Abstract
We showed previously that substitution of the first residue of the influenza hemagglutinin (HA) fusion peptide Gly1 with Glu abolishes fusion activity. In the present study we asked whether this striking phenotype was due to the charge or side-chain volume of the substituted Glu. To do this we generated and characterized six mutants with substitutions at position 1: Gly1 to Ala, Ser, Val, Glu, Gln, or Lys. We found the following. All mutants were expressed at the cell surface, could be cleaved from the precursor (HA0) to the fusion permissive form (HA1-S-S-HA2), bound antibodies against the major antigenic site, bound red blood cells, and changed conformation at low pH. Only Gly, Ala, and Ser supported lipid mixing during fusion with red blood cells. Only Gly and Ala supported content mixing. Ser HA, therefore, displayed a hemifusion phenotype. The hemifusion phenotype of Ser HA was confirmed by electrophysiological studies. Our findings indicate that the first residue of the HA fusion peptide must be small (e.g., Gly, Ala, or Ser) to promote lipid mixing and must be small and apolar (e.g., Gly or Ala) to support both lipid and content mixing. The finding that Val HA displays no fusion activity underscores the idea that hydrophobicity is not the sole factor dictating fusion peptide function. The surprising finding that Ser HA displays hemifusion suggests that the HA ectodomain functions not only in the first stage of fusion, lipid mixing, but also, either directly or indirectly, in the second stage of fusion, content mixing.
INTRODUCTION
Significant progress has been made in elucidating how the influenza virus hemagglutinin (HA) promotes membrane fusion (Hernandez et al., 1996; Hughson, 1997). For example, it is now well accepted that in response to low pH, which influenza experiences in endosomes, the fusion peptide of HA is exposed and repositioned so that it can interact, hydrophobically, with membranes to initiate fusion. Nevertheless, the precise sequence requirements, mode of interaction with the bilayer, and functions of the HA fusion peptide remain unclear (Durell et al., 1997). In addition, it is still debated whether the fusion peptide interacts with the target or the viral membrane, or both, during fusion (Kozlov and Chernomordik, 1998).
We showed previously that substitution of a glutamic acid (Glu) for the glycine (Gly) at position 1 of the fusion peptide of HA from the Japan strain (A/57) of influenza virus abolishes fusion activity, as monitored by both a content mixing assay (delivery of horseradish peroxidase) and a syncytia assay (Gething et al., 1986). Other mutations at this position have also been shown to block syncytia formation while allowing the low pH-induced conformational change (Steinhauer et al., 1995). Subsequent work with the Gly 1 to Glu mutation (in Japan HA) showed that it is impaired at an early stage of fusion, outer leaflet lipid mixing (Guy et al., 1992; Schoch and Blumenthal, 1993). To explore the molecular basis for this striking phenotype in more detail, we introduced six site-specific mutations into position 1 of the fusion peptide of HA from the X:31 strain of influenza virus. The mutations were designed to test the charge, hydrophobicity, and side-chain volume requirements of the first residue of the HA fusion peptide. The mutants were Gly 1 to Ala, Ser, Val, Glu, Gln, and Lys (Table 1). Mutants were expressed as full-length HA proteins in tissue culture cells and assessed for cell surface expression, structural features, their ability to change conformation at low pH, and then for their ability to induce membrane fusion, both lipid mixing and content mixing. Two mutants were also assessed for fusion pore formation using whole-cell patch-clamp methodology.
Table 1.
Effects of mutations at position 1 of the HA fusion peptide
| Amino acid | Side-chain volume | Hydrophobicity index | Charge | Surface expression | Trypsin cleavage | Conform. change (∼pH) | Fusion
|
||
|---|---|---|---|---|---|---|---|---|---|
| Lipid mixing | Fusion pore | Content mixing | |||||||
| Gly | 66 | 0.48 | 0 | + | + | ∼5.4 | + | + | + |
| Ala | 92 | 0.62 | 0 | + | + | ∼5.4 | + | + | ± |
| Ser | 99 | −0.18 | 0 | + | + | ∼5.4 | + | − | − |
| Val | 142 | 1.10 | 0 | + | + | ∼5.4 | − | nd | − |
| Glu | 155 | −0.74 | −1 | + | + | ∼5.4 | − | nd | − |
| Gln | 161 | −0.85 | 0 | + | + | ∼5.4 | − | nd | − |
| Lys | 171 | −1.50 | +1 | + | + | ∼5.4 | − | nd | − |
Summary of results presented in this study. Values for side-chain volumes (in Å3) and hydrophobicity indices were from Creighton (1984) and Eisenberg (1984) (normalized consensus scale), respectively.
MATERIALS AND METHODS
Mutagenesis and Vector Construction
Mutant HAs were generated using the method of Kunkle on single-stranded preparations of cDNA encoding WT HA (X:31 strain) in the plasmid pSM essentially as described previously (Qiao et al., 1998). The oligonucleotides for mutagenesis were designed to encode both the altered amino acid and an additional restriction enzyme site to facilitate mutant identification. A 1729-bp segment of HA was cut from the pSM vector with SfiI and Xba. The SfiI site was blunted. The resultant fragment was inserted into the vector pCB6 cut with HindIII and Xba. The HindIII site of pCB6 was blunted. Approximately 200 bp of all mutant cDNAs were sequenced to confirm that the desired mutations had been introduced but extraneous mutations had not. The mutants Gly1Ser and Gly1Val were sequenced in full.
Production of Stable Cell Lines Expressing WT HA and HA Mutants
Dishes (10 cm) of NIH 3T3 cells were transfected with 30–40 μg pCB6 HA cDNA (WT or mutant) using the CaPO4 technique (Hernandez and White, 1998). Cells were grown for 10 d in DMEM (Life Technologies, Gaithersburg, MD) plus 10% supplemented calf serum (Hyclone Laboratories, Logan, UT), 50,000 U penicillin, and 50,000 μg/0.5 l streptomycin (Life Technologies), an additional 146 mg/0.5 l glutamine (Life Technologies), and 600 μg/ml geneticin (Life Technologies). Individual colonies were isolated, expanded, replated in the presence of 10 mM NaButyrate (Sigma, St. Louis, MO), and screened for HA expression by Western blot analysis. Cell lines expressing large amounts of HA were chosen. Unless stated, all cell lines were treated with 25 mM NaButyrate for 14–16 h before experiments.
Transient Transfection of WT and Mutant HAs
Dishes (6 cm) of ∼40–50% confluent COS7 cells, grown in the media described above but without geneticin, were transfected with a total of 5 μg cDNA. Unless stated, the 5 μg DNA consisted of 1.25 μg (“1X”) WT or mutant HA cDNA in pCB6 and 3.75 μg carrier pCB6 (empty vector) DNA. Transfections were conducted using 12 μl of lipofectin (Life Technologies) according to the manufacturer’s instruction. The cells were incubated in the DNA/lipofectin mixture for 8 h at 37°C, at which time the mixture was replaced with DMEM containing 10% FCS. For the experiment described in Figure 6, transfections were performed with 15 μl Mirus Transit (Panvera, Madison, WI) according to the manufacturer’s instructions. The cells were incubated in the DNA/Mirus Transit mixture for 5 h at 37°C, at which time the mixture was replaced with DMEM containing 10% FCS. All transfected cells were analyzed 40–46 h post-transfection. NaButyrate (10 mM) was added during the last 16–18 h.
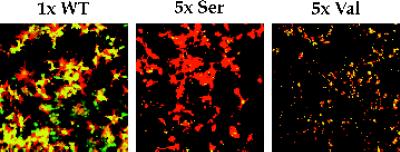
Figure 6.
Fusion activity of overexpressed Ser HA and Val HA. COS7 cells were transfected with 1× WT HA, 5× Ser HA, or 5× Val HA cDNA using the Mirus Transit reagent and processed for fusion with RBCs labeled with both R18 and calcein.
Cell Surface Biotinylation, Immunoprecipitations, and Western Blot Analyses
Dishes (10 cm) of confluent HA-expressing NIH 3T3 cells were treated with 25 mM NaButyrate as described above. The cells were then washed twice with cold PBS* (PBS with 0.5 mM MgCl2, pH 7.8) and labeled in 3 ml of PBS* containing 1 mg/ml immunopure NHS-LC-biotin (Pierce Biochemicals, Rockford, IL) for 45 min at 4°C. Excess reagent was quenched with two washes of ice-cold PBS* containing 50 mM glycine; the second wash involved an incubation for 10 min at 4°C. The cells were then washed once with PBS and treated with either 8 μg/ml l-1-tosylamide-2-phenylethyl chloromethyl ketone-trypsin (to cleave HA0) or 8 μg/ml Na-p-tosyl-l-lysine chloromethyl ketone-chymotrypsin (HA0 control) for 6 min at RT. At this time the cells were incubated for 10 min with PBS containing 50 μg/ml soybean trypsin inhibitor and lysed with a lysis buffer (50 mM HEPES, 1% NP-40, pH 7.5) containing a protease inhibitor cocktail (Qiao et al., 1998). The lysate was centrifuged at 14,000 × g in an Eppendorf centrifuge for 20 min. Approximately 800 μg of each lysate (protein concentration determined using the BCA* Protein Assay Reagent; Pierce Biochemicals, Rockford, IL) was incubated with 0.1 μg/ml Site A mAb (gift of J. Skehel, Medical Research Council, Mill Hill, England) for 60 min at RT. The mAb–HA complexes were precipitated with protein A agarose (Schleicher & Schuell, Keene, NH) for 60 min at RT and then washed four times with lysis buffer. Immune complexes were suspended in SDS gel loading buffer, heated to 95°C for 5 min, and separated by 10% SDS-PAGE. The proteins were transferred to nitrocellulose. The blot was then blocked with PBS containing 1% skim milk, 10% glycerol, 3% BSA, 1 M glucose, and 0.5% Tween 20 for 1 h at RT. After three washes with PBS-0.5% Tween 20, the nitrocellulose membrane was probed with 0.5 ng/ml streptavidin HRP (Pierce Biochemical) in PBS-0.1% Tween 20 for 60 min at RT. After three washes in PBS-0.5% Tween 20, the blot was developed with the Enhanced Chemiluminescence Reagent (Amersham, Arlington Heights, IL) according to the manufacturer’s instruction.
Metabolic Labeling
Proteins were metabolically labeled by incubating cells in media lacking cysteine and methionine (cys−/met− MEM Select Amine; Life Technologies) for 45 min at 37°C. The medium was removed and replaced with cys−/met− media containing the indicated amount of [35S] Translabel (ICN, Irvine, CA), 2% supplemented calf serum and, unless stated otherwise, 25 mM NaButyrate. The cells were then incubated for 14–18 h at 37°C before harvesting.
Proteinase K Digestion
Cell lines expressing WT or mutant HAs were metabolically labeled with [35S]-Translabel for 16 h in the presence of, unless stated otherwise, 25 mM NaButyrate. After two washes with PBS the cells were treated with 8 μg/ml l-1-tosylamide-2-phenylethyl chloromethyl ketone trypsin in PBS for 6 min at RT and then removed from their dishes with PBS containing 0.5 mM EDTA, 0.5 mM EGTA, and 50 μg/ml soybean trypsin inhibitor. Aliquots of the cells were then washed with MES-saline (30 mM MES, 100 mM NaCl) pH 7.0, and then incubated in 0.5 ml of MES-saline adjusted to the indicated pH for 15 min at 37°C. At this time the medium was reneutralized by the addition of a predetermined amount of 1.0 M Tris base. After one wash with MES-saline, pH 7.0, the cells were lysed in lysis buffer (100 mM Tris, pH 7.4, 1% NP-40) and centrifuged at 14,000 × g for 30 min at 4°C. The lysates were then digested by adding proteinase K and CaCl2 to final concentrations of 0.2 mg/ml and 2 mM, respectively, for 30 min at 37°C. The digestion was stopped by the addition of 1 μg BSA, 1 mM PMSF, and a protease inhibitor cocktail (Qiao et al., 1998). Remaining HA in the sample was immunoprecipitated with the Site A mAb and separated by 10% SDS-PAGE. The gel was dried and subjected to phosphorimager analysis.
Red Blood Cell Labeling
Red blood cells (RBCs) were labeled with either octadecylrhodamine B chloride (R18), calcein AM (10 mM; Molecular Probes, Eugene, OR), or carboxyfluorescein (CF; 2.5 mM) as described previously (Melikyan et al., 1995, 1997; Qiao et al., 1998). For the double-label experiments (see Figures 5 and 6), RBCs were first labeled with R18 and then with Calcein AM. For the experiments described in Figures 7 and 8, a lower concentration of R18 was used (2.5 μg/5 ml of 1% RBC suspension). Labeled RBCs were stored at 4°C for no longer than 2 d.
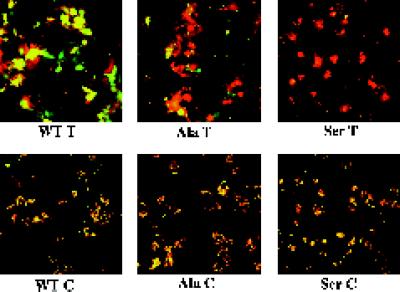
Figure 5.
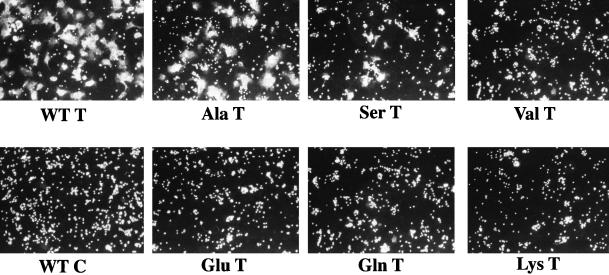
Comparison of lipid and content mixing fusion activity of WT HA and the Ala and Ser mutants. COS7 cells were transfected with plasmids (pSM) encoding WT, Ala, and Ser mutant HAs, and treated with neuraminidase and trypsin (T) or chymotrypsin (C) as described in the legend to Figure 3. The cells were incubated with RBCs labeled with both R18 (red) and Calcein AM (green), incubated in pH 5.0 fusion buffer for 2 min at 37°C, reneutralized, and observed with a confocal microscope.
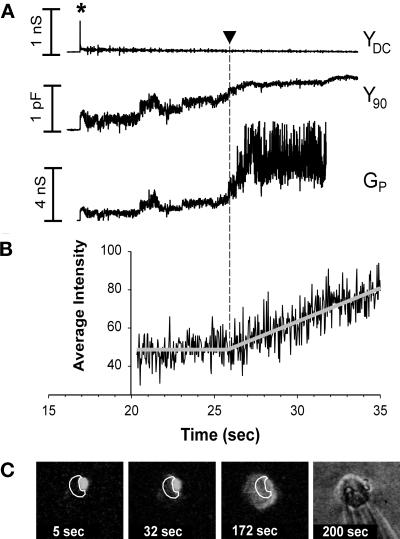
Figure 7.
Simultaneous electrical recording of fusion pores and video fluorescence measurements of R18 transfer for RBC fusion to WT HA-expressing COS7 cells. (A) The pH of the solution was lowered at 0 s. The opening of a fusion pore between a WT HA-expressing cell and an RBC was marked by a spike in YDC (at ∼17 s) and a simultaneous increment in Y90. YDC returned nearly to baseline after the spike, showing that both the COS7 and RBC membrane conductances remained small. The fusion pore conductance (Gp, bottom trace) was calculated from the Y90 trace (see MATERIALS AND METHODS). With enlargement of the fusion pore, the voltage applied across the COS7 cell membrane increasingly penetrated into the RBC, and Y90 was observed to increase. The voltage fully penetrated into the RBC once the pore enlarged sufficiently, and hence the precise value of Gp (Figure 7A) became less certain, calculated as a “noisy” Gp, once the Y90 neared saturation. (B) The time-dependent fluorescence intensities shown in C were fit with two straight lines, a constant fluorescence followed by a linearly increasing fluorescence. Their intersection determined the lag time from acidification until the onset of R18 spread (here 25.8 s, shown by arrowhead in A and B). (C) To temporally correlate the onset of membrane dye redistribution with conductance increases, for each experiment an ROI was selected for a COS7 cell (white contours), and the average intensity of fluorescence of this region, adjacent to a bound RBC, was determined on a frame-by-frame basis. The fluorescence images shown were obtained by averaging eight sequential video frames. The four images represent (from left to right) fluorescence patterns before dye spread, soon after dye spread began, after complete redistribution of R18, and the bright-field image at the conclusion of the experiment. Times after reducing the pH are indicated on all images and graphs.
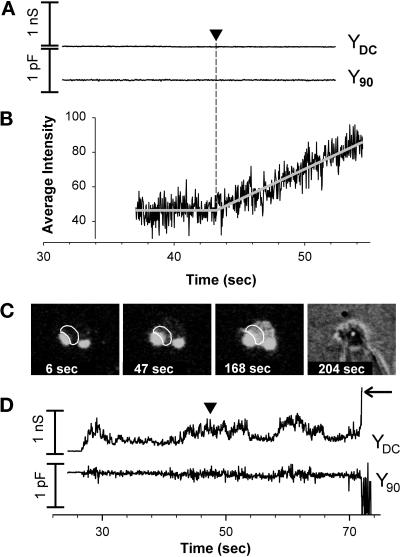
Figure 8.
Electrical and video fluorescence microscopy recordings for Ser HA-expressing cells induced to hemifuse to RBCs. (A) An experiment in which electrical changes were not detected for more than 5 min. (B) The onset of R18 spread was observed at 42.9 s. (C) Fluorescence and bright-field images for the experiment shown electrically in A. For details of quantifying dye spread, see legend to Figure 7. (D) Electrical profile of mild leakage for Ser HA-expressing cells hemifusing to RBCs. After pH was lowered, YDC increased and fluctuated and the baseline of Y90 became appreciably more noisy, but the average value of Y90 did not change. A change in YDC without a concurrent change in Y90 is the hallmark of membrane leaks, rather than fusion pore formation. R18 spread was observed (vertical arrowhead at 47.5 s) without pore formation. With time, the DC conductance became large (horizontal arrow), exceeding 50 nS. This increase in YDC resulted in an excessively noisy Y90 trace, eliminating the ability to detect whether fusion pores formed at later times. RBCs tended to swell (increased size as time progressed can be seen in fluorescent images) and could even lyse (loss of contrast in bright-field images). RBC swelling is characteristic of leaks. We did not observe any electrical activity or dye spread when either WT HA or Ser HA were not activated by cleavage with trypsin (our unpublished results).
Lipid and Content Mixing Assays
For fusion assays, we used transiently transfected COS7 cells because the stable NIH 3T3 cells expressing WT HA did not express sufficient HA to be highly fusogenic. For microscopic analysis of fusion, transiently transfected HA0-expressing cells were treated with neuraminidase (0.2 mg/ml) and 5 μg/ml either l-1-tosylamide-2-phenylethyl chloromethyl ketone-trypsin or Na-p-tosyl-l-lysine chloromethyl ketone-chymotrypsin (HA0 controls) for 6 min at RT. The medium was removed and replaced with medium containing either 50 μg/ml soybean trypsin inhibitor or 10% supplemented calf serum. Cells were washed once with PBS++ and incubated with labeled RBCs at RT (for Figures 3–6, 0.05% RBCs for 20 min; for Figures 7 and 8, 0.01% RBCs for 10 min). Unbound RBCs were removed by several washes with PBS++. After exposure to pH 5.0 fusion buffer (120 mM NaCl, 10 mM HEPES, 10 mM MES, 10 mM succinate, 0.2% glucose) for 2 min at 37°C, the medium was replaced with neutral pH medium, and the cells were observed with a fluorescence microscope. The experiments shown in Figures 7–9 were performed essentially as described above with minor modifications: at 44 h after transfection, COS7 cells were harvested from their dishes by a brief (1 min) incubation with a standard trypsin/EDTA solution. Cells were then transferred to complete growth medium, split into four 35-mm dishes, and recultured in a 5% CO2 incubator for 2–3 h. Cells were prepared for and induced to fuse as described above. After the low pH treatment, cells were placed in isotonic PBS++ supplemented with 20 mM raffinose to prevent colloidal osmotic swelling of the RBCs (Melikyan et al., 1997). The fluorescence pattern was analyzed microscopically (Laborlux D, E. Leitz, Edison, NJ). On average, ∼100 COS7 cells decorated with one to six RBCs were screened over several parts of a 35-mm dish, and the ratio of cells stained with CF to those stained with R18 was determined (Melikyan et al., 1997).
Figure 3.
Fusion activity of WT and mutant HAs: lipid mixing. COS7 cells transfected with plasmids encoding WT and mutant HAs were treated with neuraminidase (0.2 mg/ml) and either 5 μg/ml trypsin or chymotrypsin for 6 min at RT. R18-labeled RBCs (0.05%) were bound to the cells for 25 min at RT. After unbound RBCs were removed, the cells were incubated in fusion buffer, pH 5.0, for 2 min at 37°C, reneutralized, and observed with a fluorescence microscope.
Figure 9.
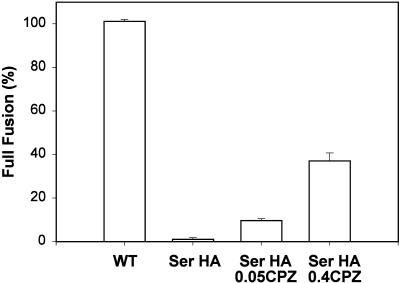
Chlorpromazine (CPZ) induces transfer of aqueous dye (CF) from RBCs to hemifused Ser HA-expressing cells. For each condition the ratio of COS7 cells stained with CF to those labeled by R18 was determined. In control experiments, after fusion was triggered by incubation for 2 min in a pH 5.0 solution at 37°C, and then incubated in PBS (supplemented with 20 mM raffinose) for 5 min at RT, virtually all WT HA-expressing cells (WT, first bar) were stained with both an aqueous dye (CF) and a membrane dye (R18). In contrast, CF spread into Ser HA-expressing cells was rarely observed, despite efficient R18 redistribution (second bar). These cells were exposed for 1 min at RT to either 0.05 mM (third bar) or 0.4 mM (fourth bar) CPZ, and then the CPZ-containing solution was replaced by PBS with raffinose. Error bars show the SE for 4–16 independent experiments.
For fluorometric analysis of lipid mixing, COS7 cells expressing either WT or mutant HAs were treated with neuraminidase and trypsin (or chymotrypsin) as described above, incubated with R18-labeled RBCs, and then washed to remove unbound RBCs. The RBC–cell complexes were then harvested in 2 ml PBS (Ca++, Mg++ free) containing 0.5 mM EDTA, 0.5 mM EGTA, and 5 mM glucose and added to 8 ml of cold pH 7 fusion buffer. The RBC–cell complexes were then centrifuged at 800 rpm for 5 min, resuspended in a small volume (200 μl) of fusion buffer, pH 7, and kept at 4°C. Fusion experiments were conducted using an LS-5B fluorimeter (Perkin Elmer-Cetus, San Jose, CA) as described previously (Danieli et al., 1996). The quantity of RBC–cell complexes that gave ∼1.0 OD (usually ∼50–100 μl) was added to 3 ml of fusion buffer in a cuvette agitated with a magnetic stirrer at RT. After a baseline was determined, the samples were brought to pH 5.2 with a predetermined amount of 1 M citric acid. The increase in fluorescence (fluorescence dequenching) was then monitored. The percentage of fluorescence dequenching was calculated relative to the total fluorescence for each sample after solubilization in 0.53% NP-40.
Patch-Clamp Analysis of Fusion Pore Formation
Transfected COS7 cells were harvested with trypsin/EDTA and cultivated for no longer than 1 h in complete growth medium on No. 1½ glass coverslips placed in a 35-mm culture dish. This procedure ensured that a significant fraction of the COS7 cells had a rounded shape and were not strongly attached to the glass. Cells were treated with neuraminidase and trypsin as described above and incubated with a dilute suspension of R18-labeled RBCs (0.002%) in PBS++ for 10 min. Using this procedure, only ∼1 of 20 COS7 cells was decorated with RBCs (usually one to three per cell). The coverslips were broken into small pieces and stored in PBS at 4°C for up to 7 h. For a measurement, a piece of glass was transferred into a patch-clamp chamber, maintained at 32–33°C by a temperature controller (20/20 Technology, Wilmington, NC), that was filled with solution containing large organic ions to minimize HA-induced conductance changes that often accompany lowering of pH (Spruce et al., 1991): 150 mM N-methylglucamine aspartate, 5 mM MgCl2, 2 mM Cs-HEPES buffer, pH 7.2. After the whole-cell configuration was established, the COS7-RBC complex was lifted from the bottom of the patch-clamp chamber (Chernomordik et al., 1997). This manipulation allowed longer recordings than possible when cells adhered to the glass substrate; suspending a cell eliminated loss of seals attributable to any drifting of the patch pipette or other movements accentuated when solutions were heated to 32°C. Fusion was triggered by ejecting with low pressure a pH 5.0 solution of the same composition but buffered with 20 mM Cs-succinate contained in another micropipette positioned ∼100 μm from the cell–RBC pair. This microperfusion of the cell was stopped after 2 min or soon after a fusion pore formed (if this came first). As the solution within the chamber mixed and diluted out the small quantity of ejected solution, the solution surrounding the cell was reneutralized. Patch pipettes were filled with 155 mM Cs glutamate, 5 mM MgCl2, 5 mM BAPTA, and 10 mM Cs-HEPES, pH 7.4.
Electrical patch-clamp experiments were performed in a conventional whole-cell, voltage-clamp mode. After the whole-cell mode was established, the cell membrane capacitance and pipette resistance were electronically compensated. For capacitance measurements, a command sine wave voltage (200 Hz, 50 mV peak to peak) superimposed on −40 mV holding potential was applied to the pipette via a PC-based computer system. The output from the patch-clamp amplifier, low pass-filtered at 5 kHz (Axopatch 2B, Axon Instruments, Foster City, CA), was digitized at 40 kHz using a 16-bit A/D board (PC-44, Innovative Integration, Westlake Village, CA). The components of admittance (current divided by voltage) that were in-phase, Y0, and out-of-phase, Y90, with the applied sine wave, as well as the DC admittance (conductance), YDC, were calculated for each period of sine wave and saved directly to a hard disk using software provided by Drs. V. Ratinov and J. Zimmerberg (National Institutes of Health) (Ratinov et al., 1998). The phase shift of the output current, introduced by the experimental system (the “phase angle”), with respect to the command voltage was adjusted by a capacitance “dithering” technique (Neher and Marty, 1982).
Fusion pore conductances were calculated off-line from changes in Y90 (Zimmerberg et al., 1994). Briefly, the final value of Y90 allows the capacitance of the RBC, CRBC, to be calculated according to Y90max = 2πfCRBC, where f is the frequency of the applied sine wave voltage. The pore conductance, Gp, was calculated as Gp = Y90max/(Y90max/Y90 − 1)1/2.
Image Processing
Simultaneously with the electrophysiological measurements, the spatial redistribution of R18 from RBCs into COS7 cells was obtained microscopically (Axiovert 100A, Zeiss). Fluorescence was monitored by a CCD-72 camera (Dage-MTI, Michigan City, IN) coupled to a multichannel plate intensifier (KS1381, Videoscope International, Washington, DC) and recorded by an S-VHS format recorder (SVO-9500MD, Sony Corporation, Park Ridge, NJ). Fluorescence and electrical measurements were synchronized by triggering a video date/time generator (Four-A Corporation, West Newton, MA) with a TTL-pulse generated by the computer system at the onset of acquiring electrophysiological data. The date/time generator superimposed a time stamp on the images, allowing each electrical time point to be precisely associated with a video frame (six to seven electrical time points per video frame for 200 Hz sinewave). Images were digitized off-line from video tape by a frame grabber (Meteor; Matrox Electronic Systems, Dorval, QC, Canada) and analyzed using locally written software using a commercial C library (Matrox Imaging Library, Matrox). When necessary, eight sequential video frames were averaged to improve signal-to-noise ratio.
To determine the onset of R18 spread into a COS7 cell, an arbitrarily shaped region of interest (ROI) was drawn to include the portion of an HA-expressing cell that was adjacent to an adhered R18-labeled RBC. For a period of ∼15 s around the time that dye began to spread, the average fluorescence intensity per ROI was obtained for every video frame. The moment R18 spread from an RBC into a COS7 cell was determined by plotting the average fluorescence intensity within an ROI as a function of time and piecewise linear-fitting the data with two straight lines. The curve fit (SigmaPlot, Jandel Scientific, San Rafael, CA) used three free parameters: the fluorescence of the ROI before fusion, the lag time between lowering pH and the onset of R18 spread, and the slope of fluorescence increase with time after R18 spread. The time between the onset of electrical signals and membrane dye spread was compared to determine their temporal relation.
RESULTS
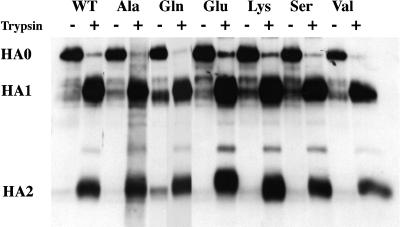
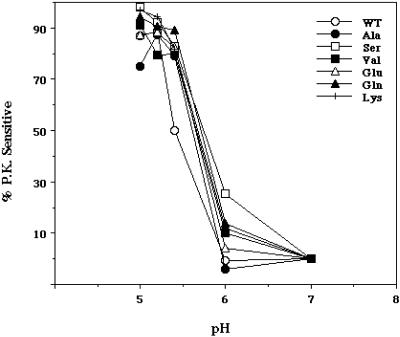
We first assessed whether the mutant HAs could be expressed at the cell surface in a fusion-permissive form (i.e., cleaved from HA0 to HA1 and HA2) and whether they could undergo a low pH-induced conformational change. As seen in Figure 1, all of the mutants were expressed at the cell surface as HA0, and they all were efficiently cleaved to HA1 and HA2 by the addition of trypsin. As seen in Figure 2, all of the mutants changed conformation at low pH; in all cases, the pH dependence of the conformational change was similar to that of WT HA.
Figure 1.
Cell surface expression and proteolytic processing of WT and mutant HAs. Stable NIH 3T3 cells expressing WT and mutant HAs were treated with NaButyrate to enhance HA expression. The cells were then labeled with the membrane impermeant reagent NHS-LC-biotin, treated with either trypsin (to cleave HA0) or chymotrypsin (HA0 control), and lysed. Equal amounts of protein from each cell lysate were precipitated with the Site A mAb, and the immune complexes were separated by reducing SDS-PAGE. The gel was transferred to nitrocellulose, probed with streptavidin-HRP, and developed by enhanced chemiluminescence.
Figure 2.
Proteinase K sensitivity of WT and mutant HAs. Cells expressing WT and mutant HAs were metabolically labeled with 35[S]-TransLabel, treated with 8 μg/ml trypsin for 6 min at RT, and then incubated at the indicated pH for 15 min at 37°C, reneutralized, and lysed in an NP-40 cell lysis buffer. Cell lysates were then digested with 0.2 mg/ml proteinase K for 30 min at 37°C. The proteins were immunoprecipitated with the Site A mAb, resolved by SDS-PAGE, and subjected to phosphorimager analysis.
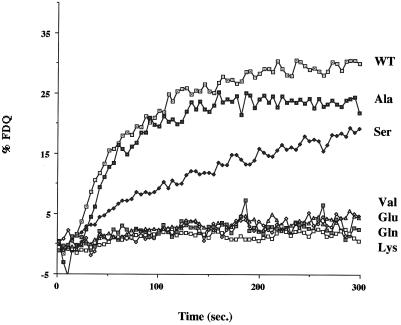
We next used a lipid mixing assay to evaluate the fusion activity of the mutant HAs. In the first experiment we used a microscopic analysis and evaluated fusion at 2 min after acidification. As seen in Figure 3, although the Ala and Ser mutants supported lipid mixing, no lipid mixing was seen with the Val, Glu, Gln, or Lys mutants. To explore further the lipid mixing activity of the mutant HAs, we conducted a fluorometric analysis. As seen in Figure 4, the Ala and Ser mutants supported considerable lipid mixing, whereas the Val, Glu, Gln, and Lys mutants did not display any such activity. The Ala mutant showed a similar time course and extent of fusion as WT HA. The fusion activity of the Ser mutant appeared slower. The reduced rate of lipid mixing of the Ser mutant and the total defects of the Val, Glu, Gln, and Lys mutants could not be attributed to poor cell surface expression. All of the mutants appeared to be equally well expressed at the cell surface as evidenced by both trypsin sensitivity (Figure 1) and RBC binding (Figure 3). Moreover, FACS analysis indicated that the Ser and Val mutants were well expressed (see below).
Figure 4.
Fusion activity of WT and mutant HAs by the R18 fluorescence dequenching assay. COS7 cells were transfected with plasmids encoding WT or mutant HAs, treated with neuraminidase and trypsin (or chymotrypsin), and then incubated with R18-labeled RBCs. The RBC–cell complexes were removed from the dish and placed in a cuvette containing fusion buffer, pH 7.0, in a fluorimeter. After attaining a baseline, the pH of the solution was lowered to 5.0 by injecting a predetermined amount of 1N citric acid, and the fluorescence was recorded. After lysis of the cells, the percentage of fluorescence dequenching was calculated, and the data were plotted with respect to time.
We next assessed the content mixing ability of the mutant HAs. In the first experiment we used doubly labeled RBCs to compare the lipid and content mixing activities of those mutant HAs that retained lipid mixing activity (Ala and Ser). As seen in Figure 5, although both mutants mediated considerable lipid mixing (delivery of R18), the Ala mutant appeared partially abolished and the Ser mutant appeared totally abolished in their ability to mediate the delivery of a small aqueous dye (calcein AM). The inability of the Ser mutant to mediate content mixing could not be attributed to inadequate cell surface expression. Repeated FACs analyses indicated that both the percentage of cells transfected (averages = 40 and 41%, respectively) and the amount of HA at the cell surface (average fluorescence intensities of 239 and 269, respectively) were virtually the same for WT HA and the Ser mutant. The striking pattern of the Ser mutant suggested that it induces hemifusion, but not full fusion, reminiscent of the behavior of glycosylphosphatidylinositol (GPI)-anchored HA (Kemble et al., 1994; Melikyan et al., 1995). As expected based on their defects in lipid mixing (Figures 3 and 4), the Val, Glu, Gln, and Lys mutants were totally abolished in their ability to mediate content mixing, although they supported good RBC binding (our unpublished results).
To further scrutinize the observations that Ser HA appears to promote hemifusion and that Val HA appears not to promote any fusion, we compared the lipid and content mixing activities of cells that express more Ser HA and Val HA than WT HA. We did this by transfecting cells with 5× Ser and 5× Val HA cDNA as compared with 1× WT HA cDNA. As seen in Figure 6, even when Ser HA and Val HA were overexpressed (average % cells transfected = 44 and 47%, respectively; average fluorescent intensities = 481 and 484, respectively) relative to WT HA (average % cells transfected = 40%; average fluorescence intensity = 239), we still observed their hemifusion and lack of fusion phenotypes, respectively.
Electrical Properties of Fusion by WT HA (X:31 strain)
When a WT HA-expressing cell was triggered to fuse to RBCs by local perfusion with a pH 5.0 solution, electrical detection showed that fusion pores formed within 30 s (Figure 7A); by a minute later the majority of these pores enlarged to conductances (Gp) too large (>10 nS) to be reliably determined by capacitance measurements (our unpublished results). The conductance patterns of fusion pores induced by WT HA (X:31 strain) were similar to those reported by others for the Japan/57 strain of HA (Spruce et al., 1989; Tse et al., 1993; Zimmerberg et al., 1994): pores opened in a stepwise manner to 0.2–1 nS where they lingered for varied times before further enlarging. In virtually every experiment for WT HA, a transient increase in DC conductance, YDC (Figure 7A, top panel, marked by an asterisk in YDC), a spike was associated with formation of a fusion pore. This transient originated from current through the fusion pore as the less negative resting potential of the RBC quickly came to the clamped holding potential, −40 mV, of the interior of the HA-expressing cell (Spruce et al., 1991). The increase in the “capacitance” trace, Y90, as well as the spike in YDC, serve as hallmarks of an electrical connection through a fusion pore between the interior of a COS7 cell and the interior of an RBC. Fusion pores were observed for every COS7 cell expressing WT HA that had bound RBCs. After the spike, YDC returned almost to the baseline and remained there well after enlargement of the pore (Figure 7A). This shows that current did not flow between the cell interiors and bath after fusion; the fusion process itself is tight, without leakage of aqueous contents.
Eventually, however, YDC did increase. For the experiment of Figure 7A, the increase in YDC was seen 140 s after pore opening (our unpublished results). The increase in YDC indicated the development of an “electrical leak” in the membrane at that point. (Such leaks are due to either loss of the high resistance “seal” between pipette and cell or increased permeability of ions through the cell membrane. HA0 had to be cleaved to observe the electrical leaks and thus the leaks had to have been HA-mediated. This is expected of membrane permeability increases but not of a loss of seal. Leaks do not signify general breakdown of the membrane, although if significant colloidal osmotic swelling occurs, lysis could follow.) Leaks observed subsequent to electrical detection of fusion pores are not unique to cells that transiently express X:31 HA; they are also observed for cells that constitutively express Japan HA (our unpublished results; J. Zimmerberg, personal communication).
While measuring the electrical signals associated with WT HA mediated fusion, we simultaneously recorded R18 movement from RBCs to COS7 cells. Figure 7C shows the fluorescence patterns corresponding to the electrical traces shown in Figure 7A. For WT HA-expressing cells, R18 always (n = 18) began to spread (Figure 7, A and B, vertical arrowhead) after fusion pore formation but never before. This result is as expected whether or not hemifusion is an intermediate stage of full fusion; electrical measures of pore formation are much more sensitive than measures of dye spread. As long as the fusion pore remained small, movement of the membrane probe was restricted; only when the pore enlarged did dye spread commence (Figure 7). This phenomenon has also been observed for cells expressing the Japan strain of HA (Tse et al., 1993; Zimmerberg et al., 1994).
Ser HA Promotes Hemifusion but Not Full Fusion
In distinct contrast to WT HA, COS7 cells expressing Ser HA did not show electrical signs characteristic of fusion pores (27 experiments); spikes in YDC or step-wise increases in Y90 were not observed (Figure 8A). (In 1 experiment that was not included in those experiments, a pore opening was observed fleetingly; it closed in <1 s, and no other pores subsequently appeared. Because lipid dye spread could have occurred through this isolated pore rather than solely as a result of hemifusion, this experiment was excluded from analysis.) As with WT HA, cells expressing Ser HA eventually developed leaks, which were shown (for a different cell pair) by increases in YDC (Figure 8D).
R18 spread was observed in 25 of the 27 Ser HA experiments. (There was only one bound RBC in the two experiments in which dye did not spread; it is possible that in these cases the RBC was not well adhered.) In 19 cases, lipid dye spread occurred after substantial electrical leakage. Here no conclusion could be drawn as to whether lipid mixing occurred through hemifusion or through pore formation. Of the six remaining experiments, four exhibited dye spread with no change in electrical signals (Figure 8, A–C), which are unequivocal demonstrations of hemifusion. In the other two cases, R18 spread, when leakage was minor enough so as not to obscure electrical signs of fusion pore formation (Figure 8D), and pores were not observed; here, hemifusion is also clearly indicated. In short, there was no instance in which lipid dye spread was shown to be a consequence of pore formation. On the other hand, it is possible that all cases of dye spread occurred through hemifusion, even when electrical leakage was substantial.
Permeability increases in membranes have long been associated with viral fusion (Carrasco et al., 1989), and it has often been questioned as to which comes first, leaks or fusion (Shangguan et al., 1996). The electrical records presented here establish unambiguously that in fusion of HA-expressing cells to RBCs, leaks are postfusion events; that is, a fusion pore forms without leaks. Electrical recordings also indicate that leakage does eventually occur for WT HA (Figure 7, 2 min after the pore formed). Because WT HA almost always causes fusion before leaks (Figure 7A, YDC) and Ser HA can induce hemifusion without leaks (Figure 8), the lytic action of HA does not appear to be essential for its ability to cause either hemifusion or fusion. In other words, true fusion of RBCs to HA-expressing cells is fundamentally not a leaky process, and any subsequent leaks, although caused by HA, are secondary to fusion. Because there are typically a few million copies of HA trimers expressed on cell surfaces, a very small percentage of the HA trimers participate in fusion. It is possible that leaks may be caused by “bystander” trimers that are not directly involved in pore formation.
Ser HA- and GPI HA-induced Hemifusion Intermediates Have Similar Stability
We next used chlorpromazine (CPZ) to explore whether the barrier that prevented transfer of aqueous dye in Ser HA cells (Figures 5 and 6) was similar to the hemifusion diaphragm created by GPI HA. CPZ is a membrane-permeable weak base that partitions preferentially into inner leaflets of cells and selectively destabilizes hemifusion diaphragms, allowing passage of aqueous dyes without significant leakage into the external medium (Melikyan et al., 1997). RBCs colabeled with CF and R18 were bound to HA-expressing cells. Fusion was triggered by lowering the pH to 5.0 for 2 min at 37°C, and CPZ was then added to the solution at neutral pH. The percentage of R18-stained HA-expressing cells that became labeled with CF (full fusion) was determined. In the absence of CPZ, virtually all WT HA-expressing cells acquired both membrane and aqueous dyes, whereas almost all Ser HA cells were stained with R18 but not CF (Figure 9). High concentrations of CPZ were required to induce transfer of CF from RBCs into Ser HA-expressing cells (Figure 9). Even at relatively high concentrations of CPZ (0.4 mM), however, only ∼40% of the Ser HA-expressing cells that were stained with R18 acquired aqueous dye. This is characteristic of the action of CPZ on hemifusion diaphragms induced by GPI HA; when GPI HA-expressing cells were treated with 0.4 mM CPZ, aqueous dye spread was seen in ∼36% of the cells that received lipid dye (Melikyan et al., 1997). These findings suggest that Ser HA and GPI HA form hemifusion diaphragms of comparable stability. In control experiments, when RBCs were bound to cells expressing Val HA and fusion conditions were established, 0.4 mM CPZ caused neither membrane nor aqueous dye mixing (our unpublished results).
DISCUSSION
The Importance of Gly1 in Fusion
The amino acid sequences of viral fusion peptides are thought to be critical for fusion to occur (Durell et al., 1997). Substitution of a Glu for the Gly at the first residue of the fusion peptide of HA of the Japan strain of influenza virus abolishes all fusion activity: lipid mixing, content mixing, and syncytia formation (Gething et al., 1986; Guy et al., 1992; Schoch and Blumenthal, 1993). Other substitutions at this position (in the X:31 influenza HA) abrogate syncytia formation (Steinhauer et al., 1995). In the present study, we used a mutagenesis approach to assess the charge, size, and hydrophobicity requirements of the first residue of the fusion peptide (in X:31 HA) at various stages of the fusion reaction: lipid mixing, fusion pore formation (probed electrophysiologically), and content mixing. We found the following: the first residue must be small (Gly, Ala, Ser) to promote lipid mixing and must be small and apolar (Gly, Ala) to form fusion pores and allow content mixing.
The fact that Gly is the optimal amino acid at residue 1 for full fusion (Ala shows reduced content mixing) (Figure 5) likely accounts for its absolute conservation among all naturally occurring influenza A viruses. Other amino acids can clearly be tolerated at this residue in the HA trimer structure (Table 2). The fact that Val, a moderately sized but highly hydrophobic residue, does not support either lipid or content mixing, emphasizes that overall hydrophobicity is not the sole criterion for fusion peptide function. Although residue 1 is conserved and X:31 HA must have a small amino acid (Gly and to some extent Ala) for full fusion, some HAs may be able to tolerate larger amino acids at this position. Thermolysin-activatable variants of an H7 influenza virus have been selected (for virus growth) that have a Leu at the first position of the fusion peptide (Orlich and Rott, 1994); however, detailed fusion phenotypes (lipid mixing, content mixing, syncytia formation) of the latter variants have not been reported.
Table 2.
Effects of mutations at position 1 of the HA fusion peptide: composite data from studies a and b
| Amino acid | Study | Lipid mixing (study a) | Fusion pores (study a) | Content mixing (study a) | Syncytia formation (study b) |
|---|---|---|---|---|---|
| Gly | a,b | + | + | + | + |
| Ala | a,b | + | + | ± | ± |
| Ser | a,b | + | − | − | − |
| Val | a | − | nd | − | nd |
| Glu | a | − | nd | − | −a |
| Gln | a | − | nd | − | nd |
| His | b | nd | nd | nd | − |
| Leu | b | nd | nd | nd | − |
| Ile | b | nd | nd | nd | − |
| Lys | a | − | nd | − | nd |
| Phe | b | nd | nd | nd | − |
Study a is the current study. Study b is Steinhauer et al. (1995). Amino acids are rank-ordered by side-chain volume; values for residues not listed in Table 1 are (in Å3) His, 167.3; Leu, 167.9; Ile, 168.8; Phe, 203.4. In study b, Steinhauer et al. (1995) reported that Gly1Ala showed reduced syncytia-forming activity and that a synthetic Gly1Ala fusion peptide showed 50% hemolytic activity. Walker and Kawaoka (1993) showed that the mutation Gly1Ala in an H5 HA showed ∼40% syncytia-forming activity.
Lack of lipid mixing, content mixing, and syncytia formation have also been observed with Gly1Glu HA from the Japan strain of influenza (Gething et al., 1986; Guy et al., 1992; Schoch and Blumenthal, 1993).
Ser HA Causes Hemifusion
The most unanticipated result of our study was that Ser HA induces hemifusion. This was concluded based on three independent assays: dye redistribution (Figures 5 and 6), electrical measurements (Figure 8), and stability of the aqueous diffusion barrier against CPZ (Figure 9). In addition to hemifusion caused by Ser HA and GPI HA (Kemble et al., 1994; Melikyan et al., 1995), there are other examples of viral protein-mediated hemifusion. Truncating the fusion protein of SV5 to eliminate its cytoplasmic tail (Bagai and Lamb, 1996), mutating glycines within the transmembrane domain of the fusion protein of vesicular stomatitis virus (Cleverley and Lenard, 1998), and adding small amounts of peptides from the coiled-coil domains of either HIV env (Munoz-Barroso et al., 1998) or SV5 F (Joshi et al., 1998) all lead to hemifusion but not full fusion. We and others have therefore posited that for viruses in general, fusion proceeds through hemifusion (Nanavati et al., 1992; Kemble et al., 1994; Melikyan et al., 1995; Bagai and Lamb, 1996; Chernomordik et al., 1998; Cleverley and Lenard, 1998).
A number of steps that are required for fusion pore formation have been identified in HA-mediated fusion. During acidification, HA trimers undergo conformational changes that lead to exposure of fusion peptides and their insertion into membranes. Several trimers then associate to form a complex that allows for hemifusion, and a fusion pore forms (Hernandez et al., 1996). The question arises as to when fusion becomes corrupted for “hemifusion mutants” such as Ser HA and those described above. It has been generally assumed that when hemifusion occurs, the process has been stopped at that point along the normal pathway, but this is not necessarily the case. For example, soon after activation mutant fusion proteins may assume conformations different from their wild-type counterparts that still induce hemifusion, but by different means.
Leakage Caused by Ser HA
Although Ser HA clearly causes hemifusion, in the majority of cases leakage precedes lipid dye spread. This is in contrast to the behavior of WT HA, for which lipid dye spread generally precedes leakage (Figure 7). Perhaps this difference relates to the observation that, although expressed at apparently equal levels, lipid dye spread for Ser HA occurs at later times than for WT HA (Figure 4). In contrast, GPI HA appears to induce hemifusion with a time course similar to that of WT HA and does so before leaks (our unpublished results). Therefore, Ser HA may be causing hemifusion by a different route than GPI HA and WT HA, and this route may readily lead to leaks. A second possibility is that leaks develop for Ser HA when “restricted hemifusion” proceeds to “unrestricted hemifusion”. Restricted hemifusion is hemifusion without spread of lipid dye; it is thought that a ring of HA trimers surrounding the site of initial hemifusion prevents dye spread and that restricted hemifusion is an intermediate of full fusion seen with WT HA (Chernomordik et al., 1998). Unrestricted hemifusion is the term used when lipid dye spread occurs in the absence of pore formation (e.g., for GPI HA). (Experimentally, the time a leak developed for Ser HA correlated well with the time lipid dye was first observed to spread [our unpublished results]. This correlation is expected if both leaks and onset of dye spread were caused by the transition from restricted to unrestricted hemifusion.) As a third alternative, because electrical leaks generally arise for WT HA well after fusion, they may arise from a postfusion conformation of HA (perhaps in “bystander HAs”) that is attained more rapidly for Ser HA. These possibilities are not mutually exclusive.
Structure and Membrane Interactions of Mutant HA Fusion Peptides
Structural studies of synthetic fusion peptides of the X:31 HA have shown that the identity of N-terminal residues can affect the conformation of the peptide and its interaction with phospholipid bilayers (Gray et al., 1996). Synthetic fusion peptides derived from nonfusogenic HA proteins (Steinhauer et al., 1995) with N-terminal sequences beginning GALF, LFGA, or ELFG have a higher ratio of β-structure to α-helix than does the synthetic WT fusion peptide whose N-terminal sequence begins GLFG. Fourier-transform infrared studies indicate that although the mutant peptides break hydrogen bonds between lipid ester carbonyls and water, the WT synthetic fusion peptide does not. This suggests that the “nonfusogenic” synthetic fusion peptides form hydrogen bonds to lipid ester carbonyls, whereas the WT fusion peptide does not (Gray et al., 1996). Whether these differences are a direct effect of hydrogen bonding to specific mutant amino acids or an indirect effect resulting from the increased amount of β-structure found for the mutant peptides is not known (L. Tamm, personal communication). Similar differences in secondary structure and lipid bonding likely occur for the mutant fusion peptides in the context of the full HA trimer. Some remodeling of the hydrogen bonding network in the lipid ester carbonyl region was also observed when BHA was bound to lipid bilayers (Gray and Tamm, 1998). An interesting possibility raised by Gray et al. (1996) is that if the nonfusogenic fusion peptides engage in interactions with lipid carbonyls, they may not be able to participate in obligate fusion peptide–fusion peptide interactions.
Although synthetic fusion peptides corresponding to the mutants described in the present study have not been studied, given the phenotype of the mutant fusion peptide that begins ELFG (Gray et al., 1996), which is equivalent to Gly1Glu, it is reasonable to propose that synthetic peptides corresponding to our nonfusogenic mutants (Gly 1 to Val, Glu, Gln, or Lys) and perhaps even to our hemifusion mutant (Gly1Ser) would also show differences in secondary structure (increased β-structure) or lipid bonding properties, as did those described by Gray et al. (1996). In addition, the mutant fusion peptides described here may interact with lipid bilayers at a different angle or to a different depth than the WT HA fusion peptide (Durell et al., 1997).
Why Does Ser HA Cause Hemifusion?
We consider two of many possibilities for why Ser HA (in X:31 HA) causes hemifusion and does not proceed to full fusion. In the first, the fusion peptide of Ser HA inserts into the bilayer and destabilizes monolayers, but in a manner different from that of WT HA. Hemifusion still occurs but Ser HA is in a molecular configuration different from WT HA and not capable of inducing pore formation. In the second, Ser HA yields hemifusion by the normal means, but the fusion peptide is involved, either directly or indirectly, in the later stages of pore formation (or pore expansion). The fusion peptide of Ser HA may interact in a different manner with the target bilayer than the WT fusion peptide, or it may be impaired in fusion peptide–fusion peptide or even fusion peptide–transmembrane domain interactions that may be needed to destabilize the hemifusion diaphragm and form a pore (Hernandez et al., 1996 [an updated version of the fusion model presented in Hernandez et al., 1996 can be found at http://www.med.virginia.edu/∼jag6n/whitelab.html]; Weissenhorn et al., 1997).
The results presented here for Ser HA provide the first example of a fusion peptide (point) mutant that blocks fusion at the stage of hemifusion. Previous studies have raised the possibility that mutations in the fusion peptide (of Japan HA) can affect fusion pore expansion. Gly4Glu shows reduced content mixing activity; Glu11Gly, although apparently similar to WT HA in lipid mixing and content mixing, shows no syncytial activity (Gething et al., 1986; Guy et al., 1992; Schoch and Blumenthal, 1993). The collective findings of these and other studies (Steinhauer et al., 1995) indicate that as larger or more polar residues are substituted for the Gly at the first position of the X:31 HA fusion peptide, fusion becomes arrested at earlier steps (Table 2).
Conclusions
In summary, our results clearly show that regardless of its precise mechanism of action, a single and very specific point mutation in the HA fusion peptide (Gly1Ser) yields a mutant HA that displays hemifusion. Also, increasing the hydrophobicity of position 1 by replacing it with larger amino acids (e.g., Gly1Val) eliminates even hemifusion. These findings suggest that position 1 of the fusion peptide is critical for determining the pathway of the fusion reaction and can affect not only the first stage of fusion, lipid mixing, but also the second stage of fusion, fusion pore formation.
ACKNOWLEDGMENTS
We thank Peter Huminski and Sofya Brenner for technical assistance, Dr. Joshua Zimmerberg for sharing unpublished results, and Dr. Lukas Tamm for helpful comments on this manuscript. The work was supported by Grants AI22470 (J.M.W.), GM27367 (F.S.C.), and GM54787 (G.B.M.) from National Institutes of Health.
Abbreviations used:
- CF
carboxyfluorescein
- CPZ
chlorpromazine
- HA
hemagglutinin
- R18
octadecylrhodamine B chloride
- RBC
red blood cell
REFERENCES
- Bagai S, Lamb R. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J Cell Biol. 1996;135:73–84. doi: 10.1083/jcb.135.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco L, Otero MJ, Castrillo JL. Modification of membrane permeability by animal viruses. Pharmacol Ther. 1989;40:171–212. doi: 10.1016/0163-7258(89)90096-x. [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic pore formation. J Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Leikina E, Frolov V, Bronk P, Zimmerberg J. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleverley DZ, Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton TE. Proteins: Structural and Molecular Principles. New York: W. E. Freeman and Co.; 1984. p. 242. [Google Scholar]
- Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Gething M-J, Doms RW, York D, White JM. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C, Tamm LK. pH-Induced conformational changes of membrane-bound influenza hemagglutinin and its effect on target lipid bilayers. Protein Sci. 1998;7:2359–2373. doi: 10.1002/pro.5560071113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C, Tatulian SA, Wharton SA, Tamm LK. Effect of the N-terminal glycine on the secondary structure, orientation and interaction of the influenza hemagglutinin fusion peptide with lipid bilayers. Biophys J. 1996;70:2275–2286. doi: 10.1016/S0006-3495(96)79793-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy HR, Durell SR, Schoch C, Blumenthal R. Analyzing the fusion process of influenza hemagglutinin by mutagenesis and molecular modeling. Biophys J. 1992;62:95–97. doi: 10.1016/S0006-3495(92)81790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Hernandez LD, White JM. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72:3259–3267. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughson FM. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- Joshi SB, Dutch RE, Lamb RA. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- Kozlov MM, Chernomordik LV. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and rearrangements. Biophys J. 1998;75:1384–1396. doi: 10.1016/S0006-3495(98)74056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, Brener SA, Ok DC, Cohen FS. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J Cell Biol. 1997;136:995–1005. doi: 10.1083/jcb.136.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, White JM, Cohen FS. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanavati C, Markin VS, Oberhauser AF, Fernandez JM. The exocytic fusion pore modeled as a lipidic pore. Biophys J. 1992;63:1118–1132. doi: 10.1016/S0006-3495(92)81679-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E, Marty A. Discrete changes of cell membrane capacitance observed under conditions of enhanced secretion in bovine adrenal chromaffin cells. Proc Natl Acad Sci USA. 1982;79:6712–6716. doi: 10.1073/pnas.79.21.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlich M, Rott R. Thermolysin activation mutants with changes in the fusogenic region of an influenza virus hemagglutinin. J Virol. 1994;68:7537–7539. doi: 10.1128/jvi.68.11.7537-7539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Pelletier S, Hoffman L, Hacker J, Armstrong RT, White JM. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratinov V, Plonsky I, Zimmerberg J. Fusion pore conductances: experimental approaches and theoretical algorithms. Biophys J. 1998;74:2374–2387. doi: 10.1016/S0006-3495(98)77946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C, Blumenthal R. Role of fusion peptide sequence in initial stages of influenza hemagglutinin-induced cell fusion. J Biol Chem. 1993;268:9267–9274. [PubMed] [Google Scholar]
- Shangguan T, Alford D, Bentz J. Influenza virus-liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry. 1996;35:4956–4965. doi: 10.1021/bi9526903. [DOI] [PubMed] [Google Scholar]
- Spruce AE, Iwata A, White JM, Almers W. Patch clamp studies of single cell-fusion events mediated by a viral fusion protein. Nature. 1989;342:555–558. doi: 10.1038/342555a0. [DOI] [PubMed] [Google Scholar]
- Spruce AE, Iwata A, Almers W. The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc Natl Acad Sci USA. 1991;88:3623–3627. doi: 10.1073/pnas.88.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC. Studies of membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse FW, Iwata A, Almers W. Membrane flux through the pore formed by a fusogenic viral envelope protein during cell fusion. J Cell Biol. 1993;121:543–552. doi: 10.1083/jcb.121.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Kawaoka Y. Importance of conserved amino acids at the cleavage site of the hemagglutinin of a virulent avian influenza A virus. J Gen Virol. 1993;74:311–314. doi: 10.1099/0022-1317-74-2-311. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Blumenthal R, Sarkar DP, Curran M, Morris SJ. Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J Cell Biol. 1994;127:1885–1894. doi: 10.1083/jcb.127.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]