yeast gene possibly involved in the Cdc42p-mediated signaling pathway, we screened for multicopy suppressors of the mating defect of an S. pombe strain (JX268) that was defective in ras1 and carried an activated form of Byr2p/Ste8p MAPKKK. JX268 cells were transformed with an S. pombe genomic library, constructed in the expression vector pART1 carrying the adh1 promoter (McLeod et al., 1987). Leu+ transformants were selected on synthetic sporulation agar plates (Egel and Egel-Mitani, 1974), and sporulation-proficient transformants were identified by staining the colonies with iodine vapor. Dark-brown colonies were picked and inspected further under the microscope to confirm sporulation. During this process, we noticed that some transformants formed spores but had round and enlarged cell morphology, suggesting that they underwent azygotic rather than zygotic sporulation. Plasmid DNA was recovered from them into Escherichia coli JA226. We eventually obtained two plasmids with overlapping inserts, one of which encompassed the other. The region essential for the suppression was delimited to a 5.8-kilobase (kb) fragment. To exclude any possible derangement of the fragment, we reisolated a corresponding 5.8-kb ScaI–SacI DNA fragment from the wild-type strain JY1. The fragment was cloned into the BamHI site of pART1 to give rise to pWT4–3 (Figure 1), which could convert JX268 to sporulation proficiency. The nucleotide sequence of the 5.8-kb fragment was determined using the Sequence Kit (United States Biochemical, Cleveland, OH) and an automated DNA sequencer (LI-COR model 4000L).
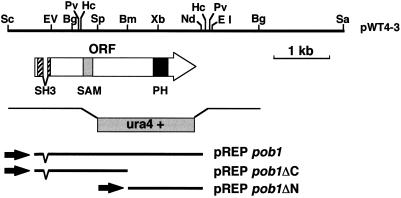
Figure 1.
Restriction map of the pob1 locus. Restriction sites on the insert of pWT4–3 are shown on the top. The extent and direction of the pob1 ORF, encoding 871 amino acids, are indicated by the arrow. The hatched box on the arrow indicates an SH3 domain, the shaded box indicates a SAM domain, and the filled box indicates a PH domain. The structure of a linear fragment carrying ura4+, which was used for disruption of the pob1 gene in vivo, is illustrated under the arrow. Inserts of three groups of subclones carrying the whole or a part of the pob1 ORF, collectively denoted either pREPpob1, pREPpob1ΔC, or pREPpob1ΔN, are also indicated. Restriction sites: Bg, BglII; Bm, BamHI; EI, EcoRI; EV, EcoRV; Hc, HincII; Pv, PvuII; Nd, NdeI; Sa, SacI; Sc, ScaI; Sp, SpeI; and Xb, XbaI.
Disruption of the pob1 Gene
A 2.5-kb PvuII fragment was eliminated from the cloned pob1 gene, and the 1.8-kb S. pombe ura4+ cassette (Grimm et al., 1988) was inserted in its place (Figure 1). A diploid strain (JZ489) was transformed with linearized DNA fragment carrying the disrupted pob1 allele. Stable Ura+ transformants were selected and analyzed by Southern blot analysis (Southern, 1979) to verify the proper replacement of one of the chromosomal pob1 alleles by the disrupted allele.
Expression of Truncated or Tagged Pob1p
Three types of pob1 subclones were constructed in the expression vectors, pREP1, pREP41, and pREP81, which carried either the weak, the medial, or the strong thiamine-repressible nmt1 promoter (Basi et al., 1993). An NdeI site was created at the initiation codon of pob1 cDNA, so that the entire pob1 ORF could be excised as an NdeI–NdeI fragment (Figure 1). One group of plasmids, collectively denoted pREPpob1, carried this NdeI–NdeI fragment in each pREP vector. Another group, denoted pREPpob1ΔC, carried the NdeI–BamHI fragment, expressing only the N-terminal half of Pob1p, and a third group, denoted pREPpob1ΔN, carried the BamHI–NdeI fragment, expressing only the C-terminal half of Pob1p.
A haploid pob1Δ strain expressing hemagglutinin (HA)-tagged Pob1p from pREP81HApob1 was recovered from progeny of the pob1Δ/pob1+ diploid strain JX645 transformed with the plasmid. Full expression of HA-Pob1p from the weak nmt1 promoter on the pREP81-based multicopy plasmid resulted in a mixture of roundish and cylindrical cells, presumably due to deviation of its copy number among cells. Typical cylindrical cells were analyzed precisely by immunostaining. We also constructed a haploid strain carrying a chromosomal gene that encoded Pob1p fused with three copies of HA at its C terminus (Pob1p-3HA), according to a standard protocol precisely described by Bähler et al. (1998). This strain, named JW100, expressed the pob1+-3HA ORF from the authentic pob1 promoter, which was integrated in the chromosome in a single copy together with the kanR gene as a drug-resistance marker, replacing the original pob1 gene.
To express green fluorescent protein (GFP)-tagged Pob1p stably, we constructed a strain JX1001, which carried a GFP-pob1+ fusion gene controlled by the authentic (strong) nmt1 promoter. The nmt1-GFP-pob1+ construct was integrated into the chromosome in a single copy, replacing the pob1 gene, by virtue of the suppression of the ade6–704 mutation by sup3.5 (Hayles et al., 1994). Because full expression of GFP-Pob1p in JX1001 cells in the complete absence of thiamine was apparently excessive and made the cells round, we cultured them in liquid MM containing 0.05 μg/ml thiamine for 20 h before microscopic observation, which generated cylindrical cells close to the wild type.
Microscopy
Staining for F-actin with rhodamine-conjugated phalloidin (Sigma Chemical, St. Louis, MO) was performed as described (Alfa et al., 1993). DNA and septa were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and Calcofluor white (Sigma), respectively, in fixed samples. For immunolocalization of Pob1p, we basically followed the protocol described by Alfa et al. (1993). Cells producing HA-tagged Pob1p were cultured at 30°C in MM to exponential phase. They were fixed with formaldehyde and stained with mouse anti-HA monoclonal antibody (Boehringer Mannheim, Indianapolis, IN). Cy3-conjugated goat anti-mouse IgG (Chemicon, Temecula, CA) was used as the secondary antibody. For costaining of F-actin, BODIPY FL phallacidin (Molecular Probes, Eugene, OR) was added when the secondary antibody was applied. DNA was counterstained with Hoechst 33342. Fluorescence microscopy was performed using an Axiophot microscope (Carl Zeiss, Thornwood, NY) with an appropriate set of filters. To examine fluorescence of GFP-Pob1p, we used a confocal microscope (Zeiss LSM510).
Construction of a pob1 Temperature-sensitive Allele
The method described by Francesconi et al. (1993) was used to generate a temperature-sensitive allele of pob1 with minor modifications. In vitro mutagenesis of pob1 was performed by using a PCR method described by Zhou et al. (1991). Briefly, a 2.4-kb HincII–HincII fragment, carrying most of the pob1 ORF including the C terminus and the essential PH-domain but lacking the N terminus and the promoter region, was cloned into pUC119. The resultant plasmid was used as a template for PCR. A pair of oligonucleotide primers for pUC119, namely HHpUC (5′-AAGCTTGCATGCCTGCA-3′) and M13–40 (5′-GTTTTCCCAGTCACGAC-3′), were used for PCR amplification. Amplified fragments were digested with PstI and EcoRI and cloned between the PstI and EcoRI sites of pUC119 carrying the ura4+ cassette. The mutagenized library thus obtained was linearized at the SpeI site within the pob1 ORF and was transformed into a haploid strain JY878. Integration of an entire plasmid at the chromosomal pob1 locus by homologous recombination was expected to result in uracil prototrophy. Ura+ transformants were selected at 25°C on an SD plate. Nine hundred Ura+ colonies were picked, and they were examined for the ability to grow at 37°C on a YPD plate. Two strains were found to be temperature sensitive, presumably carrying mutations in the pob1 gene. These Ura+ temperature-sensitive transformants were then plated on SD containing 0.5 mg/ml 5-fluoroorotic acid (5-FOA) at 25°C to segregate out the integrated plasmid. Ura− colonies were tested for temperature sensitivity, and colonies that exhibited ts growth were selected. Two ts Ura− strains were finally obtained. The ts allele of pob1 carried by one of them (JX584) was named pob1–664 and analyzed further.
Protein Assay
Cells in each cell culture (1 ml) were spun down, washed with distilled water, and dissolved in 100 μl of 1 N NaOH–2% deoxycholic acid at 32°C. The assay was done using the BCA protein assay reagent (Pierce Chemical, Rockford, IL) according to the protocol provided by the manufacturer.
Flow Cytometric Analysis
Samples for flow cytometry were prepared essentially as described previously (Imai and Yamamoto, 1994). Cells were analyzed by a flow cytometer FACScan (Beckton-Dickinson, San Jose, CA)
RESULTS
Cloning of the pob1 Gene, a Fission Yeast Homologue of S. cerevisiae BOI1 and BOI2
We isolated an S. pombe genomic clone, named pWT4–3, that could promote sporulation in a homothallic haploid strain defective in ras1 and activated in byr2, as detailed in MATERIALS AND METHODS. Although we originally intended to isolate clones that could promote mating in this strain, microscopic observation suggested that the mating defect was not suppressed by pWT4–3. The recovery of sporulation appeared to be due to aberrant diploidization induced by overexpression of the cloned gene (see below). The clone carried a 5.8-kb ScaI–SacI fragment derived from the S. pombe genome, and sequence analysis revealed an ORF on it, which potentially encoded 871 amino acid residues and was interrupted by one putative intron of 87 base pairs (bp) in length (Figure 1; the nucleotide sequence is deposited in GenBank/EMBL/DDBJ under accession number AB018044). The assigned intron was shown to be absent in mRNA by PCR analysis (our unpublished data).
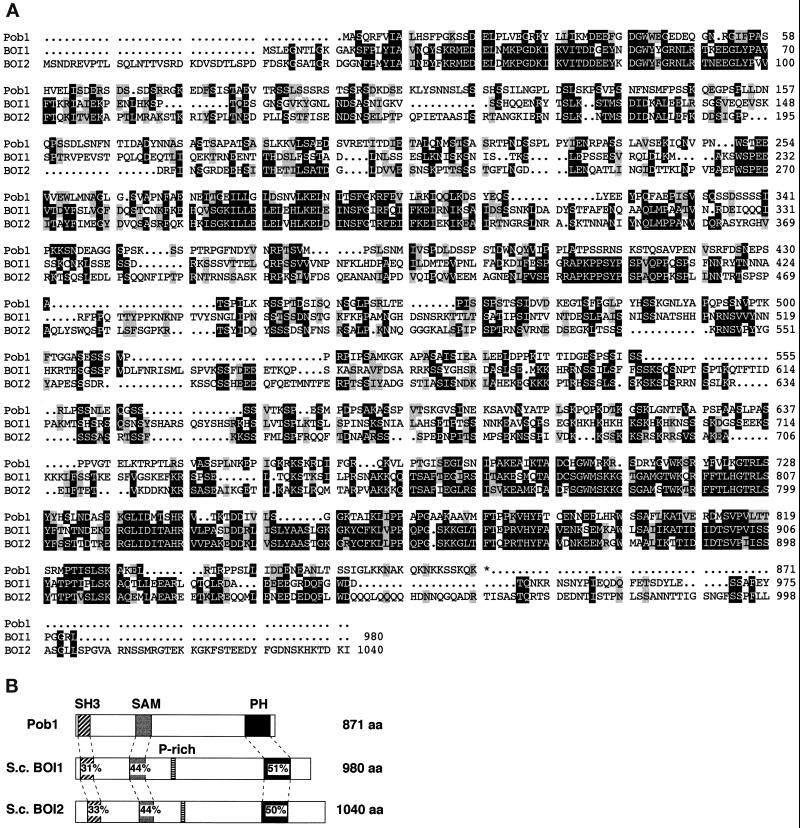
The deduced gene product was compared with database entries using the FASTA homology search algorithm (Lipman and Pearson, 1985). It was found to be highly similar to S. cerevisiae Boi1p and Boi2p, which were identified originally as Bem1p-interacting proteins (Bender et al., 1996; Matsui et al., 1996) (Figure 2). We then named the cloned gene pob1 (S. pombe BOI). Boi1p and Boi2p are essential for cell growth but are mutually redundant in function, and the boi1 boi2 double mutant is defective in bud formation and in the maintenance of cell polarity. Cells overexpressing either BOI1 or BOI2 are arrested as large, round, unbudded cells (Bender et al., 1996; Matsui et al., 1996). S. cerevisiae Boi proteins carry four characteristic motifs: an SH3 domain, a SAM domain (Schults et al., 1997), a PH domain, and a prominent proline-rich region (Figure 2B). Pob1p has an SH3 domain (residues 9–60), a SAM domain (residues 248–312),and a PH domain (residues 703–806), but a proline-rich sequence is apparently missing in Pob1p (Figure 2B).
Figure 2.
The deduced pob1 gene product. The nucleotide sequence of the pob1 gene is available from GenBank/EMBL/DDBJ under accession number AB018044. (A) Comparison of the amino acid sequences of Pob1p and S. cerevisiae Boi1p and Boi2p. Identical amino acids are shown in white against black, whereas conservative changes are shaded. Conserved amino acid substitutions are grouped as follows: L, I, V, and A; F, Y, and W; K and R; E and D; Q and N; and S and T. Three domains conserved fairly well between Pob1p and the Boi proteins, i.e., SH3 (position 9–60 of Pob1p), SAM (248–312), and PH (703–806) domains, can be seen. (B) Schematic of the structures of Pob1p and its S. cerevisiae homologues. Amino acid identity between Pob1p and either Bio1p or Boi2p is given in percentage with respect to each conserved domain. P-rich, proline-rich region.
Disruption of pob1 and Construction of a Temperature-sensitive pob1 Allele
Disruption of the pob1 ORF was carried out by insertion of an S. pombe ura4+ cassette in a diploid strain JZ489, as described in MATERIALS AND METHODS. The DNA fragment employed to disrupt pob1 is illustrated in Figure 1. Precise replacement of one of the two pob1+ alleles by pob1::ura4+ was confirmed by Southern blot analysis (our unpublished data). More than 70% of the pob1+ ORF was deleted in the pob1::ura4+ allele. Sporulation was induced in a pob1::ura4+/pob1+ strain thus obtained (JX645), and progeny asci were dissected. Each ascus generated, at most, two viable spores, and all of them were Ura−, suggesting that disruption of pob1 is lethal. This also suggested that, unlike S. cerevisiae, S. pombe probably has no second BOI homologue. Microscopic observation of spores that failed to form a colony indicated that pob1Δ spores ceased growth soon after germination. The lethality of the Ura+ spores could be rescued when a plasmid carrying the pob1 cDNA was introduced into JX645 before sporulation, confirming that pob1+ is responsible for cell viability (our unpublished data).
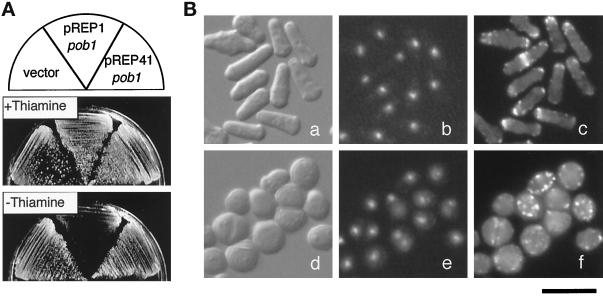
To better characterize the consequence of loss of pob1 function, we constructed a strain (JX584) carrying a temperature-sensitive allele of pob1, termed pob1–664. The procedure to obtain this allele has been described in detail in MATERIALS AND METHODS. Replacement of the original pob1+ allele by the mutant allele with no derangement was confirmed by PCR analysis in JX584 (our unpublished data). JX584 grew normally at 25°C, but its growth was severely inhibited at the restrictive temperature 37°C (Figures 3A and 4, A and B). The pob1–664 mutation was recessive in heterozygous diploids (our unpublished data). The temperature sensitivity could be rescued by a plasmid-borne pob1 gene (Figure 4, A and B), indicating that the growth defect was indeed due to the mutation that hit the pob1 gene.
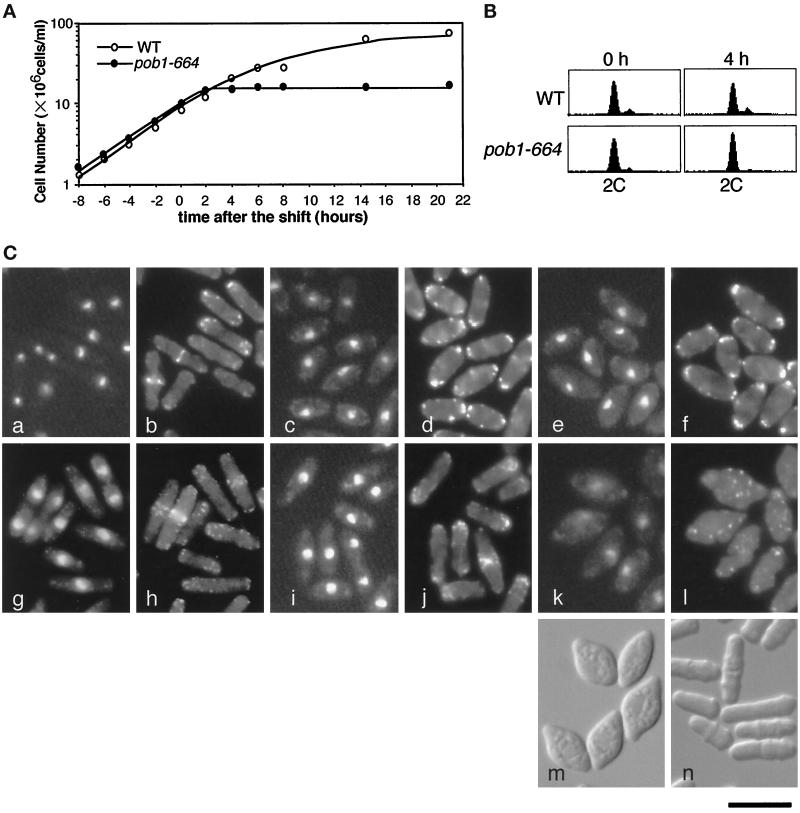
Figure 3.
Characterization of the pob1–664 temperature-sensitive mutant. (A) Growth profiles of pob1–664 (JX584) and wild-type (JY333) cells subjected to temperature shift. Cells growing exponentially in liquid YE medium at 25°C were shifted to 37°C at the concentration of ∼107 cells/ml. The cell number was chronologically measured by a Coulter counter (Coulter, Luton, UK). (B) Flow cytometric patterns of JY333 and JX584 cells cultured in panel A. Samples were taken at the time of the temperature shift and 4 h after the shift. (C) Morphological changes of JY333 and JX584 cells cultured in panel A. Samples were taken at the time of the temperature-shift and 4, 8, and 21 h after the shift. Cells were fixed and stained with Hoechst 33342 (a, c, e, g, i, and k) and rhodamine-phalloidin (b, d, f, h, j, and l). a and b, JX584, 0 h; c and d, JX584, 4 h; e and f, JX584, 8 h; g and h, JY333, 0 h; i and j, JY333, 4 h; k and l, JX584 21 h. Differential interference contrast (DIC) images of JX584 cells at 21 h after the shift (m) and at the time of the temperature-shift (n) are also shown. Bar, 10 μm.
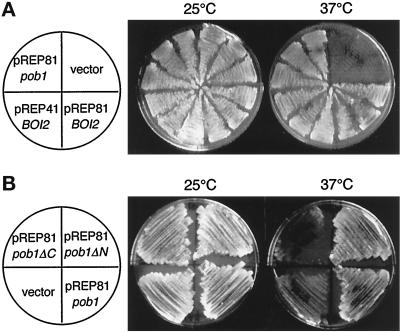
Figure 4.
Complementation of pob1 deficiency. (A) Complementation by S. cerevisiae BOI2. JX584 (pob1–664) was transformed with either pREP81pob1, the vector pREP81, pREP81BOI2, or pREP41BOI2. The latter two plasmids carry the S. cerevisiae BOI2 gene. Transformed cells were streaked on MM plates and incubated at either 25°C (left) or 37°C (right) for 4 d. (B) Complementation by truncated pob1 genes. JX584 was transformed with either pREP81pob1ΔC, pREP81pob1ΔN, pREP81pob1, or the vector pREP81. Transformants were streaked on MM plates and incubated at either 25°C (left) or 37°C (right) for 4 d.
Phenotypes Caused by Loss of pob1 Function
Possible morphological changes induced by loss of pob1 function were examined chronologically using the pob1-ts mutant JX584. Cells grown to midlog phase at 25°C in liquid YE medium were subjected to a shift to 37°C. They ceased division in ∼2 h after the shift (Figure 3A). The majority of the arrested cells displayed a single nucleus and no septum (Figure 3C, panels c and e). Cells were sampled 4 h after the shift and the amount of DNA per cell was analyzed by flow cytometry. It turned out to be 2C, where C stands for the amount of DNA in a haploid cell at G1 phase (Figure 3B), indicating that the cells remained at G2 phase in the cell cycle. However, unlike typical G2-arrest mutants such as cdc25 (Fantes, 1981), they did not show significant cell elongation after the arrest (compare Figure 3A and Table 2, especially at 4 h). This indicates that inhibition of cell elongation is a possible immediate consequence of loss of pob1 function. The arrested cells maintained high viability at the restrictive temperature: nearly 90% of cells incubated at 37°C for 15 h could resume growth when shifted down to the permissive temperature. F-actin was located in patches at both ends of the arrested cells (Figure 3C, panels d and f), and this localization persisted even after 15 h incubation at 37°C (our unpublished data). Although there was no significant increase in cell length, cells apparently continued a low level of residual protein synthesis at the restrictive temperature (Table 2), and consistently, they gradually became fatter (Figure 3C, panels c–f). After 21 h, the middle of these cells was enormously swollen, with the two tips sticking out, exhibiting a cell morphology like a lemon (Figure 3C, panel m). Actin patches were dispersed in these extremely irregular cells (Figure 3C, panel l). The same unique morphology was also observed when pob1Δ cells carrying a pob1 plasmid were led to plasmid loss under nonselective conditions (our unpublished data).
Table 2.
Residual growth of the pob1-ts mutant JX584 at 37°C
| Time after the shift to 37°C (h) | Cell length (μm) | Protein per cell (10−5 μg) |
|---|---|---|
| 0 | 8.0 ± 1.8 | 1.5 |
| 4 | 7.7 ± 1.6 | 1.4 |
| 8 | 8.3 ± 1.3 | 1.7 |
| 10 | 8.3 ± 1.4 | 2.1 |
| 15 | 8.3 ± 1.1 | 1.9 |
S. cerevisiae BOI2 Complements the Growth Defect of pob1–664 and pob1Δ
JX584 (pob1–664) was transformed with a plasmid harboring S. cerevisiae BOI2 under the control of either the weak or the medial nmt1 promoter (pREP81BOI2 or pREP41BOI2). When the promoter was activated by depleting thiamine from the medium, both pREP41BOI2 and pREP81BOI2 suppressed the temperature-sensitive growth of JX584 (Figure 4A). The growth defect of pob1Δ spores was also circumvented if the parental diploid (pob1::ura4+/pob1+) was transformed with either pREP41BOI2 or pREP81BOI2 before sporulation (our unpublished data). Most of the pob1–664 or pob1Δ cells rescued by expression of BOI2 exhibited a normal rod-like morphology. These results indicate that Pob1p is a homologue of Boi proteins not only structurally but also functionally.
The C-terminal Region of Pob1p Carrying a PH Domain Is Responsible for Cell Growth
To identify regions that are important for Pob1p function, we examined two truncated versions of Pob1p: Pob1p-ΔN, which lacked the N-terminal sequence including the SH3 and SAM domains, and Pob1p-ΔC, which lacked the C-terminal sequence including the PH domain (Figure 1). Pob1p-ΔN expressed from the weak nmt1 promoter rescued the temperature sensitivity of the pob1–664 strain, but Pob1p-ΔC did not (Figure 4B). Pob1p-ΔN could also rescue pob1Δ, although less efficiently than intact Pob1p, while Pob1p-ΔC could not (our unpublished data). Microscopic observation revealed that JX584 cells (pob1–664) rescued by Pob1p-ΔN were not normalized in morphology at the restrictive temperature. These results suggest that the C-terminal region of Pob1p is responsible for the basic function required for cell growth, and that the N-terminal region bearing the SH3 and SAM domains may augment this function so that the cells can retain the most appropriate morphology.
Intracellular Localization of Pob1p Mimics That of F-Actin Patches
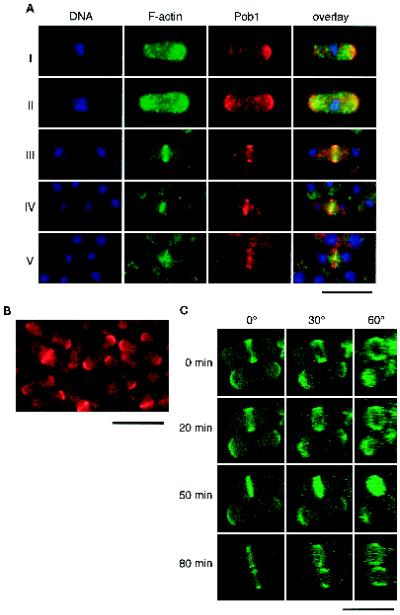
To observe intracellular localization of Pob1p, Pob1p was expressed as an HA fusion protein from the weak nmt1 promoter on pREP81, which allowed a comparable degree of transcription to the authentic pob1 promoter on the chromosome. HA-Pob1p complemented the lethality of pob1Δ cells efficiently, proving that it was functional. We stained pob1Δ cells expressing HA-Pob1p with anti-HA antibody, together with BODIPY-phallacidin and Hoechst 33342 to visualize F-actin and DNA (Figure 5A). Cells exhibiting typical rod-like shape were chosen for careful inspection. The results are shown in Figure 5A and can be summarized as below.
Figure 5.
Localization of Pob1p. (A) Comparison of intracellular localization of Pob1p and F-actin. Haploid pob1Δ cells expressing HA-Pob1p were fixed in growth phase and stained with anti-HA to detect Pob1p, BODIPY-phallacidin to detect F-actin, and Hoechst 33342 to detect DNA. The panels show from left to right: localization of nuclei (DNA) displayed in blue, localization of F-actin displayed in green, localization of Pob1p displayed in red, and overlays of these three images. (B) Detection of Pob1p-3HA expressed from a single-copy chromosomal gene. Cells of JW100 were grown asynchronously in liquid YE medium at 30°C, fixed and stained with anti-HA. (C) Chronologically chased images of growing JX1001 cells. These cells were allowed to express an appropriate level of GFP-tagged Pob1p by adding a limited amount of thiamine, so that they would assume morphology close to the wild-type. GFP fluorescence was digitized and recorded by confocal microscopy. The original view (0°) and synthesized images after rotation of either 30° and 60° are shown. Bar, 10 μm.
F-actin was observed mostly in patches at growing ends and also in probable cables in interphase cells (Figure 5A, stages I and II). The medial actin ring emerged in mitotic cells (stage III). F-actin in patches, although less obvious, could be recognized in the central region in cells undergoing cytokinesis (stages IV and V). Throughout these stages, HA-Pob1p was detected in the area where actin patches predominantly localized, implying their possible interaction. However, precise inspection revealed small but clear difference in their localization. In interphase cells, F-actin was distributed in patches near the growing ends, whereas HA-Pob1p appeared to form a layer at the very ends (stages I and II). Whereas the anti-HA antibody gave nonspecific punctate staining in the background, the layer structure was more clearly observed when GFP-tagged Pob1p was employed (Figure 5C; see below). In mitotic cells, HA-Pob1p was initially observed as a broad ring on the cell surface, which encompassed the thinner actin ring located at the center of the cell (stage III). Because of the strong fluorescence of the actin contractile ring, it was not known whether actin patches were located close to the Pob1p ring at this stage. The broad Pob1p ring was then split into two parts at an early stage of cytokinesis, presumably as a result of the onset of actin ring contraction, but it did not constrict itself (stage IV). Splitting of the Pob1p ring was more clearly visible at a later stage, and it appeared that the split rings grew centripetally, as the contractile ring shrank gradually (stage V; see below). Actin patches were visible near the division plane at this stage.
When we expressed HA-tagged Pob1p-ΔC in the wild-type strain JY333, this protein was distributed rather homogeneously throughout the cytoplasm (our unpublished data). This may suggest that the PH domain is important for the proper subcellular localization of Pob1p.
To exclude the possibility that the observed localization of Pob1p at growing tips and the middle of the cell was an abnormal outcome of the possible overproduction of HA-Pob1p in the above experimental system, we constructed a haploid strain that expressed Pob1p–HA fusion protein (Pob1p–3HA) from a single chromosomal gene driven by the authentic pob1 promoter (JW100). An asynchronous culture of this strain revealed essentially the same localization of Pob1p, i.e., at growing tips and the middle of the cell (Figure 5B), indicating that Pob1p normally alternates its location between these two sites.
Pob1p Forms Two Split Discs at Cytokinesis
Splitting of the Pob1p ring was analyzed more precisely in live cells expressing GFP-Pob1p. JX1001, which carried the GFP–pob1+ fusion gene driven by the nmt1 promoter, was cultured in the medium containing 0.05 μg/ml thiamine. The localization of Pob1p detected by GFP fluorescence was again alternating between cell tips and the center (Figure 5C). Transcription of the fusion gene was not more than 1.4 times as much as that of pob1+ under these experimental conditions (our unpublished data), supporting that the observed dynamics of the fusion protein was likely to misrepresent natural localization of Pob1p. JX1001 cells undergoing cytokinesis were examined carefully under the laser-scanning confocal microscope. Pob1p gave rise to two close but distinct bands, apparently on both sides of the cleavage furrow (Figure 5C, 50 min). Rotation of the images revealed that Pob1p formed discs at this stage. Here again, Pob1p appeared to constitute a continuous structure, unlike septum-associated F-actin patches (Balasubramanian et al., 1998). Separation of the two Pob1 bands became more evident subsequently (Figure 5C, 80 min). Curiously, however, the central region of the Pob1 disk was gone at this stage, which was seen reproducibly. These observations may imply that Pob1p is located in the area where plasma membrane should be newly synthesized and materials for the cell wall or septum deposited.
Overexpression of pob1 Inhibits Cell Growth and Causes Round Cell Shape
To observe the effects of pob1 overexpression, we connected the pob1 ORF to a series of nmt promoters on pREP vectors, as described in MATERIALS AND METHODS, and expressed it ectopically in a wild-type strain JY333 by depleting thiamine from the medium. Growth of JY333 cells, examined on plate, was severely inhibited by the expression of pob1 from the strong nmt1 promoter, but only moderately inhibited by the expression from the medial nmt1 promoter (Figure 6A). These cells exhibited round morphology as a terminal phenotype (our unpublished data; see below). To confirm that rounding of cells occurred as an immediate consequence of pob1 overexpression, we grew the nmt1-GFP-pob1+ strain JX1001 at 25°C in the presence of 0.05 μg/ml thiamine so that the cells assume cylindrical morphology (Figure 6B, panel a). Distribution of F-actin appeared normal in these cells (Figure 6B, panel c). Exponentially growing cells were transferred to the medium containing no thiamine with appropriate dilution and incubated for 23 h, a time span necessary and sufficient to derepress the nmt1 promoter (Balasubramanian et al., 1998). The cells became round (Figure 6B, panel d), in which F-actin displayed random and punctate distribution (Figure 6B, panel f). Occasionally binucleate cells were seen among them (Figure 6B, panel e). These results indicate that overexpression of pob1 in S. pombe causes cellular depolarization, as was the case with overexpression of BOI1 or BOI2 in S. cerevisiae (Bender et al., 1996; Matsui et al., 1996).
Figure 6.
Effects of overexpression of Pob1p. (A) Growth of wild-type cells carrying three different plasmids. Cells of JY333 transformed with either the vector pREP1, pREP1pob1, or pREP41pob1 were streaked on MM plates with or without 2 μM thiamine and incubated for 4 d at 30°C. (B) Morphological change induced by overexpression of Pob1p. JX1001 cells grown in the presence of a limited amount of thiamine assuming rod-like shape (a, b, and c) were shifted to the medium with no thiamine and incubated for 23 h at 25°C (d, e, and f). Cells were fixed and stained with Hoechst 33342 and rhodamine- phalloidin. DIC images (a and d), fluorescence of Hoechst-stained DNA (b and e), and F-actin stained with rhodamine-phalloidin (c and f) are shown. Bar, 10 μm.
Pob1p Is Required for Cell Separation in Addition to Cell Elongation
pob1–664 Cells incubated at the restrictive temperature arrested with 2C DNA content, as described above. These cells displayed the interphase array of microtubules (our unpublished data), indicating that they were indeed in G2 phase. This observation implies that pob1 function may be directly required for the G2–M transition. Alternatively, it may be that these cells arrest in G2 due to checkpoint control. Because S. pombe cells cannot enter M phase until they reach a critical size (Nurse, 1975), it is possible that pob1–664 cells fail to elongate up to this size at the restrictive temperature. To test this possibility, we designed an arrest-release experiment, in which hydroxyurea (HU), an inhibitor of DNA synthesis, was used to enlarge cells above the critical size. Here we mainly describe observations obtained employing the wee1 mutant, which was assumed to hurdle the checkpoint control by size more easily (Figure 7).
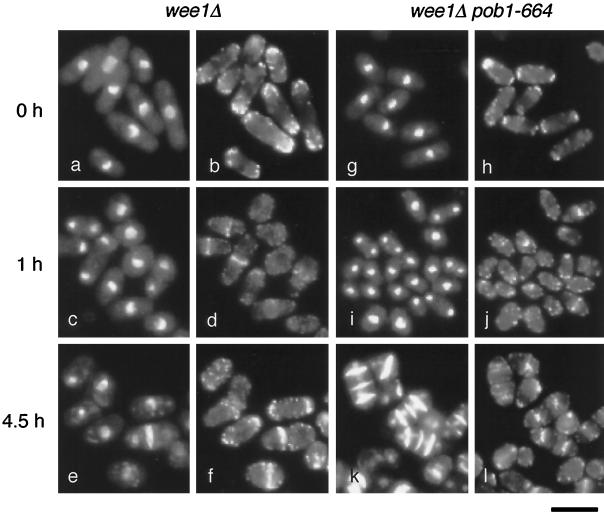
Figure 7.
Requirement of Pob1p for cell separation. Cells of JX646 (wee1Δ pob1–664) and JZ1005 (wee1Δ pob1+) as a control were cultured in liquid YE supplemented with 12 mM HU at 25°C for 3 h. They were then sifted to 37°C, and 2 h later, HU was washed out (time = 0). Incubation was continued at 37°C for indicated periods. Sampled cells were fixed and stained with DAPI, rhodamine-phalloidin, and Calcofluor white. Nuclear DNA stained with DAPI and septa stained with Calcofluor white are shown together in panels a, c, e, g, i, and k, whereas F-actin stained with rhodamine-phalloidin is shown in panels b, d, f, h, j, and l. Bar, 10 μm.
A wee1Δ pob1–664 double mutant JX646 and a control strain JZ1005 (wee1Δ pob1+) were cultured in the presence of HU for 3 h at the permissive temperature. Under these conditions, both JZ1005 and JX646 accumulated unseptated cells with a single nucleus, the size of which exceeded the minimum that would be required to enter mitosis. These cells were shifted to the restrictive temperature, and HU was washed out after 2 h. The cells were incubated further at the restrictive temperature. One hour after the washout of HU, both JZ1005 and JX646 cells began to form contractile rings (Figure 7, d and j). In the case of JX646, many septated cells accumulated 4.5 h after the washout, some of which showed even three septa (Figure 7k). Thus, these cells underwent contraction of actin rings apparently normally but were blocked at cell separation. Cells with three septa could be explained as products of two rounds of mitosis and septation, with no cell separation in between. Measurements of cell length suggested that these cells were long enough to sustain two rounds of mitosis. Binucleate compartments were rarely observed, indicating that mitosis and cytokinesis proceeded normally in these cells. JZ1005, examined as a control, underwent cell separation after septation and eventually assumed the typical wee cell shape. In similar experiments using wee1+ strains, septated but unseparated cells were generated by the wee1+ pob1–664 mutant but not by the wee1+ pob1+ strain, although the frequency of appearance of septated cells was much lower than that for the wee1 pob1–664 mutant (our unpublished data). From these results we conclude that pob1-defective cells are potentially capable of mitosis and cytokinesis but are impaired in cell separation. It remains to be seen whether the septum formed in the absence of functional Pob1p is completely normal or not.
DISCUSSION
S. cerevisiae Boi1p and Boi2p were originally identified as Bem1p-binding proteins, and they have been suggested to interact with Cdc42p directly or indirectly (Bender et al., 1996). Curiously, however, the interaction of Boi proteins with Bem1p, which occurs through their N-terminal region, has no apparent significance in the wild-type genetic background, and their C-terminal region alone can fulfill the function specific to Boi proteins (Bender et al., 1996; Matsui et al., 1996). Overproduction of Boi1p or Boi2p in S. cerevisiae cells causes growth arrest and rounding of cell shape (Bender et al., 1996; Matsui et al., 1996), which can be suppressed by overproduction of Cdc42p (Bender et al., 1996). Double disruption of BOI1 and BOI2 results in impaired morphogenesis and poor viability, causing cells to become large and round or lysed with buds (Bender et al., 1996; Matsui et al., 1996). This growth defect can be suppressed by overproduction of Rho3p, or its relative Rho4p (Bender et al., 1996; Matsui et al., 1996). These observations indicate that S. cerevisiae Boi proteins participate in the maintenance of cell growth and polarity in cooperation with Rho family proteins including Cdc42p and Rho3p/Rho4p.
Despite these findings, the molecular function of Boi proteins has not been unveiled. Based on physical and genetic interactions among gene products and cellular physiology caused by loss or gain of each gene function, Bender et al. (1996) speculated the following. Provided that Cdc42p functions mainly in bud emergence and Rho3p functions mainly in bud growth, Boi1p/Boi2p may help either to position an activator of Rho3p at a site where Cdc42p had acted previously, or to displace Cdc42p or a regulator of Cdc42p from a site where Rho3p is to act subsequently. Evidence confirming this idea such as proper localization of Boi1p/Boi2p at cell cortex, however, has not been reported to date.
This study has shown that Pob1p is likely to be the singular counterpart of S. cerevisiae Boi1p/Boi2p in S. pombe. In addition to the ability of Boi2p to replace Pob1p functionally, these proteins are similar to each other in that their N-terminal region, including the SH3 and SAM domains, is not essential for their basic function, and that overproduction of them causes growth arrest and rounding of the cell. Furthermore, as is the case with S. cerevisiae, overexpression of a member of the rho gene family can suppress the pob1–664 ts mutation (Toya, Nakano, Mabuchi, and Yamamoto, unpublished results). Thus, although Boi proteins are mainly required for bud growth whereas Pob1p is required for cell elongation, this apparent difference should be taken to reflect only the morphological difference between the two yeast species, and it is plausible that the proteins execute essentially the same molecular function.
The above considerations suggest that comparative studies of Pob1p and Boi1p/Boi2p may be invaluable for the understanding of the molecular function of this protein family. In this regard, our study has provided the following new information. 1) Subcellular localization of Pob1p during the cell cycle has been visualized and shown to be closely related to that of actin patches. 2) Use of a temperature-sensitive mutant has enabled us to conclude that loss of Pob1p function leads to an instant cessation of cell elongation. 3) Pob1p is essential for not only cell elongation but also cell separation, suggesting that it is involved in completion of the septum. These points are discussed in more detail below.
Observed localization of Pob1p at growing cell tips in interphase and along the division plane during cytokinesis is consistent with its involvement in both cell elongation and cell separation. However, most of the pob1–664 cells shifted to the restrictive temperature arrested in G2 phase rather than at cell separation. This will be explained by the fact that only a small portion of cells stays in M phase at a given time in an asynchronous culture. A noteworthy phenotype of the pob1 mutant is that actin patches persist at the cell ends even after cellular elongation has been severely suppressed and that the cells eventually assume a unique lemon-like morphology.
Wild-type S. pombe cells halted growth and became round and swollen when pob1 was overexpressed in them. Such cells were often binucleate and occasionally carried misformed septa, suggesting that they were defective in positioning a division plane and executing cytokinesis properly. This would account for why the host strain JX268 carrying a pob1 plasmid diploidized without conjugation and produced spores in our original screen.
It has been shown that the binding of Boi1p/Boi2p to Bem1p is mediated by the proline-rich region of Boi1p/Boi2p and the second SH3 domain of Bem1p (Bender et al., 1996; Matsui et al., 1996). We have been unsuccessful thus far in proving physical interaction between Pob1p and Scd2/Ral3p by two-hybrid analysis. This may be because Pob1p does not carry a prominent proline-rich region like Boi1p and Boi2p, and hence may interact with Scd2/Ral3p only weakly through PXXP motifs, the potential binding sites for SH3 domains (Alexandropoulos et al., 1995). Alternatively, it may be that Pob1p and Scd2/Ral3p have no natural physical interaction but can manage to fulfill the same function as S. cerevisiae Boi1p/Boi2p and Bem1p. Indeed, the direct contact of Boi1p/Boi2p to Bem1p does not seem to be essential for their function, because their N-terminal half carrying the proline-rich region is dispensable for the maintenance of cell viability (Bender et al., 1996; Matsui et al., 1996). As demonstrated in this study, the N-terminal half of Pob1p is similarly dispensable for cell viability.
Cells either carrying an active form of Cdc42p (G12V or Q61L) or overexpressing its activator Scd1/Ral1p show a similar phenotype to pob1-overexpressing cells (Miller and Johnson, 1994; Kitayama and Yamamoto, unpublished). Cdc42p and Scd1/Ral1p both function downstream of Ras1p in a cascade and regulate cell morphology and the ability to mate (Chang et al., 1994). Analysis of cells defective in either cdc42 or pak1/shk1, the latter of which encodes a protein kinase thought to function downstream of Cdc42p, has shown that these gene products are essential for polarized cell growth (Marcus et al., 1995; Ottilie et al., 1995). Thus, it will be possible that Pob1p plays a role in close association with the Scd1/Ral1p-Cdc42p-Pak1/Shk1p pathway in fission yeast cells. Consistently, Scd2/Ral3p, which is an S. pombe homologue of Bem1p, is also relevant to this pathway (Chang et al., 1994). So far, however, our preliminary analysis has revealed no two-hybrid interaction between S. pombe Cdc42p and Pob1p (our unpublished results), while S. cerevisiae Cdc42p displayed a two-hybrid interaction with Boi1p (Bender et al., 1996). This may mean either that the interaction between Cdc42p and Boi1p is mediated by a third protein(s) in budding yeast cells, or that fission yeast Pob1p, for some reason, has lost a direct interaction with Cdc42p, as well as with Scd2/Ral3p. More work is obviously required to clarify the link between the Boi family proteins and the Rho family small GTPases.
Entry to mitosis was blocked in pob1–664 cells at the restrictive temperature and they stayed in G2 phase. S. pombe cells should reach a critical size in order to acquire the capacity to enter mitosis (Nurse, 1975). Our experiments using the wee1Δ pob1–664 strain revealed that Pob1p is not required for mitosis per se, supporting the idea that pob1–664 cells fail to undergo mitosis because they cannot attain the necessary cell size (or a certain size/shape-related parameter) to do so.
Pob1p is dispensable for cytokinesis but not for cell separation. Although septation takes place in the absence of Pob1p function, it is unknown whether the septa formed under these conditions are intact or not. It is generally accepted that septation in fission yeast proceeds in two steps (Robinow and Hyams, 1989). First, the primary septum is generated centripetally at the cleavage furrow after the contraction of the actin ring. Then the secondary septa are constructed on both sides of the primary septum. After their completion, the primary septum undergoes degradation so that the daughter cells can separate. According to this model, it appears that the formation of the primary septum is not affected in the pob1 mutant. This strain is likely to be defective in cell separation either because the secondary septa are not properly formed, or because the primary septum is not degraded.
Through all stages of the cell cycle, Pob1p localized in the same area as F-actin patches. However, whereas F-actin was concentrated in patches at the cell tips or along the division plane, Pob1p appeared to form a structure more like a layer. In mitotic cells, Pob1p first gathered to form a broad ring at the cell center, which was then split into two, each part developing into a disk as the contractile ring constricted. These observations suggest that Pob1p is likely to be located on growing plasma membrane, possibly via the function of actin patches, and may recruit proteins required for de novo synthesis of cell wall or the secondary septum to the growing site. Consistent with this idea, Pob1p contains a PH domain in the C terminus, the proposed function of which is to target proteins to membranes. Our preliminary analysis has shown that localization of Pob1p is randomized in cells defective in the profilin gene cdc3, in which actin is disorganized (Balasubramanian et al., 1994). In contrast, the addition of latrunculin A, which depolymerizes F-actin, has had little immediate influence on the intracellular distribution of Pob1p (Toya and Yamamoto, unpublished results). Thus, how Pob1p interacts and cooperates with actin remains an interesting subject for future study.
During revision of this article, Katayama et al. (1999) introduced an S. pombe protein that exhibits subcellular localization highly similar to Pob1p. This protein, called Mok1p, forms a layer at growing cell tips, translocates to the division area, and eventually produces two split layers. Mok1p (identical with Ags1p) functions as an α-glucan synthase (Hochstenbach et al., 1998; Katayama et al. 1999). Although neither loss of function nor overproduction of Mok1p appears to lead to the same morphological alteration as was seen with Pob1p, these observations strongly suggest that Pob1p and Mok1p coexist in a bulky structure responsible for cell wall biosynthesis.
Defects in cell separation have been observed in S. pombe cells overexpressing either rho1 (Arellano et al., 1996; Nakano et al., 1997) or pkc1/pck2 (Mazzei et al., 1993; Toda et al., 1993), or in mutant cells deficient in pmk1/spm1 (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997). Rho1p is an activator of (1–3)β-d-glucan synthase involved in cellular morphogenesis (Arellano et al., 1996). pkc1/pck2 encodes a protein kinase C homologue, and pmk1/spm1 encodes a MAP kinase, which regulates cell integrity and functions coordinately with the Pkc1/Pck2p pathway (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997). It remains to be seen whether Pob1p is functionally related to either of these gene products, which are apparently involved in cell wall synthesis and/or degradation.
ACKNOWLEDGMENTS
We thank Dr. Y. Matsui for the gift of an S. cerevisiae BOI2 clone. This work was supported by a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (07283102).
REFERENCES
- Alexandropoulos K, Cheng G, Baltimore D. Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Arellano M, Duran A, Pérez P. Rho1 GTPase activates the (1–3)beta-d-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15:4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M, Hirani B, Burke J, Gould K. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M, McCollum D, Chang L, Wang K, Naqvi N, He X, Sazer S, Gould K. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M, McCollum D, Gould K. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bender L, Lo H, Lee H, Kokojan V, Peterson V, Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in Yeast. J Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E, Barr M, Wang Y, Jung V, Xu H, Wigler M. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- Fantes P. Isolation of cell size mutants of a fission yeast by a new selective method: characterization of mutants and implications for division control mechanisms. J Bacteriol. 1981;146:746–754. doi: 10.1128/jb.146.2.746-754.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S, Park H, Wang T. Fission yeast with DNA polymerase delta temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 1986;44:329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Yamamoto M. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to. ras1−. Mol Gen Genet. 1988;215:26–31. doi: 10.1007/BF00331298. [DOI] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R, editor. Handbook of Genetics. Vol. 1. New York, NY: Plenum Publishing; 1974. pp. 395–446. [Google Scholar]
- Hakuno F, Hughes D, Yamamoto M. The Schizosaccharomyces pombe mra1 gene, which is required for cell growth and mating, can suppress the mating inefficiency caused by a deficit in the Ras1 activity. Genes Cells. 1996;1:303–315. doi: 10.1046/j.1365-2443.1996.27029.x. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Hochstenbach F, Klis FM, van den Ende H, van Donselaar E, Peters PJ, Klausner RD. Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc Natl Acad Sci USA. 1998;95:9161–9166. doi: 10.1073/pnas.95.16.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes & Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- Johnston G, Prendergast J, Singer R. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Hirata D, Arellano M, Pérez P, Toda T. Fission yeast α-glucan synthase Mok1 requires the actin cytoskeleton to localize the sites of growth and plays an essential role in cell morphogenesis downstream of protein kinase C function. J Cell Biol. 1999;144:1173–1186. doi: 10.1083/jcb.144.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D, Pearson W. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Marcus S, Polverino A, Chang E, Robbins D, Cobb M, Wigler M. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J, Hyams J. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- Mata J, Nurse P. tea1 And the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Matsui R, Akada R, Toh-e A. Yeast src homology region 3 domain-binding proteins involved in bud formation. J Cell Biol. 1996;133:865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Toh-e A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and. BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May K, Watts F, Jones F, Hyams J. A type II myosin involved in cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil Cytoskeleton. 1997;38:1–12. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Mazzei G, Schmid E, Knowles J, Payton M, Maundrell K. A Ca2+-independent protein kinase C from fission yeast. J Biol Chem. 1993;268:7401–7406. [PubMed] [Google Scholar]
- McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P, Johnson D. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1990;194:795–826. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Motegi F, Nakano K, Kitayama C, Yamamoto M, Mabuchi I. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 1997;420:161–166. doi: 10.1016/s0014-5793(97)01510-x. [DOI] [PubMed] [Google Scholar]
- Nakano K, Arai R, Mabuchi I. The small GTP-binding protein Rho1 is a multifunctional protein that regulates actin localization, cell polarity, and septum formation in the fission yeast Schizosaccharomyces pombe. Genes Cells. 1997;2:679–694. doi: 10.1046/j.1365-2443.1997.1540352.x. [DOI] [PubMed] [Google Scholar]
- Nielsen O, Davey J, Egel R. The ras1 function of Schizosaccharomyces pombe mediates pheromone-induced transcription. EMBO J. 1992;11:1391–1395. doi: 10.1002/j.1460-2075.1992.tb05184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–51. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottilie S, Miller P, Johnson D, Creasy C, Sells M, Bagrodia S, Forsburg S, Chernoff J. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice H. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow CF, Hyams JS. General cytology of fission yeast. In: Nasim A, Young P, Johnson B, editors. Molecular Biology of the Fission Yeast. San Diego, CA: Academic Press; 1989. pp. 273–331. [Google Scholar]
- Schults J, Ponting C, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Methods in Yeast Genetics: Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. pp. 163–167. [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Toda T, Dhut S, Superti-Furga G, Gotoh Y, Nishida E, Sugiura R, Kuno T. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol Cell Biol. 1996;16:6752–6764. doi: 10.1128/mcb.16.12.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Two novel protein kinase C-related genes of fission yeast are essential for cell viability and implicated in cell shape control. EMBO J. 1993;12:1987–1995. doi: 10.1002/j.1460-2075.1993.tb05848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes & Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu H, Riggs M, Rodgers L, Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, White M, Marcus S, Wigler M. Concerted action of RAS and G proteins in the sexual response pathways of Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:50–58. doi: 10.1128/mcb.14.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitsevskaya-Carter T, Cooper J. Spm1, a stress-activated MAP kinase that regulates morphogenesis in S. pombe. EMBO J. 1997;16:1318–1331. doi: 10.1093/emboj/16.6.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Ebright R. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]