Abstract
Polo kinases execute multiple roles during cell division. The fission yeast polo related kinase Plo1 is required to assemble the mitotic spindle, the prophase actin ring that predicts the site for cytokinesis and for septation after the completion of mitosis (Ohkura et al., 1995; Bahler et al., 1998). We show that Plo1 associates with the mitotic but not interphase spindle pole body (SPB). SPB association of Plo1 is the earliest fission yeast mitotic event recorded to date. SPB association is strong from mitotic commitment to early anaphase B, after which the Plo1 signal becomes very weak and finally disappears upon spindle breakdown. SPB association of Plo1 requires mitosis-promoting factor (MPF) activity, whereas its disassociation requires the activity of the anaphase-promoting complex. The stf1.1 mutation bypasses the usual requirement for the MPF activator Cdc25 (Hudson et al., 1990). Significantly, Plo1 associates inappropriately with the interphase SPB of stf1.1 cells. These data are consistent with the emerging theme from many systems that polo kinases participate in the regulation of MPF to determine the timing of commitment to mitosis and may indicate that pole association is a key aspect of Plo1 function. Plo1 does not associate with the SPB when septation is inappropriately driven by deregulation of the Spg1 pathway and remains SPB associated if septation occurs in the presence of a spindle. Thus, neither Plo1 recruitment to nor its departure from the SPB are required for septation; however, overexpression of plo1+ activates the Spg1 pathway and causes transient Cdc7 recruitment to the SPB and multiple rounds of septation.
INTRODUCTION
Mutation of the polo gene in Drosophila melanogaster confers a pleiotropic phenotype initially observed to include defects in spindle formation and chromosome segregation (Sunkel and Glover, 1988; Llamazares et al., 1991) and more recently cytokinesis defects (Carmena et al., 1998). The protein kinase encoded by polo has now been found to have a counterpart in a wide range of eukaryotes. Evidence is accumulating to suggest that these polo-like kinases (Plks) play roles in diverse mitotic functions, including spindle pole maturation, regulation of bipolar spindle formation, and the activity of anaphase-promoting complex (APC) ubiquitin protein ligase (Glover et al., 1998; Nigg, 1998). Consistent with these multiple roles, Plks have been localized in animal cells to the spindle poles and centromeres early in mitosis and to the central spindle and midbody during telophase and cytokinesis (Golsteyn et al., 1995; Lee et al., 1995; Logarinho and Sunkel, 1998; Wianny et al., 1998).
There is also growing evidence for a role for polo kinases in the commitment to and exit from mitosis. Mitosis-promoting factor (MPF) p34cdc2 cyclin B is regulated by a balance between the activities of inhibitory Wee1/Mik1 kinases and the opposing Cdc25 phosphatase. When a critical portion of the p34cdc2 becomes dephosphorylated, it triggers the activation of Cdc25 by at least one intermediary kinase, thus amplifying the initial MPF activity in a positive feedback loop (Hoffmann et al., 1993; Dunphy, 1994; Izumi and Maller, 1995; Kuang et al., 1994). The Xenopus Plk, Plx1, was suggested as a potential component of this amplification loop because it was found to associate with and phosphorylate Cdc25 (Kumagai and Dunphy, 1996). This has been substantiated by studies of the G2/M transition in Xenopus oocytes or cell-free systems derived from activated eggs (Abrieu et al., 1998; Qian et al., 1998a). Immunodepletion or immunoinhibition of Plx1 blocks the conversion of p34cdc2 into its mitotic form. Conversely, microinjection of Plx1 into oocytes accelerates activation of both Cdc25 and MPF. The recent identification of a polo kinase kinase in Xenopus, xPlkk, suggests either that multiple protein kinases participate in the feedback loop or that polo kinases can be activated independently of MPF activation (Qian et al., 1998b).
p34cdc2 is inactivated after the metaphase–anaphase transition by the proteolytic degradation of its regulatory subunit, a B-type cyclin. It has been proposed that Plks have a role in activating the APC, a ubiquitin protein ligase that is responsible for cyclin degradation. The budding yeast Plk, Cdc5p, is required to activate the APC and degrade the B type cyclin, Clb2, before itself becoming a substrate of the APC (Charles et al., 1998; Cheng et al., 1998; Shirayama et al., 1998). Moreover, Descombes and Nigg (1998) have presented evidence that Xenopus Plx1 is required for the degradation of APC targets for cytostatic factor-arrested extracts to exit M phase, and mammalian Plk will phosphorylate and inactivate APC components in vitro (Kotani et al., 1998).
Although the Saccharomyces cerevisiae cdc5.1 mutant arrests in the late stages of nuclear division with an elongated spindle, deletion of the fission yeast gene plo1+ leads to defects in two quite different stages of the division cycle: the formation of the spindle and execution of cytokinesis. Cells lacking the essential Plo1 protein accumulate in mitosis with a monopolar rather than a bipolar spindle and are neither able to form the prophase actin ring that is required to initiate cytokinesis nor to organize any septal material (Ohkura et al., 1995). Recent studies have identified conditional plo1 mutations that alter the selection of the site for prophase actin ring (Bahler et al., 1998). Complementary experiments showed that strong overexpression of the plo1+ gene resulted in both defects in spindle formation and inappropriate septation in interphase cells. Inappropriate septation during interphase can also be driven by manipulation of a regulatory network that involves the G protein Spg1 (Gould and Simanis, 1997; Schmidt et al., 1997). The GTP-bound form of Spg1 recruits the Cdc7 protein kinase to the spindle pole bodies (SPBs), and by the time of cytokinesis the active form of the complex becomes restricted to one of the two SPBs (Fankhauser and Simanis, 1994; Sohrmann et al., 1998). The position of Plo1 in this regulatory cascade is currently under study (Gould and Simanis, 1997).
In this work we describe a series of experiments that examine the role played by recruitment of Plo1 to the SPB during mitosis. We show that Plo1 localizes to the SPB upon commitment to mitosis and that this association diminishes after APC activation but still persists until the end of mitosis. Plo1 undergoes premature SPB association in the Cdc25 bypassing mutant stf1.1 (Hudson et al., 1990; Bridge et al., 1998). These observations are discussed in the context of a potential role for Plo1 in the regulation of MPF at the commitment to mitosis.
MATERIALS AND METHODS
Cell Culture and Strains
Cell culture and maintenance were carried out according to Moreno et al. (1991). All Plo1 asynchronous localization experiments were carried out in rich medium (yeast extract [YES]). When promoter repression or derepression was required, cells were grown in appropriately supplemented EMM2. Repression of the nmt1 promoter (Maundrell, 1993) was carried out by the addition of 4 μM thiamine to the minimal media. All synchronization experiments were conducted in appropriately supplemented EMM2. Matings were carried out on MSA plates (Egel et al., 1994).
Synchronous cultures were generated by size selection either after centrifugation on a lactose gradient as described previously (Carr et al., 1995) or by centrifugal elutriation (Mitchison and Carter, 1975). The strains used in this study are listed in Table 1.
Table 1.
Strains used in this study
| Strain number | Genotype | Source |
|---|---|---|
| IH109 | h−cut4.533 | Hirano et al., 1986 |
| IH110 | h− nuc2.633 leu1.32 | Hirano et al., 1986 |
| IH111 | h− cdc14.118 | Nurse et al., 1976 |
| IH112 | h− cdc15.140 | Nurse et al., 1976 |
| IH113 | h− cut2.120 | Hirano et al., 1986 |
| IH119 | h− cut8.563 | Hirano et al., 1986 |
| IH132 | h− cdc25.22 | Nurse et al., 1976 |
| IH136 | h− cut7.24 leu1.32 | Hagan and Yanagida, 1995 |
| IH154 | h− cut9.665 leu1.32 | Hirano et al., 1986 |
| IH164 | h− cdc2.33 | Nurse et al., 1976 |
| IH183 | h− cdc12.112 | Nurse et al., 1976 |
| IH259 | h− cut1.RB5.leu1.32 | Lab stock |
| IH280 | h− cut12.1 leu1.32 | Bridge et al., 1998 |
| IH295 | h−/+ plo1::ura4/plo leu1.32/leu1.32ura4.d18.ura4.d18 ade6,N219.ade6,216 his2/HIS2 | Ohkura et al., 1995 |
| IH365 | h− ura4.d18 leu1.32 | Lab stock |
| IH366 | h+ ura4.d18 leu1.32 his2 | Lab stock |
| IH376 | h− cdc10.v50 | Lab stock |
| IH385 | h− cut11.1 leu1.32 | Lab stock |
| IH572 | h−/+ cut12::ura4/cut12 leu1.32/leu1.32 ura4.d18/ura4.d18 ade6.M210/ade6.216 his1/his1.102 | Bridge et al., 1998 |
| IH666 | h+ stf1.1 leu1.32 | Hudson et al., 1990 |
| IH738 | h+ cdc7.A20 ura4.d18 | Schmidt et al., 1997 |
| IH1292 | h− ura4.d18 pINT5 spg1 | Schmidt et al., 1997 |
| IH1297 | h− cdc16.116 ura4.d18 | Nurse et al., 1976 |
| IH1298 | h− cdc11.H1 ura4.d18 leu1.32 | A. Poziemba and I. Hagan, and unpublished data |
| IH1308 | h− ura4.d18 | This study |
| IH1313 | h90ura4.d18 leu1::ura4+pl41plo1.NHA | This study |
| IH1314 | h90ura4.d18 leu1::ura4+pl41Plo1.NHAG | This study |
| IH1319 | h− stf1.1 ura4.d18 | This study |
| IH1353 | h− stf1.1 ura4.d18 leu1::ura4+pl41 Plo1.NHA.G | This study |
| IH1123 | h− leu1.32 ura4.d18 p41plo1.NHA | This study |
| IH1367 | h+ cdc25.22 his2 ura4.d18 leu1::ura4+pl41plo1.NHA | This study |
| IH1366 | h+ cdc25.22 his2 ura4.d18 | This study |
| IH1319 | h− plo1::ura4 leu1.32 ura4.d18 ade6.M210 p41plo1.NHA | This study |
| IH1409 | h− ura4.d18 leu1.32 p41plo1 | This study |
| IH1410 | h− ura4.d18 leu1.32 p41plo1.NHA | This study |
| IH1548 | h− cdc25.22 leu1.32 pHN 204 | Ohkura et al., 1995 |
Molecular Genetic Manipulations
An NotI site containing oligonucleotide, designed to keep the gene in frame, was inserted at the Nru1 site located 40 bp from the 3′ end of the plo1+ gene in pHN205. An NotI DNA fragment from pDMHA encoding three of the hemagglutinin (HA) epitopes that are recognized by the mAb 12CA5 was inserted at the novel NotI site to create p41plo1.CHA. An NdeI–SmaI fragment containing the plo1+ gene was isolated from the plasmid pHN205 (Ohkura et al., 1995) and ligated into pREP41HA (Craven et al., 1998) to give plasmid p41plo1.NHA. Plasmid p41plo1.NHA was transformed into diploid IH295, in which one copy of the plo1+ gene has been completely replaced with the ura4+ gene. The resulting strain was sporulated, and spores were plated onto minimal media lacking uracil (to select for growth of cells containing the plo1.d1 allele) and leucine (to select for those containing p41plo1.NHA). Many ura+ leu+ colonies were obtained, whereas none were obtained in control transformation in which pREP41HA was used. This showed that p41plo1.NHA is able to complement the plo1.d1 deletion.
An HA-tagged version of plo1+ for integration into the genome was made by excising the XhoI–SacI plo1+ fragment from p41plo1.NHA and cloning it into pINT541 (a gift from M. Sohrmann and V. Simanis, ISREC, Lausanne, Switzerland) to produce plasmid pDM14. Because pINT541 lacks a termination and polyA addition sequence, this was reintroduced by cutting a SacI fragment from p41plo1.NHA (which contained the nmt1+ polyA addition sequences) and inserting this fragment into pDM14 to generate pI41plo1.NHA. This DNA was integrated into the S. pombe genome as described previously (Fankhauser and Simanis, 1994).
A version of plo1+ fused to both HA and green fluorescent protein (GFP) at its amino terminus was created by excising an NdeI fragment containing the GFP encoding DNA from pGEMT-GFP (Craven et al., 1998), and inserting it into the NdeI site of pI41plo1.NHA. The resulting plasmid was called pI41plo1.NHAG. This DNA was integrated into the Schizosaccharomyces pombe genome at the leu1 locus as described previously (Fankhauser and Simanis, 1994).
Immunological Techniques
Western Blotting.
For each preparation, 2 × 107 cells were harvested from a log phase culture by centrifugation at 2000 × g for 2 min. They were then washed with 1 ml of Stop buffer (Simanis and Nurse, 1986) and transferred to a 2-ml microfuge tube. This was then centrifuged at 13,000 rpm for 1 min, the buffer was removed, and the cells were then either processed directly or snap-frozen in liquid nitrogen and stored at −80°C. The cells were resuspended in 50 μl of HE buffer containing 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 mM PMSF (Simanis and Nurse, 1986). The cells were then disrupted with 1 ml of acid-washed glass beads by agitation for 3 s in a Hybaid Ribolyser at power setting 6. The cell extract was spun out through a hole in the bottom of the tube. Boiling SDS-PAGE sample buffer was added to the protein extract, and the mix was incubated at 100°C for 3 min.
Western blot detection was carried out after equal amounts of protein samples were run on a 10% SDS-PAGE gel. Final protein loadings for cell cycle Western blot analysis were determined after developing one blot for TAT1 and adjusting the loading volumes accordingly. Detection was carried out using the Pierce Super Signal Substrate (Pierce, Rockford, IL) (see Figure 1) or alkaline phosphatase reagents (see Figures 3 and 7) and using standard immunoblotting protocols (Harlow and Lane, 1988). To quantitate the intensity of the signals in the cell cycle Western blots, the blots were scanned, and the signal density in the bands was measured using NIH image.
Figure 1.
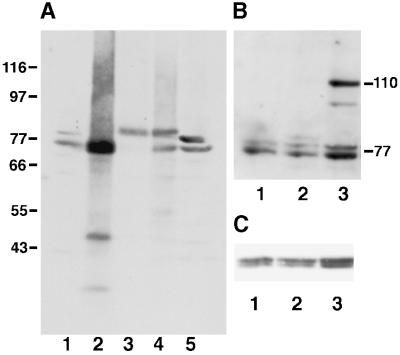
Characterization of HN184 antibody. Protein extracts from different S. pombe strains were analyzed by immunoblots and probed with HN184 antisera (A, B) or the anti-α tubulin antibody TAT1 as a loading control (C). (A) A wild-type cell (IH365) extract shows a Plo1 doublet of the expected size of ∼77 kDa (lane 1). The intensity of the lower band increased in wild-type cells, which expressed plo1+ from a multicopy plasmid under the control of the nmt41 promoter (IH1409) (lane 2). In a strain from which the genomic copy of plo1+ has been deleted, and the cells were kept alive by the expression of an HA-tagged plo1+ gene from the multicopy plasmid p41plo1.NHA, the two wild-type bands were replaced by a single band at 80 kDa (IH1391) (lane 3). As predicted from lanes 1–3, two bands were seen in a wild-type strain overexpressing the HA-tagged protein (IH1289), a bottom wild-type band and an upper HA-tagged band (lane 4). The intensity of the upper band in wild-type cells was increased in extracts prepared in the presence of 0.5 μM okadaic acid (lane 5). Blotting with TAT1 showed similar loading levels in each lane. (B) Lane 1, IH365 extract. Lane 2, extract of IH1313 showing the faint additional HA-tagged plo1 band at 80 kDa. Lane 3, extract of IH1314 showing an additional GFP- and HA-tagged band of Plo1 at 110 kDa. C shows the same filter as depicted in B, probed with TAT1 to demonstrate consistency of loading.
Figure 3.
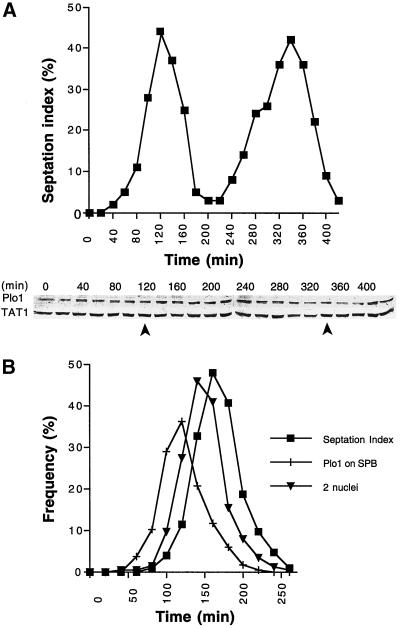
Fluctuations in Plo1 protein levels do not account for the cell cycle specificity of Plo1 localization to the SPB. (A) A cell cycle-synchronized population of wild-type cells (IH365) was generated by elutrient centrifugation at 25°C in appropriately supplemented EMM2. The small G2 cells were harvested at the beginning of the experiment, and samples were then taken every 20 min for Western blot analysis and scoring the septation index (n = 200 for each time point). The top panel shows the septation index, which indicates good cell cycle synchrony in the culture, and the bottom panel shows a Western blot of whole-cell extracts. The blot was cut into two, and the top portion was probed with antibodies to Plo1 and the bottom portion was probed with antibodies to a tubulin (TAT1). The tubulin blot acts as a loading control. The position of the peaks seen in the septation index are indicated by arrows beneath the blots. Quantitation of the ratio of the TAT1 to Plo1 signals for two independent blots for each of two independent experiments (each of which had two complete cell cycles) led to the conclusion that there is no apparent fluctuation in Plo1 levels as cells progress through the cell cycle. (B) Anti-Plo1 immunofluorescence analysis of an elutrient centrifugation synchronized population of wild-type cells growing in EMM2 at 25°C. Small G2 cells were harvested at the beginning of the time course at time point 0 and processed every 20 min for anti-Plo1 immunofluorescence microscopy. The septation index of each sample was determined by calcofluor staining before the cells were processed for immunofluorescence microscopy. Plo1 association with the SPB precedes anaphase B by ∼20 min and septation by ∼30 min. n = 200 for each data point. A repeat of this experiment gave similar profiles.
Figure 7.
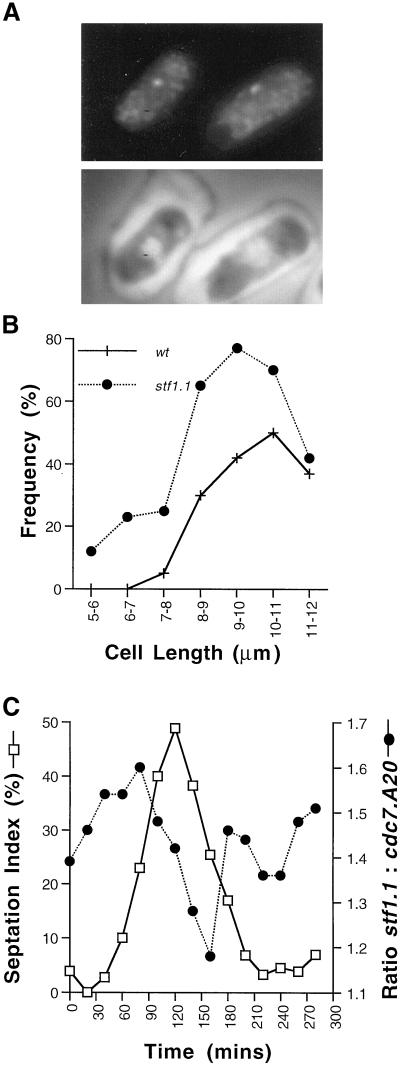
Premature recruitment of Plo1 to the SPBs in stf1.1 mutant cells. (A) An asynchronous stf1.1 culture was fixed and stained using HN184 antibodies and DAPI. Plo1 localized to the SPB of small interphase cells. The cells in A have shrunk after processing for immunofluorescence. This is because a very short fixation regime is used for Plo1 staining, and the cell structure has contracted in the 2 d before this image was captured. All statistical analysis throughout this article were conducted on freshly stained cells before shrinkage occurred. (B) Asynchronous wild-type IH365 and stf1.1 cells were processed for immunofluoresence using HN184 antibodies and DAPI. The presence or absence of Plo1 staining was noted, and the frequency of a positive score was plotted as a functionof the cells’ length. This was carried out for a minimum of 30 cells for each length category. Plo1 localizes to smaller cells and a higher proportion of stf1.1 cells than wild-type. (C) A culture of stf1.1 cells was synchronized by elutrient centrifugation. Samples were taken every 20 min and mixed with asynchronous cdc7. A20 mutant cells that had been at the restrictive temperature for 4–6 h. These samples were stained with HN184 antibodies and DAPI. The inclusion of the septation mutant provided an internal control for the quality of sample preparation in the sample because the long multinucleate cdc7. A20 cells were easily distinguishable from the shorter stf1.1 cells. Thus the ratio of Plo1 SPB association in stf1.1 relative to cdc7. A20 staining gives a measure of Plo1 association with the stf1.1 SPB in interphase. The level of SPB association in cdc7. A20 remained constant throughout the experiment, but the relative staining on the SPBs of stf1.1 cells varied. Two hundred cells of each strain were scored at each time point. The ratio of Plo1 association in stf1.1 cells relative to cdc7. A20 cells is presented (circles) alongside the septation index (open squares). Although this complex experiment was only performed once with a synchronous culture, data from the analysis of mixed asynchronous populations result in similar conclusions (see RESULTS).
HN184 sera were used at a concentration of 1:100, and TAT1 anti-α tubulin antibodies were used at 1 in 100 (Woods et al., 1989).
Fluorescence Microscopy.
For GFP autofluorescence, microscopy cells were fixed in 3.7% formalin for 10 min, and the DNA was stained using DAPI as described in Moreno et al. (1991). Calcofluor staining to score septation indices was also according to Moreno et al. (1991).
Immunofluorescence microscopy was performed according to Hagan and Hyams (1988), with the exception that for Plo1 immunofluorescence glutaraldehyde was omitted and fixation was limited to 10 min. Affinity-purified rabbit anti-Sad1 antibodies, AP9.2 (Hagan and Yanagida, 1995), were used at a dilution of 1:25, whereas sheep affinity-purified anti-Sad1 antibodies (K. S. Sheldrick and I. M. Hagan, unpublished data) were used at 1:50; the anti-tubulin antibody, TAT1 (Woods et al., 1989), was used at 1:80; anti-GFP antibodies (generous gift from Ken Sawin, ICRF, London) were used at 1:50; and the Plo1 antibodies (HN184) were used at a dilution of 1:20. Appropriate secondary antisera were from Sigma (Poole, Dorset, UK). For double-labeling, cells were incubated with both antibodies simultaneously. Images (250), which were captured with a Hamamatsu SIT camera (Hamamatsu, Bridgewater, NJ), were integrated into one final image with pixel pipeline frame grabber in NIH image. Merging different images to produce color overlays and the generation of final figures were both done in Adobe Photoshop v4.0. Cell-length measurements were determined using the appropriate NIH image tool.
RESULTS
Reagents for the Analysis of Plo1 Protein
Localization of a protein to specific regions of the cell or to discrete organelles can play a key role in its participation in specific cellular events. We therefore wished to determine the distribution of Plo1 protein kinase at all stages of cell cycle progression.
The generation of the anti-Plo1 antibody (HN184) has been described previously (Ohkura et al., 1995). A number of additional reagents were generated to verify the validity of the results obtained with HN184. Expression of a gene in which the HA epitope tag was 24 amino acids before the end of the plo1 coding sequence (plo1.CHA) from the Rep41 nmt1 promoter was unable to support the growth of spores lacking plo1+. In contrast, the expression of either the wild-type gene (plo1+) or a version in which three HA epitopes were fused at the N terminus in frame with the plo1 ATG (plo1.NHA) complemented the deletion, indicating that plo1.NHA encoded a functional protein. Western blot analysis using HN184 confirmed these results and demonstrated the specificity of this anti-Plo1 antisera (Figure 1A). A doublet was detected at 77 kDa in wild-type cells (Figure 1A, lane 1). The intensity of the upper band was enhanced relative to the lower band when the phosphatase inhibitor okadaic acid was included in the cell extraction buffer (Figure 1A, lane 5). The intensity of the lower band was markedly increased in extracts prepared from wild-type cells in which plo1+ was expressed from the nmt1 promoter of pREP41 (Figure 1A, lane 2). Both bands were absent from extracts of plo1.d1 cells, which were kept alive by the expression of plo1.NHA (Figure 1A, lane 3). In these cells the 77-kDa doublet was replaced by a novel band at 80 kDa that corresponded to the expected increase in mass arising from fusion with the tag. In both cases, where Plo1 was overproduced a single rather than a double band was seen. The relative intensity of this Plo1NHA band was markedly reduced when expression was driven from a version of the gene that had been integrated at the leu1 locus and regulated by the weaker nmt41 promoter (Figure 1B, lane 2). To facilitate protein localization, the cDNA encoding GFP was fused to plo1.NHA in pINT541, and the construct was integrated into the genome. Induction of this gene resulted in the appearance of a novel band at 110 kDa (Figure 1B, lane 3). Together these data show that the HN184 sera specifically recognized the Plo1 protein.
Plo1 Associates with the Mitotic but Not Interphase SPB
In a first approach we observed the autonomous fluorescence of a chimeric protein in which the coding sequence for GFP was fused in frame to the amino terminus of Plo1. This fusion protein was integrated into the genome at leu1 locus and regulated by the nmt81 promoter, which gives a very low level of expression. Illumination of living or formalin-fixed cells with the appropriate wavelength to excite GFP revealed one or two dots in cells that were in the constant length and division stages of the cell cycle (Figure 2A) (Mitchison and Nurse, 1985). No signal could be detected in interphase cells. Immunolocalization of the fusion protein with antibodies against GFP confirmed the pattern given by intrinsic GFP fluorescence and permitted counterstaining with antibodies to the SPB component Sad1. Early mitotic cells showed one or two dots of GFP–Plo1 that also stained with anti-Sad1 antibodies, confirming that they represented the SPB; however, although Sad1 was associated with interphase SPBs, Plo1 was not (Figure 2B).
Figure 2.
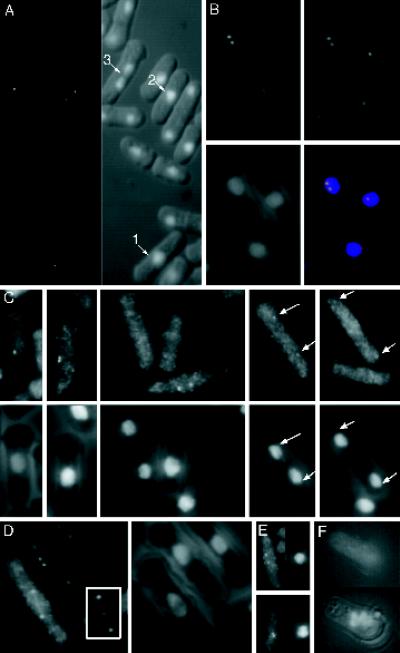
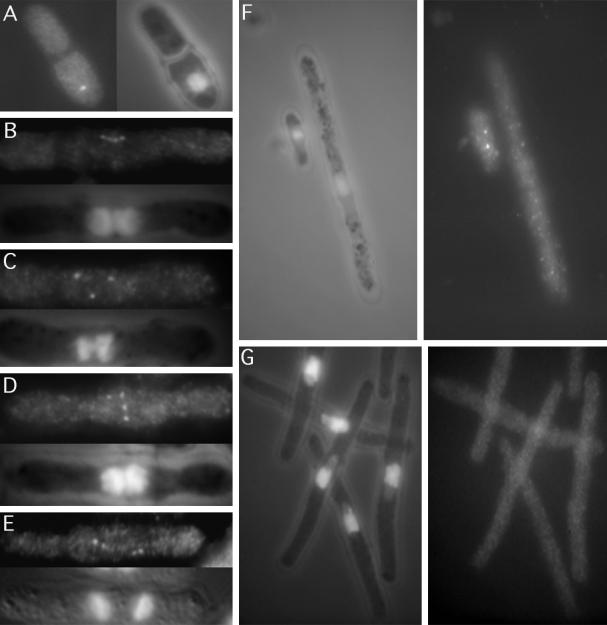
Plo1 localizes to the SPBs and mitotic spindle in a cell cycle-specific manner. (A) GFP autofluorescence in formalin-fixed cells of the strain IH1314 grown at 25°C in EMM2 supplemented with leucine to fully induce the nmt41 promoter. Two dots were seen in mitotic cells up to early anaphase B (cell 1, 2, 3). (B) Immunofluorescence staining of these cells with anti-GFP antibodies (B, top left panel) and Sad1 (B, top right panel) antibodies. Sad1 (top right, and red in bottom right) and Plo1 (top left, and green in bottom right) colocalized to give a yellow signal in the mitotic cell in the merged image (bottom right panel). DAPI staining (blue, bottom right) along with phase contrast (B, bottom left panel) shows chromatin localization with respect to the outline of the cell. (C) Plo1 immunofluorescence staining of the wild-type strain IH365 using HN184 antisera (top panels) and the same cells stained with DAPI and coincidentally viewed with phase contrast (bottom panels) to reveal the location of chromatin relative to the cell periphery. The panels are arranged from left to right to show successive stages as mitosis progresses. The earliest localization of Plo1 was to a single SPB. It then localized as two bright and distinct dots as the SPBs separated, and as anaphase progressed the localization to the SPBs continued, but the intensity was greatly reduced (arrows in the final panels). The images in Figure 4C were processed to the same extent, to highlight the variations in Plo1 intensity rather than to give the best qualitative image. (D) HN184 stained short spindles of wild-type cells until the start of anaphase, when very briefly it was at its brightest after the chromatin had separated. After this the staining could no longer be seen. (E) Microtubules of IH365 cells were depolymerized by incubation in an ice bath for 20 min. Left-hand panels show HN184 staining; right-hand panels show DAPI phase-contrast images of the same cells. The top cell was stained before cooling, and the bottom cell was stained after cooling. Plo1 SPB association was unaffected by microtubule depolymerization. (F) HN184 staining of germinating spores from which plo1+ has been deleted. The top panel shows no HN184 signal, whereas the DAPI phase-contrast image in the bottom panel shows the condensed chromatin, which is characteristic of mitotically arrested cells.
Direct detection of endogenous Plo1 with the HN184 antiserum gave similar patterns (Figure 2C). The first sign of Plo1 staining with these antibodies was a single dot on the side of the nucleus. The frequency of cells with single dots in asynchronous cultures (0.8% at 25°C, 1.2% at 36°C [n = 500]) suggests that Plo1 associates with the SPB for a significant period before the appearance of two distinct juxtaposed dots of Plo1 staining during spindle formation. SPB staining was bright and persisted until early anaphase B, whereupon its intensity diminished abruptly. SPB staining was still detected until spindle breakdown, however, but it was extremely faint and barely detectable above background levels (Figure 2C). We also detected association of Plo1 with the spindle in some cells. This association was seen from the beginning of spindle formation until the time in anaphase B when the SPB staining diminished radically. The specificity of HN184 immunostaining was indicated by the fact that the mitotic SPBs of germinating spores that lacked plo1+ were not recognized by the antibodies (Figure 2F). We are therefore confident that the fluorescence signals seen using HN184 resulted from detection of Plo1 protein. Given that both the immunofluorescence and GFP fluorescence data independently showed a similar cell cycle variation in Plo1 signals at the SPB, we conclude that this variation was not due to processing artifacts, fluorescence quenching, or epitope masking but reflected a genuine cell cycle regulation of Plo1 association with the SPB.
Proteins may either associate directly with the SPB or be transported there by microtubule motor proteins. Given that polo kinases associate with motor proteins (Lee et al., 1995; Adams et al., 1998) and that we observed spindle association of Plo1 (Figure 2E), we determined whether Plo1 would associate with the SPB after microtubule depolymerization by cold shock (Hagan and Yanagida, 1995). Although microtubules were completely depolymerized by incubation on ice, Plo1 remained bound to the SPB (Figure 2E).
Plo1 Association with the SPB Is Not Determined by Fluctuations in the Level of the Protein
One possible explanation for the lack of any detectable association of the Plo1 with the SPB during the majority of the cell cycle is that the protein is present only during mitosis. We therefore monitored Plo1 protein levels in whole-cell extracts from a wild-type culture that had been synchronized with respect to cell cycle progression by elutrient centrifugation. Large fluctuations in Plo1 protein levels did not accompany cell cycle progression; rather, protein levels appeared constant (Figure 3A). Measurements of the ratio of the intensity of the Plo1 doublet to the α tubulin doublet recognized by TAT1 indicated that the ratio of Plo1 to α tubulin levels did not fluctuate. Most significantly, Plo1 was present throughout interphase. Because Plo1 associated only with the SPBs of mitotic cells in a similarly synchronized culture (Figure 3B), we conclude that the cell cycle variation in Plo1 association with the SPB is not generated by alteration of the stability of the protein.
Plo1 Association with the SPB and the Regulation of Septation
Loss of Plo1 protein blocks septation, whereas excessive overproduction of the protein drives septation during interphase (Ohkura et al., 1995). Because several regulators of septation associate with the SPB early in mitosis (Sohrmann et al., 1998), it is possible that the SPB association of Plo1 is an integral part of its role as a regulator of septation. To explore this possibility further we determined the subcellular distribution of Plo1 in strains that had been genetically manipulated to either block septation or drive septation at inappropriate stages.
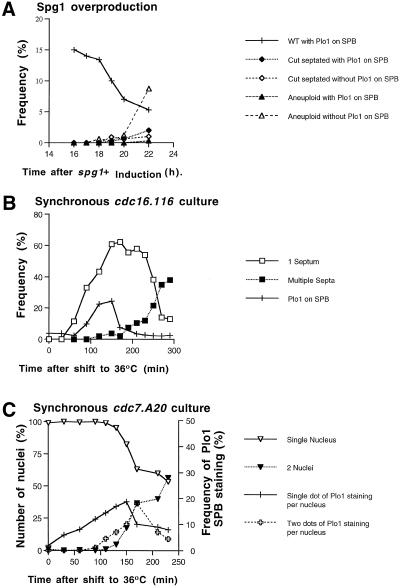
Multiple septation events can be driven during interphase in single cells either by dramatically increasing the levels of the G protein Spg1 or by reducing the activity of the Cdc16/Byr4 complex that provides the GAP activity for Spg1. The latter can be achieved by shifting the temperature of a culture of cdc16.116 cells to the restrictive temperature (Minet et al., 1981; Schmidt et al., 1997; Furge et al., 1998). We observed that the frequency of Plo1 localization to the SPB decreased upon induction of Spg1 (Figure 4A). This reduction may arise indirectly from the block of mitotic commitment that accompanies the DNA damage or the reduction in cell volume resulting from septation in interphase (Nurse, 1990). To examine the relationship of SPB association of Plo1 to mitotic progression in cells destined to undergo repeated septation without an intervening mitosis, we first selected G2 cells from a cdc16.116 culture by elutrient centrifugation. We then shifted the temperature of the culture of these G2 cells to the restrictive temperature and followed SPB association of Plo1, mitosis, and septation. Septation in some cells was induced within 30 min of the temperature shift (Minet et al., 1981), but the association of Plo1 with the SPB occurred later in a single peak at the normal time expected for a mitotic event (Figure 4B). Plo1 did not associate with the SPBs during the generation of the second septum in the arrested cells. Thus, when septation is induced by inappropriate Spg1 activation, either through overexpression of Spg1 or inactivation of Cdc16, it occurs without previous association of Plo1 with the SPB.
Figure 4.
Plo1 localization to the SPB is not driven by the premature activation of Spg1 that drives inappropriate septation but is affected in cdc7. A20 mutants. (A) Induction of cytokinesis by overexpressing spg1+ from an integrated version of the gene that was controlled by the nmt1 promoter (IH1292). Samples were taken for immunofluorescence every hour after gene induction by removal of thiamine from the medium and stained with HN184 antibodies and DAPI. Overproduction of Spg1 led to a reduction in the number of cells with Plo1 on their SPB. (B) cdc16.116 cells were synchronized with respect to cell size by elutrient centrifugation and immediately incubated at 36°C at time 0 on the graph in B. Correlative HN184 immunofluoresence, DAPI, and calcofluor staining showed that the septation started to occur within 1 h of the shift to the restrictive temperature. In contrast, Plo1 association with the SPB correlated with the normal timing of mitosis in such an experiment and did not rise once the second round of septation was initiated (filled squares). (C) cdc7. A20 cells were synchronized with respect to cell size by elutrient centrifugation, and the isolated small G2 cells were immediately cultured at 36°C. Samples were taken for immunofluoresence and stained with HN184 and DAPI. Scoring for one or two dots of intense Plo1 staining showed one dot of Plo1 staining on the nuclei of G2 cells much earlier than in wild-type cells. The appearance of two dots per nucleus coincided with the normal timing of mitosis. n = 200 for each data point on the graphs. Each experiment gave similar results when repeated.
Although these data show that septation does not require previous Plo1 association with the SPB, they do not clearly establish whether it is necessary for Plo1 to be lost from the mitotic SPB before septation can occur. Septation will take place in cells blocked in mitosis as a result of mutations in cut4 and cut9, genes that encode components of the APC, in cut7, which encodes a kinesin-related protein needed for spindle formation, and in cut8, which encodes a protein required for sister chromatid separation (Hirano et al., 1986; Hagan and Yanagida, 1990; Samejima and Yanagida, 1994a,b, Yamashita et al., 1996). We therefore incubated cut7.24, cut4.533, cut8.563, and cut9.665 mutant cells at the restrictive temperature for 3 h to induce cytokinesis in mitotically arrested cells and examined Plo1 localization. In each case Plo1 was clearly associated with the SPB in all septating mitotic cells. One such example of a cut7.24-arrested cell is shown in Figure 5A. These experiments showed that the disassociation of Plo1 from the SPB was not required for septation. Moreover, they also show that Plo1 disassociation from the SPB requires APC function.
Figure 5.
Anti-Plo1 immunofluorescence localization of Plo1 in cdc and cut mutants. (A) Cut7.24 cells that arrest in mitosis but continue to execute cytokinesis regardless of the mitotic defect still had strong Plo1 staining at the time of this abnormal cytokinesis. Anti-Plo1 immunofluorescence microscopy with HN184 revealed that Plo1 shows strong association with the mitotic SPBs of the early septation mutants: (B) cdc7. A20, (C) cdc11. H1, (D) cdc14.118, (E) cdc15.140. Images of cells in the early stages of spindle formation in the first division after cytokinesis that had been inhibited by incubation at the restrictive temperature are shown. Thus, each of the two nuclei had two SPBs that stained with Plo1 antibodies. Plo1 did not localize in (F) cdc25.22 (long cell in A) or (G) cdc2.33 mutant cells after incubation of the temperature-sensitive mutants for 4 h at the restrictive temperature. Wild-type cells (small cell in F) were mixed with cdc25.22 cells to show that the lack of Plo1 staining was not due to a technical problem. Because the smaller wild-type cells stained normally, we conclude that there was no association of Plo1 with the SPB.
Activation of Spg1 recruits Cdc7 kinase to the SPB (Sohrmann et al., 1998). In the absence of Spg1 or Cdc7 function, the Dmf1 and F-actin rings form, but the subsequent events leading to septation do not occur. In addition to mutations in spg1+ and cdc7+ mutation in a number of other genes, including cdc14+ and cdc11+ have identical septation defects, and genetic data suggest that they function in conjunction with Spg1 and Cdc7 to regulate cytokinesis (Gould and Simanis, 1997; Schmidt et al., 1997). Because the intimate relationship between Plo1 and septation suggests an interaction of Plo1 with the Spg1 pathway, we localized Plo1 in mixed cultures of these cytokinesis-regulating mutants containing wild-type cells as morphologically distinct processing controls. Plo1 was strongly present at the SPBs of both the wild-type processing control and the multinucleate cells of the mutants cdc7.A20, cdc11.IH1, and cdc14.118 (Figure 5B–D). The HN184 antibody also stained the mitotic SPBs of cdc15.140 cells, which lack a structural component of the actin ring and are blocked in cytokinesis at a similar stage (Figure 5E) (Nurse et al., 1976; Fankhauser et al., 1997). cdc7.A20 and cdc7.24 were unique among these mutants in that twice as many cdc7ts as cdc7+ cells had strong SPB staining at the restrictive temperature. In contrast, essentially the same proportion of mutant and wild-type cells had Plo1 on the SPB in mixed cultures of cdc11.IH1/cdc11+ and cdc14.118/cdc14+ at the restrictive temperature.
To determine any cell cycle specificity of this additional SPB association in cdc7ts mutant backgrounds, Plo1 localization was monitored in a cell cycle-synchronized cdc7.A20 culture that had been shifted to the restrictive temperature in early G2 phase. Plo1 associated with the SPB in ∼10% of G2 cells after the temperature shift, rising to a maximum in anaphase (Figure 4C). Scoring cells with single or double dots showed a predominance of single-dot staining in interphase cells.
We conclude from these experiments that although there was some influence of mutations in the Cdc7 protein kinase upon the recruitment of Plo1 to the SPB, activation of the Spg1 cascade did not recruit Plo1 to the SPB.
Plo1 Overexpression Activates the Spg1 Cascade
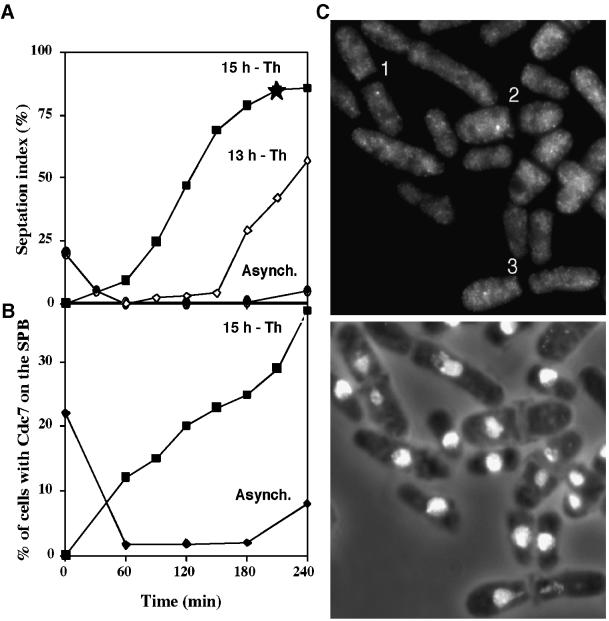
Because plo1+ overexpression drives septation, it is possible that it may do so by activating the Spg1 pathway (Ohkura et al. 1995). To test this possibility plo1+ was overexpressed in temperature-sensitive cdc25.22 cells, which are blocked in interphase at the restrictive temperature because of an inability to activate MPF. This results in septation without mitosis (Ohkura et al. 1995). After a culture of cdc25.22 cells was induced to overexpress plo1+ from a multicopy plasmid, it was split into two, and two independent synchronous cultures were generated 13 and 15 h later and incubated at 36°C. Septation was induced with different kinetics in these two cultures (Figure 6A). Analysis of the culture in which Plo1 had been induced for 15 h showed that Plo1 overexpression resulted in Cdc7 recruitment to the SPBs (Figure 6, B and C). The kinetics of Cdc7 association suggest that the protein cycles on and off the SPB with every round of septation. Initially the proportion of cells with Cdc7 on the SPB exceeds the septation index, showing that it is recruited before septation. The proportion of cells with Cdc7 on the SPB in the later stages of the experiment did not reach the same level as the septation index, showing that once a septum is generated, in these conditions, Cdc7 leaves the SPB.
Figure 6.
Overproduction of Plo1 resulted in the association of Cdc7 with the SPB in G2-arrested cdc25.22 cells. (A) Two independent synchronous cultures were generated by elutrient centrifugation 13 and 15 h after the removal of thiamine from IH1548 (h− cdc25.22 leu1.32 pHN 204) cells cultured in EMM2 medium at 25°C to induce overexpression of the plo1+ gene. The temperature of each synchronous culture was immediately shifted to the restrictive temperature of 36°C after its generation. Although inappropriate septation was induced in each culture, many more septa were seen in the 15-h culture (filled squares) than the 13-h culture (open diamonds) as each was followed into the cdc25.22 arrest. As a control the temperature of an uninduced asynchronous culture was shifted to 36°C (filled circles). Septation was inhibited in this culture for 4 h, after which some cells began to leak past the cdc25.22 arrest point. (This is a common feature of an asynchronous culture. In contrast to the synchronized cells, some members of this population had been at the cdc25.22 arrest point for the full 4-h time course of the experiment.) Staining of the 15-h and control culture showed that Spg1 was activated to recruit Cdc7 to the interphase SPBs as a consequence of Plo1 overproduction. (C) Cdc7 and DAPI/phase-contrast images of the same field of cells taken from the 210-min time point of the 15-h culture (indicated by a star in A). Cells 1–3 have Cdc7 staining of their SPBs in nuclei, which have been bisected by the inappropriately initiated septum.
SPB Association of Plo1 Requires Cdc25 and Cdc2 Activation
Commitment to fission yeast mitosis is regulated by the activity of the MPF regulators Wee1 and Cdc25. Analysis of the Xenopus polo-like kinase Plx1 suggests that Plo1 may participate in the activation of MPF and that this role may involve regulation of Cdc25 activity. Because Plo1 association with the SPB appeared to be a very early event in mitosis, we were keen to determine whether it required Cdc25 or Cdc2 activity. We examined the ability of Plo1 to associate with the SPB in temperature-sensitive cdc25 and cdc2 mutants in which cell cycle progression was blocked because of incubation at the nonpermissive temperature. Plo1 did not associate with the SPBs cdc25.22, which had been arrested by incubation at 36°C for 4 h (Figure 5F). The fact that Plo1 was clearly visible on the SPBs of wild-type cells that had been mixed in with these cdc25.22 mutants before fixation showed that the lack of staining was not an artifact of sample preparation in this particular experiment (Figure 5F). Thus, Plo1 association with the SPB required Cdc25 activity. Anti-Plo1 immunofluorescence microscopy of cdc2.33, which had been incubated at the restrictive temperature for 4 h to inactivate p34cdc2, again showed that Plo1 was not associated with the SPB (Figure 5G). Thus, association of Plo1 with the SPB is an early mitotic event that follows activation of p34cdc2
Premature Recruitment of Plo1 to the SPB in the cdc25 Bypassing Mutant stf1.1
Dominant mutations in the stf1/cut12+ gene, which encodes an essential SPB component, can bypass the normally essential requirement for Cdc25 in the activation of MPF (Hudson et al., 1990; Bridge et al., 1998). Given the association of Plo1 with the SPB at the onset of mitosis and its potential role in MPF activation (see INTRODUCTION), it was of interest to examine the localization of Plo1 in the stf1.1 mutant background.
In contrast to wild-type cells, the SPBs of small interphasic as well as mitotic stf1.1 cells showed strong Plo1 staining (Figure 7A). Cell length is a measure of cell cycle progression in fission yeast. It is clear from the plot of the frequency of Plo1 association with the SPB against cell length in Figure 7B that Plo1 associated with the SPBs of small G2 cells in stf1.1 but not wild-type cells. One possible explanation for the variation in the localization of Plo1 to the SPB in stf1.1 mutants is a variation in the processing conditions between different experiments. To control for such possible variations in processing between experiments, asynchronous cultures of either wild-type or stf1.1 cells were mixed with cdc7.A20 arrested cells before immunofluorescence microscopy. After processing for immunofluorescence microscopy, it was trivial to distinguish the cdc7.A20 stf1+ long multinucleate cdc7.A20 cells from the shorter, mononucleate or binucleate cdc7+ stf1.1 cells and to score the frequency of cells with Plo1 on the SPB of each strain independently. Using this data we established that Plo1 associated with the SPB in 2.9 times as many cells in a stf1.1 culture as in the isogenic stf1+ control. In parallel samples double-stained with antibodies to Plo1 and Sad1, Sad1 antibodies stained all SPBs, indicating that the difference in staining of the SPBs with the Plo1 antibodies did not reflect a general reduction in antibody accessibility to the SPB. To rule out the possibility that the variation in Plo1–SPB association reflected a variation in epitope exposure, the fluorescence emerging from the fusion protein in which Plo1 is tagged at its amino terminus with GFP was monitored in living stf1+ control and stf1.1 strains. Of the stf1.1 cells, 9% had a single spot of SPB fluorescence, whereas only 1% were seen in wild-type cells (n = 400). Moreover, 10% of stf1.1 cells in which cell cycle progression had been arrested by treatment with hydroxyurea for 3 h had strong SPB staining with Plo1 antibodies, whereas isogenic stf1+ controls had no staining. We conclude that Plo1 associated prematurely with the SPB in a stf1.1 mutant background.
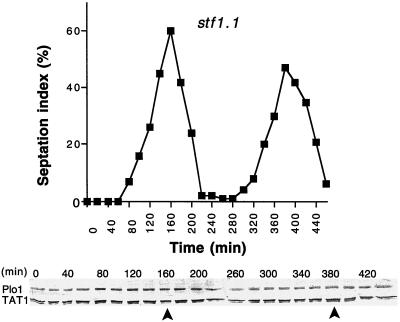
To determine the kinetics of SPB association of Plo1 in stf1.1 cells, a culture of stf1.1 cells that had been synchronized with respect to cell cycle progression by elutrient centrifugation were processed for immunofluorescence microscopy. To control for variations in processing between different time points, asynchronous cell cycle-arrested cdc7.A20 cells were mixed with the synchronized cells before processing for immunofluorescence. The ratio of Plo1 association with the SPBs of the small mononucleate or binucleate stf1.1 cells relative to that in the long multinucleate cdc7.A20 cells varied with cell cycle progression. SPB association increased in G2 and was maximal just before maximal septation. It subsequently declined to its lowest level after septation before increasing once again early in the next cell cycle (Figure 7C). At all stages, more cdc7+ stf1.1 cells had Plo1 associated with the SPB than the cdc7.A20 stf1+ cells.
A trivial explanation for the increased affinity for the SPB in stf1.1 mutants could be that the interphase levels of Plo1 protein are elevated in stf1.1 cells. We therefore analyzed Plo1 protein levels in a culture of stf1.1 cells that had been synchronized with respect to cell cycle progression. Plo1 protein levels remained constant throughout the cell cycle (Figure 8). Therefore we conclude that Plo1 is prematurely recruited to the SPB in strains harboring the stf1.1 mutation.
Figure 8.
Plo1 protein levels remain constant as stf1.1 cells progress through the cell cycle. A cell cycle-synchronized population of stf1.1 cells (IH666) was generated by elutrient centrifugation at 25°C in supplemented EMM2. The small G2 cells were harvested at the beginning of the experiment, and samples were then taken every 20 min for Western blot analysis and scoring the septation index (n = 200 for each time point). The top panel shows the septation index, which indicates good cell cycle synchrony in the culture, whereas the bottom panel shows a Western blot of whole-cell extracts. The blot was cut into two, and the upper portion was probed with antibodies to Plo1 and the lower portion with antibodies to a tubulin (TAT1). The tubulin blot acts as a loading control. The position of the peaks seen in the septation index are indicated by arrows beneath the blots. The relative intensity of the TAT1 and Plo1 bands for each time point were determined for three independent Western blots of both this culture and another synchronous stf1.1 culture. Plo1 levels showed no significant variation during the time course of either experiment (a total of four distinct cell cycles).
Plo1 and Stf1 Associate with the SPB Independently of the Presence of Each Other
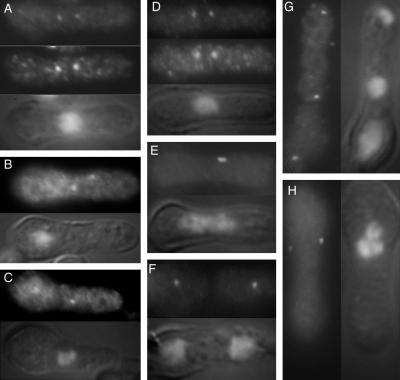
One possible explanation for the premature recruitment of Plo1 to the SPB in stf1.1 cells could be that Plo1 binds directly to Stf1/Cut12 and that the stf1.1 mutation results in inappropriate promotion of this association in a proportion of cells. A prediction of this model is that Plo1 should not bind to the SPB in the absence of Stf1/Cut12 protein; however, Plo1 associated with both of the mitotic SPBs of cells containing the recessive cut12.1 temperature-sensitive mutation at the restrictive temperature. Plo1 also localized to both mitotic SPBs of germinating spores in which the stf1/cut12 gene had been replaced with ura4+ (Bridge et al., 1998) (Figure 9A–C). Conversely Stf1/Cut12 localized to the SPBs of germinating spores in which the plo1+ gene had been replaced with ura4+ (Ohkura et al., 1995) (Figure 9D–H). Thus Plo1 does not appear to require functional Stf1/Cut12 protein to associate with the mitotic SPB and vice versa.
Figure 9.
Plo1 associates with the SPB of stf1/cut12 mutants and Stf1/Cut12 associates with the SPB of cells lacking Plo1. (A–C) Germinating spores from a homozygous ura4.d18 diploid strain in which one copy of the stf1/cut12+ gene had been replaced with the ura4+ gene were inoculated into media lacking uracil and incubated at 30°C for 20 h before processing for immunofluorescence microscopy. The top panels show anti-Plo1 staining, whereas the middle panel shows the result of simultaneous staining with a sheep anti-Sad1 antibody. The bottom panel in each image is a combined DAPI/Nomarski image of the cell in the other panels. The cells in B and C were stained only with Plo1 antibodies. Plo1 localizes to each SPB in all cases, regardless of their lack of Stf1/Cut12 and the inherent inability to execute mitosis. (D–H) Germinating spores from a homozygous ura4.d18 diploid strain in which one copy of the plo1+ gene had been replaced with the ura4+ gene were inoculated into media lacking uracil and incubated at 30°C for 14 h before processing for immunofluorescence microscopy. The top panels in D–F and the left-hand panels in G and H show anti-Stf1/Cut12 staining, whereas the bottom panel in D–F and the right-hand panels in G and H show DAPI DIC images. The middle panel in D shows the result of simultaneous staining with a sheep anti-Sad1 antibody. The cells in E–H were stained only with Stf1/Cut12 antibodies. Stf1/Cut12 localizes to each SPB in all cases, regardless of their lack of Plo1.
DISCUSSION
The original Drosophila polo1 mutation resulted in highly pleiotropic phenotypes (Sunkel and Glover, 1988). This appears to be because the hypomorphic nature of the mutation reveals roles for the enzyme at more than one stage of cell division. This is accentuated by differences in cell cycle regulation in the embryo, in imaginal cells, and in meiosis (Carmena et al., 1998). Recent studies of the function of Plks in budding yeast, Xenopus, and mammalian cells point to roles in the regulation of commitment to mitosis, spindle formation, pole maturation, the regulation of APC activity, and in cytokinesis (see INTRODUCTION) (for review, see Glover et al., 1998; Nigg, 1998). Our present characterization of the localization of the fission yeast polo-like kinase Plo1 suggests that recruitment of Plo1 to the SPB may be of key importance in regulating the timing of mitosis and possibly spindle formation.
Plo1 Association with the Mitotic SPB
We have shown that native Plo1 and Plo1 GFP fusion proteins associate with the fission yeast SPB during the early stages of mitosis. Our results extend and are consistent with those of Bahler et al. (1998). They showed that the endogenous fluorescence of a fusion protein in which the coding sequence for GFP had been fused to the 3′ end of the plo1+ ORF resulted in the appearance of one or two nuclear associated dots and that a line was often seen connecting two dots. They found that the dots disappeared during anaphase B. It is most likely from our observations that these dots are the SPBs and that the line between them represents the spindle. Bahler et al. (1998) also recorded a ring of Plo1GFP fluorescence that is seen in the middle of the cell at the time when the prophase actin ring would be forming. Our failure to detect such a ring would be consistent with the demonstration that the signal of this ring was disrupted by the fixation protocols that we have used.
SPB association of Plo1 is strong from mitotic commitment until just after the initiation of anaphase B. A Plo1 signal is still seen at the SPB until spindle dissolution, although the signal is very faint. It remains to be determined whether all of the Plo1 in the cell is associated with the equatorial ring and SPB at these stages or whether there is an additional pool of Plo1 kinase executing additional functions at other cellular locations. If there is more Plo1 present in the cell, it is unclear whether the Plo1 that we detect on the SPB is anchored there from the time that the signal is first seen until it goes away again. The staining of the SPB with GFP Plo1 fusion proteins and with antibodies that recognize Plo1 may have identified Plo1 molecules transiently associating with the SPB. In this scenario Plo1 may require SPB association for activation before diffusing away to execute specific functions at distal sites. In such a situation the SPB would merely be a loading site for the formation of an active complex so that controlled biochemical events at the SPB would have global consequences. This speculation would be testable by photobleaching of the GFP-tagged Plo1 at the SPB to determine whether the SPB remains dim throughout the remainder of mitosis.
Stability of Plo1
In budding yeast, Cdc5p activates the APC and is degraded by APC at the end of anaphase (Charles et al., 1998; Shirayama et al., 1998). Similarly, Plk protein levels fluctuate in mammalian cells, although it is unclear whether this is due to cell cycle-regulated degradation or to the constitutive turnover of proteins that are transcribed periodically; however, Fang and coworkers (1998) report that in Xenopus extracts Plk1 is an APC substrate. In contrast, in S. pombe Plo1 levels do not undergo drastic fluctuations at any cell cycle stage, suggesting that regulation is mediated by posttranslational modification, compartmentalization, or association with regulatory subunits; however, it is difficult to rule out the possibility that discrete subpopulations of Plo1 undergo destruction at specific locations.
Anaphase Redistribution as a Common Theme for Polo Kinases
The redistribution of Plo1 during anaphase and the need for APC function to accomplish this rearrangement is reminiscent of other systems. In S. cerevisiae, Cdc5p is associated with the SPB until APC activation (Shirayama et al., 1998). In mammals and Drosophila, Plks redistribute from the spindle poles and the kinetochores to the interzone microtubules during anaphase to finally partition to the midbody during telophase (Golsteyn et al., 1995; Lee et al., 1995; Adams et al., 1998; Logarinho and Sunkel, 1998). This redistribution is possibly mediated in part by the MKLP kinesin-related protein, which is the CHO1 antigen in mammalian systems and the product of the pavarotti gene in Drosophila (Nislow et al., 1992; Lee et al., 1995; Adams et al., 1998). Polo associates with this kinesin-related protein in both systems, and mammalian Plk can phosphorylate MKLP1. Mutation of pavarotti results in a considerable reduction in the number of interzone microtubules and abolishes Polo relocation to the interzone. Conversely, polo mutants fail to relocate Pavarotti protein to the interzone of late anaphase and telophase spindles in male meiosis. Mutation of either gene leads to defects in cytokinesis (Adams et al., 1998; Carmena et al., 1998). These data suggest that Polo redistribution to the midzone on microtubules is required for it to correctly regulate cytokinesis in higher systems. Moreover, Polo may also regulate its own relocation by controlling the activity of the MKLP1 motor.
The maintenance of the central position of the septum when the interzone has wandered >10 μm from the centroid (Hagan and Hyams, 1988) indicates that cytokinesis in fission yeast is independent of the central overlap zone of the fission yeast mitotic spindle. Moreover, Plo1 is only detected at the site of cytokinesis much earlier during mitosis (Bahler et al., 1998); however, despite these differences in the organization and execution of cytokinesis and Plo1 distribution, the fact that polo kinases are required for cytokinesis in such diverse systems suggests that although the spatial coordination of polo kinase function may be different in different systems, the downstream effectors in these diverse systems may be similar.
SPB Association and the Control of Septation
The localization of the septation regulator Spg1 to the SPBs raises the possibility that Plo1 recruitment to the SPB may be a crucial event in the regulation of septation. Activation of Spg1 results in a physical interaction with the protein kinase Cdc7 and the recruitment of Cdc7 to the SPB (Sohrmann et al., 1998). Thus, Plo1 association with the SPB at the beginning of mitosis may enable the Spg1 pathway.
If SPB-associated Plo1 does indeed regulate the commitment to septation by activating the SPB-bound Spg1 pathway, it should be possible to bypass this requirement by activating the downstream Spg1 cascade independently of SPB-bound Plo1. We show that this does in fact happen. Spg1 overexpression or Cdc16 inactivation drives inappropriate septation in interphase in the absence of previous SPB association of Plo1. The ability of Plo1 to both drive septation and recruit Cdc7 to the SPB upon overexpression would also put Plo1 upstream of Spg1; however, whether this is a direct or indirect effect is currently still unclear. There must be some feedback, however, from the Spg1 pathway because Plo1 was inappropriately recruited to the interphase SPB by inactivation of Cdc7 protein kinase. Thus, loss of the function of the Spg1 effector increases the affinity of Plo1 for the SPB in interphase; however, there are some problems with such a straightforward interpretation because it is unclear why the cdc11 mutant does not have an equivalent phenotype. Genetic data suggest that cdc11+ acts antagonistically to cdc16+ (Marks et al., 1992). Thus, loss of Cdc11 function might be expected to have the same consequences as loss of Cdc7. A simple explanation is that the influence of Cdc7 on Plo1 recruitment to the pole is a functionally distinct role compared with its previously characterized role in Spg1 association.
It is equally possible that association of Plo1 with the SPB may be entirely independent of its function in septation. In this case, its role at the SPB may only be required until chromosome segregation is initiated, and consequently it is lost from the SPB after activation of the APC, as we have observed. The reduction of the affinity of Plo1 for the SPB could also require a specific function of Cdc7, which also undergoes reorganization at the SPBs at this time. Whichever model emerges as reflecting the in vivo status, it is significant that Plo1 can remain at the SPB in cells in which mitosis is blocked as a result of a number of mutations and yet cytokinesis still occurs. This indicates that disassociation of Plo1 from the SPB is not a requirement for cytokinesis.
Localization of Plo1 to the SPB and Commitment to Mitosis
It has been established previously that the formation of the fission yeast mitotic spindle occurs in the presence of an intact cytoplasmic microtubule cytoskeleton (Hagan and Yanagida, 1995). Because this array persists throughout interphase, it suggests that full commitment to mitosis is not an instantaneous process in fission yeast. It is therefore significant that Plo1 associates with the SPB to give a single dot of fluorescence for a considerable time before the presence of two distinct SPBs signal the formation of the prophase spindle. Thus, there is an intact interphase cytoskeleton when there is a single spot of Plo1 on the SPB. Because association of Plo1 with the SPB is dependent on previous activation of MPF, it is the earliest biochemical or cytological mitotic event that has been determined for fission yeast to date.
Commitment to mitosis involves the activation of a positive feedback loop whereby a trigger level of MPF is amplified by a downstream kinase through the activation of p34cdc2 phosphatase Cdc25 (Hoffmann et al., 1993; Kuang et al., 1994; Izumi and Maller, 1995; Kovelman and Russell, 1996). Data from the analysis of Plx1 support the notion that polo kinases play a key role in this feedback loop (Kumagai and Dunphy, 1996; Abrieu et al., 1998; Qian et al., 1998a). The timing of Plo1 association with the SPB would be coincident with the activation of this feedback loop. It has been reported previously that semidominant mutants in the SPB component encoded by the stf1/cut12+ gene bypass the requirement for Cdc25 activation of MPF (Hudson et al., 1990; Bridge et al., 1998). If Wee1 is inhibited at the same time that Cdc25 is activated during the amplification loop, it is entirely possible that cells harboring the stf1.1 mutation no longer require Cdc25 function because they are inappropriately triggering the feedback loop to inhibit Wee1 in the absence of previous stimulation by MPF. If this were the case, premature mitotic commitment would be inhibited by a dominant size control that probably acts through the regulation of Wee1 (Nurse, 1975; Fantes, 1977; Fantes and Nurse, 1977). Once the restraint of this size control is removed in stf1.1 mutants, the inappropriate activation of the feedback loop would drive cells into mitosis.
The ability of Plo1 to associate with the SPBs of cells in which the stf1/cut12+ gene has been mutated recessively to abolish its function suggests that the effect of Stf1/Cut12 on Plo1 recruitment is unlikely to be mediated by a direct interaction between the two proteins, or if it is, that Plo1 interacts with multiple SPB components. The recently identified polo kinase kinase that is distinct from p34cdc2 in Xenopus could be a candidate for the kind of molecule that could bridge an indirect link between Stf1/Cut12 and Plo1 function (Qian et al., 1998b). Whatever the molecular basis underlying this phenomenon, the premature localization of Plo1 to the SPB in stf1.1 mutants reveals a close relationship between Plo1 recruitment to the SPB and mitotic commitment. Together the data that we present support the notion that association with the spindle poles could be a key mechanism through which Plks regulate mitosis and cytokinesis.
ACKNOWLEDGMENTS
We thank Professor K. Gull and Dr. K. Sawin for the generous gifts of TAT1 and GFP antibodies, respectively. We thank Dr. Kayoko Tanaka for stimulating discussions and Anna Poziemba for cdc11.IH1. This work forms part of a collaboration between the Dundee and Manchester groups and was supported by the Cancer Research Campaign. H.O. is a Wellcome senior research fellow for biomedical science.
Abbreviations used:
- APC
anaphase promoting complex
- GFP
green fluorescent protein
- HA
hemagglutinin
- MPF
mitosis promoting factor
- SPB
spindle pole body
REFERENCES
- Abrieu A, Brassac T, Galas S, Fisher D, Labbé JC, Dorée M. The Polo-like kinase Plx1 is a component of the MPF amplification loop at the G2/M-phase transition of the cell cycle in Xenopus eggs. J Cell Sci. 1998;111:1751–1757. doi: 10.1242/jcs.111.12.1751. [DOI] [PubMed] [Google Scholar]
- Adams RR, Tavares AM, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Steever AB, Wheatley S, Wang Y-L, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Morphew M, Bartlett R, Hagan IM. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Riparbelli MG, Minestrini G, Tavares A, Adams R, Calliani G, Glover DM. Drosophila polo kinase is required for cytokinesis. J Cell Biol. 1998;143:659–671. doi: 10.1083/jcb.143.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM, Moudjou M, Bentley NJ, Hagan IM. The Chk1 pathway is required to prevent mitosis following cell-cycle arrest at start. Curr Biol. 1995;5:1179–1190. doi: 10.1016/s0960-9822(95)00234-x. [DOI] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Cheng L, Hunke L, Hardy CFJ. Cell cycle regulation of the Saccharomyces cerevisiae Polo-like kinase Cdc5p. Mol Cell Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJF, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Descombes P, Nigg E. The polo-like kinase Plk1 is required for M phase exit and destruction of mitotic regulators in Xenopus egg extracts. EMBO J. 1998;17:1328–1335. doi: 10.1093/emboj/17.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG. The decision to enter mitosis. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- Egel R, Willer M, Kjaerulff S, Davey J, Nielsen O. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 1994;10:1347–1354. doi: 10.1002/yea.320101012. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase promoting complex in mitosis and G1. Mol Cell. 1998;2:163–71. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1997;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Simanis V. The Cdc7 protein-kinase is a dosage-dependent regulator of septum formation in fission yeast. EMBO J. 1994;13:3011–3019. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P. Control of cell size and cell cycle time in Schizosaccharomyces pombe. J Cell Sci. 1977;24:51–67. doi: 10.1242/jcs.24.1.51. [DOI] [PubMed] [Google Scholar]
- Fantes P, Nurse P. Control of cell size at division in fission yeast by growth-modulated size control over nuclear division. Exp Cell Res. 1977;107:377–386. doi: 10.1016/0014-4827(77)90359-7. [DOI] [PubMed] [Google Scholar]
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two component GTPase activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Glover DM, Hagan IM, Tavares A. Polo kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell-cycle regulation of the activity and subcellular-localization of Plk1, a human protein-kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell-division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hirano T, Funahashi S, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human Cdc25-C by Cdc2 cyclin-B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JD, Feilotter H, Young PG. stf1: non wee mutations epistatic to cdc25 in the fission yeast Schizosaccharomyces pombe. Genetics. 1990;126:309–315. doi: 10.1093/genetics/126.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Maller J. Phosphorylation and activation of Xenopus Cdc25 phosphatase in the absence of Cdc2 and Cdk2 kinase activity. Mol Biol Cell. 1995;6:215–226. doi: 10.1091/mbc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani S, Tugendreich S, Fujii M, Jorgensen PM, Watanabe N, Hoog C, Hieter P. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- Kovelman R, Russell P. Stockpiling of Cdc25 during a DNA-replication checkpoint arrest in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:86–93. doi: 10.1128/mcb.16.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang J, Ashorn CL, Gonzalea-Kuyvenhoven M, Penkala JE. Cdc25 is one of the MPM-2 antigens involved in the activation of maturation-promoting-factor. Mol Biol Cell. 1994;5:135–145. doi: 10.1091/mbc.5.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Purification and molecular-cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- Lee KS, Yuan YO, Kuriyama R, Erikson R. Plk is an M-phase specific protein kinase and interacts with a kinesin-like protein. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. Polo encodes a protein-kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- Logarinho E, Sunkel CE. The Drosophila POLO kinase localizes to multiple compartments of the mitotic apparatus and is required for the phosphorylation of MPM2 reactive epitopes. J Cell Sci. 1998;111:2897–2909. doi: 10.1242/jcs.111.19.2897. [DOI] [PubMed] [Google Scholar]
- Marks J, Fankhauser C, Simanis V. Genetic interactions in the control of septation in Schizosaccharomyces pombe. J Cell Sci. 1992;101:801–808. doi: 10.1242/jcs.101.4.801. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Minet M, Nurse P, Thuriaux P, Mitchison M. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1981;137:440–446. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM, Carter BLA. Cell cycle analysis. In: Prescott DM, editor. Methods in Cell Biology. New York: Academic; 1975. pp. 201–219. [PubMed] [Google Scholar]
- Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic-analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Polo-like kinases: positive regulators of cell division from start to finish. Curr Opin Cell Biol. 1998;10:776–783. doi: 10.1016/s0955-0674(98)80121-x. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase Plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Qian YW, Erikson E, Li C, Maller JL. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol Cell Biochem. 1998a;18:4262–4271. doi: 10.1128/mcb.18.7.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y-W, Erikson E, Maller JL. Purification and cloning of a protein kinase that phosphorylates and activates the polo-like kinase Plx1. Science. 1998b;282:1701–1703. doi: 10.1126/science.282.5394.1701. [DOI] [PubMed] [Google Scholar]
- Samejima I, Yanagida M. Bypassing anaphase by fission yeast cut9 mutation: requirement of cut9+ to initiate anaphase. J Cell Biol. 1994a;127:1655–1670. doi: 10.1083/jcb.127.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima I, Yanagida M. Identification of cut8+ and cek1+, a novel protein-kinase gene, which complement a fission yeast mutation that blocks anaphase. Mol Cell Biol. 1994b;14:7683–7683. doi: 10.1128/mcb.14.9.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Sohrmann M, Hofmann K, Woolard A, Simanis V. The Spg1 GTPase is an essential dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V, Nurse P. The cell-cycle control gene cdc2+ of fission yeast encodes a protein-kinase potentially regulated by phosphorylation. Cell. 1986;45:261–268. doi: 10.1016/0092-8674(86)90390-9. [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Wianny F, Tavares A, Evans MJ, Glover DM, Zernicka-Goetz M. Mouse polo-like kinase 1 associates with the acentriolar spindle poles, meiotic chromosomes and spindle midzone during oocyte maturation. Chromosoma. 1998;107:430–439. doi: 10.1007/s004120050327. [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, Macrae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal-antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature. 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]