Abstract
Glycosphingolipids are ubiquitous constituents of cells. Yet there is still room for improvement in the techniques for analyzing glycosphingolipids. Here we report our highly sensitive and convenient analytical technology with imaging mass spectrometry for detailed structural analysis of glycosphingolipids. We were able to determine detailed ceramide structures; i.e. both the sphingosine base and fatty acid, by MS/MS/MS analysis on a PVDF membrane with 10 pmol of GM1, with which only faint bands were visible by primuline staining. The limit of detection was approximately 1 pmol of GM1, which is lower than the value in the conventional reports. (10 pmol)
Keywords: Thin-layer chromatography, Matrix-Assisted Laser Desorption/Ionization, Glycosphingolipid
1. Introduction
Glycosphingolipids (GSLs), consisting of a hydrophilic carbohydrate chain and a hydrophobic ceramide moiety, are amphipathic molecules and ubiquitous constituents of cell membranes, where they appear to be involved primarily in biological processes such as cell proliferation, differentiation, cell-to-cell recognition, and signaling[1–5]. The human brain contains many GSLs, especially gangliosides, and it is thought that GSLs play important roles in brain functions. Many brain diseases, for example, Tay-Sachs disease[6–10] have been shown to be related to GSLs. To investigate the mechanism of brain function in more detail, it is necessary to understand the composition and function not only of proteins as we have reported previously[11–14] but also of GSLs. However, a more highly sensitive method than the existing ones is required to investigate the total GSL composition in the brain.
Thin-layer chromatography (TLC) is routinely used for separation and partial characterization of neutral and acidic GSLs and phospholipids in mixtures[15–17]. However, even under optimized TLC conditions, TLC characterization of individual GSLs does not yield unambiguous structural information. The information obtained for the composition of these highly complex mixtures remains limited; namely, GSLs that contain the same sugar may migrate to different positions owing to differences in their ceramide structures. Unless combined with additional methods, e.g., with immunoassays, species of similar structure and mobility cannot be differentiated.
Mass spectrometry (MS) is an indispensable tool for the structural characterization of GSLs. Both electrospray ionization (ESI)[18] and matrix-assisted laser desorption/ionization (MALDI)[19] mass spectrometry have been applied for the analysis of GSLs[20–23]. MALDI MS can ionize GSLs even in the presence of contaminated substances, and the introduction of samples into the MS instrument is simpler in MALDI MS than in ESI MS[24–26]. However, it is difficult to identify individual molecular species by MS alone. Mass spectrometry cannot fully supply sugar sequences because metastable decay of sialic acid yields degenerate spectra with the base peak being the monosialo or asialo peak, nor can it differentiate isobaric species; however, multi-stage MS (MSn) analysis can supply information on each ceramide structure. Although TLC offers information on sugar sequences, only one broad band could be detected with each GSL, which contains distinct ceramide moieties. Combining these data, we could finally obtain total information on GSLs. We consider that both TLC and MS are useful methods to analyze GSLs.
Several methods involving a combination of TLC and MS have been reported. One is the indirect method where identified silica bands are scraped off the TLC plate, and subsequently, the analyte molecules are extracted from the silica gel and purified[27]. This method would be applied to either MALDI MS or ESI MS with high mass accuracy. However, it is not a sufficient method because it is time-consuming work that involves the potential loss of materials, where closely spaced narrow bands that similarly migrate are difficult to scrape off separately. Furthermore, it requires more than one microgram for a single species of GSL to be analyzed.
Another method that combines TLC and MS is direct TLC-MS. This method largely eliminates the disadvantages of the indirect method, thus increasing the utility of TLC for structural characterization[28–33]. Direct TLC-MS presents difficulties with regard to complete retrieval of the target compound from the silica coated on the plate and could also cause contamination of silica to the MS instrument. These difficulties were largely resolved in a method that we previously introduced for the transfer of separated lipids from a high-performance TLC (HPTLC) plate onto a polyvinylidene difluoride (PVDF) membrane; we designated this method as TLC-Blot (Far-Eastern blot)[34]. In a previous study, lipid bands that had been transferred onto a PVDF membrane were cut and introduced onto the target stage for secondary ion MS (SIMS)[33], however the analytical sensitivity and resolution of this method were inadequate. In TLC-MALDI MS or TLC-Blot-MALDI MS, problems related to obtaining good mass accuracy and resolution arise due to the difficulties associated with the desorption of analytes from the rough surface. Recently, remarkable advances achieved in the development of MALDI MS ion trap instruments have enabled TLC-MALDI MS analysis to be performed with high mass accuracy and sensitivity despite the irregular surface of the plate[35]. Some instruments include an external ion trap, such as a Fourier transform ion cyclotron resonance ion trap (FTICR)[28,29] and an orthogonal extracting ion trap[30,31] could also apply to TLC-MALDI MS. Developed GM1 was difficult to be identified even by using highly sensitive TLC-FTICR MS at the amount of 120 fmol[28]. TLC coupled with orthogonal (o)-time-of-flight (TOF) MS was also used to determine molecular masses and partial sugar sequences of gangliosides with high resolution; in particular, the attachment positions of the sialic acids were determined[30].
In another study, GSLs developed on a TLC plate were transferred to a PVDF membrane then coupled to MALDI MS; this was called the TLC-Blot-MALDI MS method[35–37]. There have been only a few recent reports on TLC-Blot-MALDI MS, because the additional work, i.e., the transfer from TLC plates to PVDF membranes, requires, and the orthogonal configuration of the instrument allows for direct desorption of samples from the irregular surface of the TLC plate without impairment of either mass accuracy or resolution[38]. However Costello et al. have mentioned in their report that the spectral quality is better on PVDF membranes than on TLC plates, in terms of sensitivity, mass resolution, and background interference[36]. We showed that GSLs were concentrated on the reverse surface of the transferred membrane[39]. The membrane is available for further biochemical processes, e.g. enzyme reactions, and is easier to handle than the silica-coated plate. Moreover, it has an advantage in terms of storage: GSLs on PVDF membranes are stable at room temperature for a longer time. Thus, we considered that TLC-Blot-MALDI MS is better than TLC-MALDI MS for analysis of minor components in small amounts of biologically complex mixtures.
The aim of the previous study was to analyze the visible GSLs with the conventional staining method; i.e., GSLs were analyzed based on the combined information from TLC and MS. Due to the similarity of the properties of the procedures, the limits of detection of these studies were the same as that of the conventional staining method; 10 pmol[30,31]. However, we tried to detect far more minor components to fully identify the components of the sample, regardless of the detection limit of staining, by the use of the imaging mass spectrometry method.
Imaging mass spectrometry (IMS) (or mass microscopy) that utilizes MALDI technology can provide a molecular ex vivo view of dissected organs or whole-body sections from an animal, making possible the label-free tracking of both endogenous and exogenous compounds with sufficient spatial resolution and molecular specificity of targets[40–49]. Though there has been a report in which distribution of glycolipids species was analyzed by raster scan of TLC bands[30], the work did not realize a detailed structural analysis of that particular ceramide species. Recently we reported the system of imaging mass spectrometry by MALDI-quadrupole ion trap (QIT)-TOF MS. This procedure enabled us to identify molecular species with high-resolutional MSn analyses from mixture ions[48].
There are plural molecular species in GSLs, depending on the differences in the chains of fatty acids and sphingoids in ceramides, but it is impossible to differentiate and detect all of these species by TLC. In the present work, we performed not only imaging of GM1 on a PVDF membrane to visualize and obtain the relative to front (Rf) value of each molecular species, but also structural analyses of the oligosaccharide chain and identification of molecular species of ceramides by conducting MS/MS and MS/MS/MS analyses. Because the detection limit of GSLs depends on the MS sensitivity in this case, the sensitivity is expected to be increased compared to the detection using the conventional staining method. As a result, we demonstrated that as little as about 1 pmol GM1 is detectable, which has not been the case thus far. By combining the MS information with the imaging technology, we are now able to visualize analytes and obtain the Rf values of different GSLs in ceramide moieties that have been impossible to discriminate with the previous detection methods. Indeed, imaging of the GM1 band that appeared singular with primuline staining successfully discriminated each molecular species, and even the structural analysis of ceramide molecular species-i.e., that of the sphingosine base and fatty acid- is possible by including the result of MS/MS/MS analysis. This procedure is very easy to conduct and needs no specific equipment to discriminate analytes or specific antibodies to detect them; usually, only one-time TLC and the transfer to a PVDF membrane make it possible to separate, visualize, and identify GSLs at a low picomole level. It is useful to detect minor components of GSLs and each molecular species and to identify ceramide structures in detail. We consider that this system will be useful in fully investigating GSL compositions including minor components.
2. Experimental
2.1. Chemicals
GM1 was purchased from Wako Pure Chemical Industries (Osaka, Japan). All solvents used for MS were of HPLC grade and were purchased from Kanto Chemical Co., Inc. (Tokyo, Japan). Bradykinin, angiotensin-II, and ACTH were obtained from Sigma-Aldrich Japan (Tokyo, Japan) and used as calibration standards. 2, 5-Dihydroxy benzoic acid (DHB) obtained from Bruker Daltonics (Leipzig, Germany) was used as the matrix.
2.2. Thin-layer chromatography
GM1 dissolved in chloroform/methanol (1/1, v/v) was applied as 5-mm spots to silica gel 60 HPTLC plates (Merck, Darmstadt, Germany). Plates were developed with a solvent system of chloroform/methanol/0.2% aqueous CaCl2, (55/45/10, v/v/v). After developing of GM1, the HPTLC plate was sprayed with primuline reagent until the plate became wet and then was air-dried thoroughly. GM1 bands were visualized under UV light at 365 nm.
2.3. TLC-Blot
The HPTLC plate was dipped in the blotting solvent (2-propanol/0.2% aqueous CaCl2/methanol, 40/20/7, v/v/v) for 20 s, after which the HPTLC plate was immediately placed on a flat glass plate. First, a PVDF membrane, a Teflon membrane, and then a glass fiber filter sheet were placed over the plate. This assembly was pressed evenly for 10–15 s with a TLC thermal blotter (ATTO, Tokyo, Japan). The PVDF membrane, removed from the HPTLC plate, was then air-dried. Samples were transferred to the PVDF membrane facing the HPTLC plate, but the GSLs were exclusively located on the reverse side.
2.4. MALDI-QIT-TOF MS
MALDI-QIT-TOF MS analyses were performed using AXIMA-QIT mass spectrometer (Shimadzu, Kyoto, Japan) in negative ion mode. Ionization was performed with a 337-nm pulsed N2 laser. The ion-trap chamber was supplied with two separate and independent gases, helium and argon. A continuous flow of helium gas was used for collisional cooling. The pulsed gas, argon, was used for collision-induced fragmentation[50]. Precursor and fragment ions obtained by collision-induced dissociation (CID) were ejected from the ion trap and analyzed by a reflectron TOF detector.
The MS spectra were calibrated externally using a standard peptide calibration mixture containing 10 pmol/µl each of bradykinin peptide fragment (amino acid residue 1–7) ([M-H]−, m/z 755.40), human angiotensin-II peptide fragment ([M-H]−, m/z 1044.54), and human ACTH peptide fragment (amino acid residue 18–39) ([M-H]−, m/z 2463.19). 50 mg/ml DHB in methanol/0.1% TFA solution (1/1, v/v) was used as the matrix. DHB solution (totaling 10–20 µl) was deposited sufficiently to GSLs on the PVDF membrane, which was transferred by the heat iron method from TLC plates. The crystallization process was accelerated under a gentle stream of cold air. To achieve good mass spectra, we pressed the PVDF membrane thoroughly so as to flatten its surface and remove the excess matrix crystals. The PVDF membrane was attached to a MALDI sample plate with electrified double-adhesive tape to reduce charge-up of the plate.
2.5. Imaging mass spectrometry
The raster scan of the PVDF membrane was performed automatically. The number of laser irradiations was 5 shots in each spot. The analyzed area was approximately 8×4 mm2; it was scanned in both the X and Y directions to produce an array of 81×41 (3321 profiles): the interval of data points was 100 µm. A data converter (Axima2Analyze, Novartis, Basel, Switzerland) was developed to generate a BioMap readable data file from imaging data of AXIMA-QIT. Two-dimensional ion density maps were created using the image reconstruction software (BioMap, Novartis).
2.6. Analysis of glycosphingolipids obtained from the brain of a patient with Tay-Sachs disease
Crude GSL extract was prepared from the brain sample (75 g) of a patient with Tay-Sachs disease variant B. The GSL extract was dissolved in 70 ml of chloroform/methanol/water (30/60/8, v/v/v) and was applied onto a Q-Sepharose column (2.5×75 cm) that had been equilibrated with the same solvent. After the column was washing the abovementioned solvent, the gangliosides were eluted with a linear gradient of sodium acetate. The flow rate was 1.0 ml/min, and 7-ml fractions were collected. The detailed procedure has been described previously[51]. Fractions 186–196 were pooled and developed by TLC, the solution of the pooled fractions was designated as fraction X. The solvent system was chloroform/methanol/0.2% aqueous CaCl2, (55/45/10, v/v/v), and the GSLs were visualized by primuline staining.
3. Results and discussion
3.1. The detection of GM1 by TLC-Blot-MALDI-QIT-TOF MS
Gangliosides belong to a family of complex glycosphingolipids (GSLs) and consist of an oligosaccharide head group containing at least one sialic acid residue, attached to a hydrophobic ceramide anchor. This class of compounds presents a challenge for a mass spectrometric analysis because of the ready elimination of sialic acid[52] and the high degree of heterogeneity that frequently characterizes both the carbohydrate and the ceramide portions[53]. Although the glycoepitope is known to be involved in the GSL properties, the ceramide structure is also implied as it may influence the glycosylation pattern of the GSL, their presentation at the cell surface, the degree of clustering of receptor molecules, and the signaling pathways[54–56]. Many studies have shown the complexity of the ceramide moiety of GSLs. A previous report showed that bovine brain GM1 contains equal amounts of two major bases, sphingenine (d18:1) and eicosasphingenine (d20:1), and other bases at less than 5%[57,58]. Carbohydrate sequences of bovine brain GM1 and its major ceramide molecular species are shown in Fig. 1(A) and (B), respectively.
Fig. 1. Molecular structures of GM1 used in this study.
Abbreviations correspond to the nomenclature of Svennerholm et al.[59]. It has been shown that major molecular species of GM1 from the bovine brain were d18:1/18:0 shown in (A) and d20:1/18:0 shown in (B)[57,58].
Gangliosides are routinely analyzed in the negative ion mode in order to reduce metastable fragmentation of sialic acid. As we conducted MS analysis of GM1 bands on PVDF membranes transferred from TLC plates, we observed simple spectra in which the [M-H]− ion was detected as the only major component, m/z 1544 (d18:1/18:0) and m/z 1572 (d20:1/18:0) (Fig. 2(A), negative ion mode spectra). Therefore, the system would be proper for the analysis of complex mixtures. Despite irregular surfaces of the GM1 band on PVDF membranes, the ion trap helped to minimize the mass error between the predicted and the observed masses, and it was revealed that the mass accuracy was highly preserved even on PVDF membranes (Table 1). However, as Table 1 shows, there is one more possible molecular species in m/z 1572 of GM1; whether m/z 1572 originates from d20:1/18:0 or d18:1/20:0 could not be determined merely by mass spectra. While m/z 1572 in this case is already known to be d20:1/18:0 from the previous reports[57,58], molecular identification of the corresponding mass spectra in unknown samples would require more detailed MS/MS and MS/MS/MS analyses in practice.
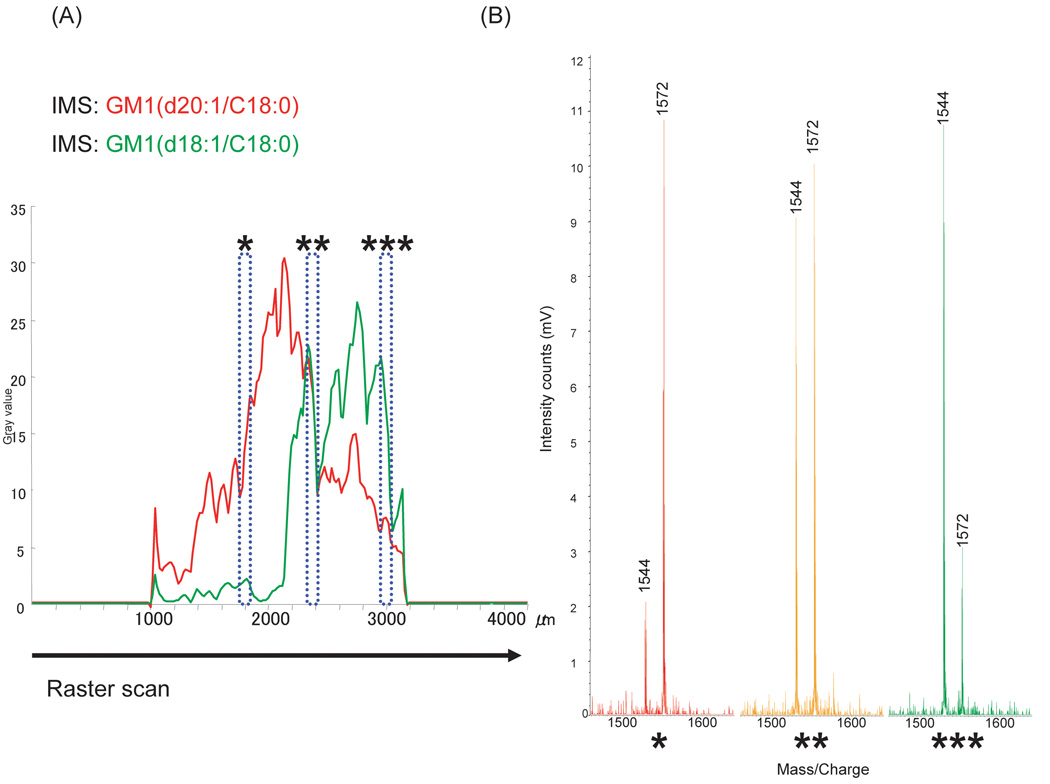
Fig. 2. Imaging mass spectrometry of GM1 by TLC-Blot-MALDI-QIT-TOF MS.
(A) The overall spectrum obtained from a GM1 band on the PVDF membrane. (B) An optical image of GM1 on thin-layer chromatogram stained with primuline. (C) and (D) Ion images obtained from GM1 of different ceramide moieties, m/z 1572 and 1544, respectively. (E) Ion images were merged with m/z 1572 shown in red and m/z 1544 in green. The arrow shows the direction of the raster scan.
Table 1.
Theoretical mass and observed mass of major ceramide species of GM1 from the bovine brain in negative ion mode desorbed directly from the PVDF membrane.
| Ganglioside | Predicted m/z (M-H)− | Observed m/z |
|---|---|---|
| GM1 (d18:1/18:0) | 1544.8 | 1544.8 |
| GM1 (d20:1/18:0) | 1572.9 | 1572.8 |
| GM1 (d18:1/20:0) | 1572.9 | 1572.8 |
3.2. Imaging mass spectrometry of GM1 by TLC-Blot-MALDI-QIT-TOF MS
It is impossible to distinguish the molecular species of GM1, m/z 1544 and m/z 1572, when detected by staining methods on TLC plates or PVDF membranes. Practically, GM1 was revealed as a concentrated band by primuline staining. The molecular species in this band were not distinguished quantitatively (Fig. 2(B)). We attempted in this work to precisely discriminate those molecular species, by applying the technique of IMS. We expected that applying IMS would visualize GSLs and distinguish the minute patterns of GM1 moiety that are developed on TLC based on the information of their mass sizes or hydrophobicity, and enable us to discriminate molecular species in greater detail than the previous procedure.
In the present study, we performed imaging of m/z 1544 (d18:1/18:0) and m/z 1572 (d20:1/18:0) as target molecules of GM1. The result is shown in Fig. 2. Fig. 2(B) shows the optical image of primuline staining. GM1 was observed as a single, concentrated band. Fig. 2(C) is the imaging result of m/z 1572; the observed band appeared concentrated especially in its upper part. By contrast, Fig. 2(D), the imaging result of m/z 1544, showed a stronger intensity in its lower part. Merging of Fig. 2 (C) and (D) gave rise to an image clearly separating these two molecular species between the upper and lower part (Fig. 2(E)), both of which have seemingly appeared as a single band of GM1. The localization of m/z 1572 in the upper part in the TLC development would be explained by the fact that sphingosine base in m/z 1572 is longer than that in m/z 1544 by two carbon chains and that m/z 1572 has relatively higher hydrophobicity.
We conducted IMS with a 100-µm interval between data points, yielding a total of 3321 profiles.Fig. 3(A) shows the densitometric analysis of the ion images. The X-axis indicates the distance from the top to the bottom; the total length of the image area is 4000 µm. The arrow shows the direction of the raster scan. The detection limits was fixed by setting a threshold at gray value 10. As the results show, m/z 1572 was detected in the area of 1700–2800 µm and m/z 1544 was detected in the area 2200–3000 µm from the top. In the area around 2300–2400 µm, m/z 1572 and m/z 1544 were equally detected. By the use of the IMS method, we could analyze the minute patterns of GM1 on the µm-order with fine resolution because we could select an appropriate interval of data points, while the conventional TLC method could not separate and detect each GSL distribution as well. The mass spectrum in each area is shown in Fig. 3(B). The red spectrum in the area nearby 1800 µm from the top exhibited a higher intensity for m/z 1572 than m/z 1544, whereas the yellow spectrum in the area nearby 2400 µm from the top exhibited comparable intensities for m/z 1572 and m/z 1544, and the green spectrum in the area nearby 3000 µm from the top showed a lower intensity for m/z 1572 than m/z 1544. These mass intensities in conjunction with the area information as well showed how each of the molecular species was distributed in the single GM1 band. It seems that IMS method is useful for GSL analyses of a concentrated mixed sample which could not be separated only by TLC. The capability of discriminating the molecular species was examined and proved not only for gangliosides but also for neutral GSLs or phospholipids (data not shown).
Fig. 3. Distributional analysis of GM1 by TLC-Blot-MALDI-QIT-TOF MS.
(A) The densitometric analysis of imaging data acquired from the raster scan of the GM1 band on the PVDF membrane (red, m/z 1572; green, m/z 1544). The X-axis indicates the distance from the top to the bottom pixel. The arrow shows the direction of the raster scan. (B) Semi-quantitative mass spectra acquired from the area with asterisks in (A). Each area of the rectangles with asterisks shows a parallel ratio between m/z 1572 and m/z 1544 in the mass spectrum with the same number of asterisks. (*; m/z 1572 was mainly observed, **; m/z 1572 and 1544 were equally detected, ***; m/z 1544 was mainly observed.)
There have been previous reports in which the detailed distribution of molecular species in a band of glycolipid[30] or phospholipid[38] was shown using mass spectra alone. But the present report on detection of GM1 is quite new in that such a result was visualized by the imaging data with µm-order resolution, and further that structural analyses by MS/MS and MS/MS/MS analyses demonstrated the precise structure of these molecular species.
3.3. The detection limit of GM1 by TLC-Blot-MALDI-QIT-TOF MS
The detection limit of GSLs in TLC-MALDI MS or TLC-Blot-MALDI MS reported thus far is about 10 pmol, which is because the detection of GSLs was at first conducted with primuline staining and then only those detected were used for the analyses. In the present work, the detection sensitivity solely depends on MS, because the visualization of GM1 distribution was attempted with use of IMS, rather than staining. This allowed us to expect a higher sensitivity in detection than that by the conventional methods. Results of the primuline staining of the various-concentration GM1 bands are shown in Fig. 4(A). As the results show, the detection limit of GM1 in this staining method is about 10 pmol, and we were hardly able to detect a band at 1 pmol. We also measured the detection limit of our method by varying the concentration of GM1 applied; namely, 1 nmol (lane 1), 100 pmol (lane 3), 10 pmol (lane 5), and 1 pmol (lane 7). As a result, we were barely able to detect the GM1 species of m/z 1544 (d18:1/18:0) at the level of about 1 pmol (Fig. 4(E)). The signal-to-noise ratios (S/N) were approximately 243, 80, 24, and 5, respectively. Thus, TLC-Blot-MALDI-QIT-TOF MS has almost 10 times higher sensitivity compared to the detection by the conventional staining method and is considered to be a very efficient way to detect ultra trace amounts of GSLs on PVDF membranes.
Fig. 4. TLC-Blot-MALDI-QIT-TOF MS spectra of primuline-stained GM1 on the PVDF membrane (negative ion mode).
(A) Thin-layer chromatogram stained with primuline. Lanes 1–8 contain bovine brain GM1: (lane 1–2) 1 nmol, (lane 3–4) 100 pmol, (lane 5–6) 10 pmol, and (lane 7–8) 1 pmol. The plate was developed with chloroform/methanol/0.2% aqueous CaCl2, (55/45/10, v/v/v), and GM1 spots were detected with primuline staining then transferred to a PVDF membrane. Each GM1 was directly analyzed by TLC-Blot-MALDI-QIT-TOF MS. (B), (C), (D), and (E) are spectra derived from 1 nmol (lane 1), 100 pmol (lane 3), 10 pmol (lane 5), and 1 pmol (lane 7) of GM1, respectively. The detection limit of GM1 was estimated at 1 pmol. The number of laser irradiations was 2 shots in arbitrarily 100 spots per band.
3.4. Analyses of molecular species of GM1 by TLC-Blot-MALDI-QIT-TOF MS
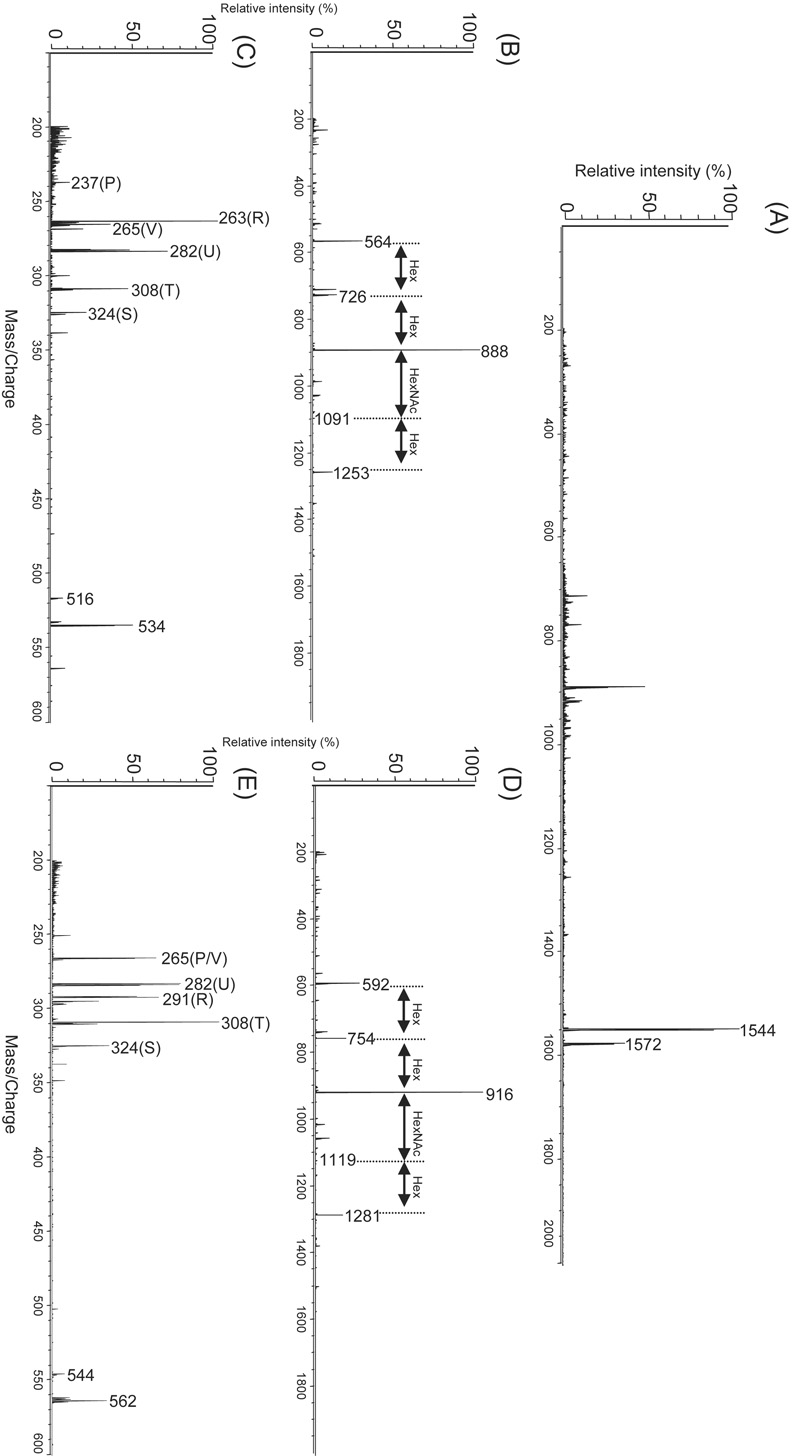
The fact that the difference in molecular weight between m/z 1544 and m/z 1572 of bovine brain GM1 is m/z 28, together with results of some previous reports[57,58], allow us to assume that they are molecular species of a sphingosine base, i.e., d18:1/18:0 and d20:1/18:0, respectively. However, because GM1 with altered fatty acid, d18:1/20:0, shows the same molecular weight of m/z 1572 (Table 1), analyses of the fragmentation pattern of ceramide together with analyses of the fragment ion (S, T, U, V, and P, R) (Fig. 5) that originates from the ceramides shown in Table 2 are needed to definitively identify them as molecular species.
Fig. 5. Nomenclature for cleavages of precursor ions of ceramides.
Annotations reported by Ann et al. are cited[60].
Table 2.
Major fragment ions observed for each molecular species of GM1.
| Negative ions | ||||||||
|---|---|---|---|---|---|---|---|---|
| M | ceramide | Sphingoid chain | Fatty acid | |||||
| Molecular species | [M-H]− | [M-H]− | P[M-H]− | R[M-H]− | S[M-H]− | T[M-H]− | U[M-H]− | V[M-H]− |
| GM1(d18:1/18:0) | 1544 | 564 | 237 | 263 | 324 | 308 | 282 | 265 |
| GM1(d20:1/18:0) | 1572 | 592 | 265 | 291 | 324 | 308 | 282 | 265 |
| GM1(d18:1/20:0) | 1572 | 592 | 237 | 263 | 352 | 336 | 310 | 293 |
The MS analysis of about 1 nmol of GM1 was developed by TLC-Blot-MALDI-QIT-TOF MS (Fig. 6(A)) and the subsequent MS/MS analysis of each of the m/z 1544 and m/z 1572 obtained provided peaks that originated from the discrimination of oligosaccharide chains. These also allowed us to know the sequence information of the oligosaccharide chains (Fig. 6(B) and (D)). We were able to detect peaks that originated from the ceramide, though they were only represented in small amounts (m/z 564 and m/z 592). Then, further MS/MS/MS analysis of these ions as precursors would exhibit fragment patterns specific to each sphingosine base and fatty acid listed in Table 2. Indeed, MS/MS/MS analysis of the ceramide m/z 564 that originates from m/z 1544 exhibited P and R ions originating from d18:1 (m/z 237 and 263, respectively) and S, T, U, and V ions originating from C18:0 (m/z 324, 308, 282, and 265, respectively), while MS/MS/MS analysis of the ceramide m/z 592 that originates from m/z 1572 exhibited P and R ions originating from d20:1 (m/z 265 and 291, respectively) and S, T, U, and V ions originating from C18:0 (m/z 324, 308, 282, and 265, respectively). These facts surely revealed that d18:1/18:0 and d20:1/18:0 are the major molecular species included in bovine brain GM1, quite as the previous reports demonstrated. It is considered that the variations in matrix crystallization alter the ionization efficiency. However, our method is undoubtedly reproducible because it provides homogeneous conditions for sample-analyte crystallization and uses purified samples obtained after development by TLC. In particular, crude samples can cause the variations in the ionization efficiency due to their complex constitutions, thus the use of these samples results in poor efficiency of reproducibility. However, excessive processing for purification can result in sample loss. In this regard, our method enables reproducible and sensitive measurement because it facilitates in situ MS analysis on the transfer membrane. The ionization efficiency of the method depends on the molecular constitutions of the samples; high reproducibility of ionization efficiency could be achieved by our method with same samples. Thus, we consider that this method also enables quantitative approach between the d18:1/18:0 and d20:1/18:0 species of GM1 because these molecules were almost identical in their molecular constituents.
Fig. 6. TLC-Blot-MALDI-QIT-TOF MS, MS/MS, and MS/MS/MS spectra of GM1 to identify ceramide moieties.
(A) MS spectrum; (B) MS/MS spectrum obtained with the precursor ion at m/z 1544; (C) MS/MS/MS spectrum obtained with the second precursor ion at m/z 564. (D) MS/MS spectrum obtained with the precursor ion at m/z 1572; (E) MS/MS/MS spectrum obtained with the second precursor ion at m/z 592.
The analyses in the previous study of TLC-MALDI MS were only MS analysis in general. No work has been reported that involves ceramide analysis for precise identification of the molecular species using MS/MS/MS analysis. In this study, we were able to detect to a limit of almost 1 pmol on MS analysis (Fig. 4). In addition, we measured the detection limit of MS/MS/MS analysis (Fig. 7). Finally we could detect each unique fragment ion of the sphingoid chain (P and R) and fatty acid (S, T, U, and V) to identify the ceramide structure of molecular species in MS/MS/MS analysis to as little as 10 pmol of GM1 (Fig. 7(C)) but could not detect at a lower picomole level (data not shown). This suggests the possibility of even identifying the molecular species of a sample, with only a limited amount of it, barely stained on TLC by primuline staining.
Fig. 7. Sensitivity test of TLC-Blot-MALDI-QIT-TOF MS for the analysis of the ceramide structure at 10 pmol of GM1.
(A)MS spectrum; (B) MS/MS spectrum obtained with the precursor ion at m/z 1544; (C) MS/MS/MS spectrum obtained with the second precursor ion at m/z 564. The detection limit of TLC-Blot-MALDI-QIT-TOF MS/MS/MS for structural identification of the ceramide moiety was found to be almost 10 pmol.
3.5. Analyses of GSLs derived from the brain of a human patient with Tay-Sachs disease
Tay-Sachs disease is a classical GSL storage disease. Although the genetic and biochemical bases for massive cerebral accumulation of the ganglioside GM2 have been well established, the mechanism underlying neural dysfunction in this disease remains elusive. In this study, we used the method described above to analyze GSLs derived from the brain of a Tay-Sachs patient in order to obtain more information regarding the detailed composition of GSLs. In this manner, we hoped to achieve a breakthrough with regard to identifying the mechanisms operating in Tay-Sachs disease.
First, the tissue extract was column fractionated. Further one of the fractions, designated as fraction X (obtained by pooling fractions 186–196), was developed along with GSL standards (GM1, GD1a, GD1b, and GT1b). Fig. 8A shows the result of TLC of standards and fraction X visualized by primuline staining. Fraction X seemed to contain various GSLs. We arbitrarily selected 4 bands, namely, bands (a), (b), (c), and (d) for TLC-Blot-MALDI-QIT-TOF MS analysis.
Fig. 8. TLC-Blot-MALDI-QIT-TOF MS analyses of GSLs extracted from a Tay-Sachs patient.
(A) Thin-layer chromatogram stained with primuline: (lane 1) GSLs standards (GM1, GD1a, GD1b, and GT1b); (lane 2) GSLs derived from the brain of a patient with Tay-Sachs disease and pooled to produce fraction X. The plate was developed with chloroform/methanol/0.2% aqueous CaCl2, (55/45/10, v/v/v), and spots were detected on the TLC plate by primuline staining and were then transferred onto a PVDF membrane. (a), (b), (c), and (d) represent the spectra derived from the band produced by fraction X, and (e) represents the spectrum corresponding to the GD1a standard. Each band was directly analyzed by TLC-Blot-MALDI-QIT-TOF MS and bands (a), (b), (c), and (d) were identified as sulfatide, GM2, GM1, and GalNAc-GD1a, respectively.
MS analysis revealed that bands (a), (b), (c), and (d) corresponded to sulfatide, GM2, GM1, and GalNAc-GD1a, respectively. Bands (b) and (c) were very faint, therefore we were unable to identify the compounds therein solely by the primuline staining. However MS analysis of each band successfully produced the precise spectrum with sufficient sensitivity, and the molecules corresponding to these bands were identified as GM2 and GM1, respectively. The results demonstrated that our method is more sensitive than previous staining methods for detection in patient samples. Moreover, based on its Rf value, the band of (d) was estimated as GD1a, however, TLC-blot-MALDI-QIT-TOF MS analysis suggested this molecule to be GalNAc-GD1a. We speculated the molecular structure by MS analysis in this study helped by literatures. Further MS/MS analysis compared with standard GalNAc-GD1a will serve reliability. These results showed that our method can enable the analysis of glycolipids on PVDF membranes with high sensitivity and accuracy.
4. Conclusion
Here we described a procedure involving a combination of TLC-Blot-MALDI-QIT-TOF MS and IMS (designated as TLC-Blot-MALDI-QIT-TOF IMS) for highly sensitive and convenient detection as well as detailed structural analysis of GSLs. The positions of GSL spots on the PVDF membrane were determined by primuline staining using the conventional method, the limit of detection of which is almost 10 pmol. In this report, we could identify GM1 on PVDF membranes directly subjected to MALDI-QIT-TOF MS at only 1 pmol. Furthermore, MS, MS/MS, and MS/MS/MS spectra of GM1 revealed simple and informative fragmentation patterns and allowed structural characterization of the ceramide structures as well as the sugar sequences. Thus, no additional procedure is required to separate, visualize, and identify GSLs but TLC-Blot-MALDI-QIT-TOF imaging mass spectrometric analysis in this system.
The results demonstrated that TLC-Blot-MALDI-QIT-TOF IMS would be a powerful tool for sensitive identification of minor species and structural identification of novel structures. Therefore, the method can potentially be applied for a detailed screening/mapping of complex GSL mixtures from tissue sources, which are often available in only very limited amounts.
Acknowledgements
This work was supported by a Grant-in-Aid under the SENTAN program of the Japan Science and Technology Agency, a Center of Excellence Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology to M.S.; and NIH Grant Number R01NS009626 from the National Institute of Neurological Disorders and Stroke to Y.-T. L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannun YA, Bell RM. Science. 1989;243:500. doi: 10.1126/science.2643164. [DOI] [PubMed] [Google Scholar]
- 2.Hakomori S, Igarashi Y. J. Biochem. 1995;118:1091. doi: 10.1093/oxfordjournals.jbchem.a124992. [DOI] [PubMed] [Google Scholar]
- 3.Hakomori S, Handa K, Iwabuchi K, Yamamura S, Prinetti A. Glycobiology. 1998;8:11. doi: 10.1093/oxfordjournals.glycob.a018822. [DOI] [PubMed] [Google Scholar]
- 4.Ohanian J, Ohanian V. Cell. Mol. Life Sci. 2001;58:2053. doi: 10.1007/PL00000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakomori S. Curr. Opin. Hemotol. 2003;10:16. doi: 10.1097/00062752-200301000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Itoh T, Li YT, Li SC, Yu RK. J. Biol. Chem. 1981;256:165. [PubMed] [Google Scholar]
- 7.Krivit W, Desnick RJ, Moller J, Wright F, Sweely CC, Snyder PD, Jr, Sharp HL. Am. J. Med. 1972;52:763. doi: 10.1016/0002-9343(72)90082-4. [DOI] [PubMed] [Google Scholar]
- 8.Saifer A, Robin M, Volk BW. J. Neurochem. 1963;10:577. doi: 10.1111/j.1471-4159.1963.tb05055.x. [DOI] [PubMed] [Google Scholar]
- 9.Sandhoff K, Christomanou H. Hum. Genet. 1979;50:107. doi: 10.1007/BF00390234. [DOI] [PubMed] [Google Scholar]
- 10.Svennerholm L. Biochem. Biophys. Res. Commun. 1962;27:436. doi: 10.1016/0006-291x(62)90030-x. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. Cell. 2000;103:569. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 12.Setou M, Nakagawa T, Seog DH, Hirokawa N. Science. 2000;2000:1796. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 13.Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Nature. 2002;417:83. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- 14.Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, Fukuda Y, Chiba T, Morone N, Yuasa S, Inokuchi K, Ohtsuka T, MacGregor GR, Tanaka K, Setou M. Cell. 2007;130:943. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muthing J. J. Chromatogr. A. 1996;720:3. doi: 10.1016/0021-9673(95)00499-8. [DOI] [PubMed] [Google Scholar]
- 16.Van Echten-deckert G. Methods Enzymol. 2000;312:64. doi: 10.1016/s0076-6879(00)12900-3. [DOI] [PubMed] [Google Scholar]
- 17.Yu RK, Ariga T. Methods Enzymol. 2000;312:115. doi: 10.1016/s0076-6879(00)12903-9. [DOI] [PubMed] [Google Scholar]
- 18.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 19.Karas M, Hillenkamp F. Anal. Chem. 1988;60:2299. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 20.Juhasz P, Costello CE. Rapid Commun. Mass Spectrom. 1992;7:343. doi: 10.1002/rcm.1290070508. [DOI] [PubMed] [Google Scholar]
- 21.Metelmann W, Muthing J, Peter-Katalinic J. Rapid Commun. Mass Spectrom. 2000;14:543. doi: 10.1002/(SICI)1097-0231(20000415)14:7<543::AID-RCM908>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 22.Vukelic Z, Metelman W, Muthing J, Kos M, Peter-Katalinic J. J. Biol Chem. 2001;382:259. doi: 10.1515/BC.2001.033. [DOI] [PubMed] [Google Scholar]
- 23.O'Connor PB, Mirgorodskaya E, Costello CE. J. Am. Soc. Mass Spectrom. 2002;13:402. doi: 10.1016/S1044-0305(02)00351-3. [DOI] [PubMed] [Google Scholar]
- 24.Shaler TA, Wickham JN, Sannes KA, Wu KJ, Becker CH. Anal. Chem. 1996;68:576. doi: 10.1021/ac9502662. [DOI] [PubMed] [Google Scholar]
- 25.Amini A, Dormady SJ, Riggs L, Regnier FE. J. Chromatogr. A. 2000;894:345. doi: 10.1016/s0021-9673(00)00798-6. [DOI] [PubMed] [Google Scholar]
- 26.Bajuk A, Gluch K, Michalak L. Rapid Commun. Mass Spectrom. 2001;15:2383. doi: 10.1002/rcm.521. [DOI] [PubMed] [Google Scholar]
- 27.Meisen I, Peter-Katalinic J, Muthing J. Anal. Chem. 2004;76:2248. doi: 10.1021/ac035511t. [DOI] [PubMed] [Google Scholar]
- 28.Ivleva VB, Elkin YN, Budnik BA, Moyer SC, O'Connor PB, Costello CE. Anal. Chem. 2004;76:6484. doi: 10.1021/ac0491556. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor PB, Budnik BA, Ivleva VB, Kaur P, Moyer SC, Pittman JL, Costello CE. J. Am. Soc. Mass Spectrom. 2004;15:128. doi: 10.1016/j.jasms.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Dreisewerd K, Muthing J, Rohlfing A, Meisen I, Vukelic Z, Peter-Katalinic J, Hillenkamp F, Berkenkamp S. Anal. Chem. 2005;77:4098. doi: 10.1021/ac048373w. [DOI] [PubMed] [Google Scholar]
- 31.Ivleva VB, Sapp LM, O'Connor PB, Costello CE. J. Am. Soc. Mass Spectrom. 2005;16:1552. doi: 10.1016/j.jasms.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura K, Suzuki Y, Goto-Inoue N, Yoshida-Noro C, Suzuki A. Anal. Chem. 2006;78:5736. doi: 10.1021/ac0605501. [DOI] [PubMed] [Google Scholar]
- 33.Taki T, Ishikawa D, Handa S, Kasama T. Anal. Biochem. 1995;225:24. doi: 10.1006/abio.1995.1102. [DOI] [PubMed] [Google Scholar]
- 34.Taki T, Kasama T, Handa S, Ishikawa D. Anal Biochem. 1994;223:232. doi: 10.1006/abio.1994.1579. [DOI] [PubMed] [Google Scholar]
- 35.Wilson ID. J. Chromatogr. A. 1999;24:429. doi: 10.1016/s0021-9673(99)00618-4. [DOI] [PubMed] [Google Scholar]
- 36.Guittard J, Hronowski XL, Costello CE. Rapid Commun. Mass Spectrom. 1999;13:1838. doi: 10.1002/(SICI)1097-0231(19990930)13:18<1838::AID-RCM726>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 37.Mehl JT, Hercules DM. Anal. Chem. 2000;72:68. doi: 10.1021/ac990003y. [DOI] [PubMed] [Google Scholar]
- 38.Rohlfing A, Muthing J, Pohlentz G, Distler U, Peter-Katalinić, S J, Berkenkamp S, Dreisewerd K. Anal. Chem. 2007;23 doi: 10.1021/ac070633x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa D, Taki T. Methods Enzymol. 2000;312:145. doi: 10.1016/s0076-6879(00)12905-2. [DOI] [PubMed] [Google Scholar]
- 40.Caprioli RM, Farmer TB, Gile J. Anal. Chem. 1997;69:4751. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 41.Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Anal. Chem. 2004;76:1145. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 42.Shimma S, Furuta M, Ichimura K, Yoshida Y, Setou M. Surf. Int. Anal. 2006;38:1712. [Google Scholar]
- 43.Shimma S, Furuta M, Ichimura K, Yoshida Y, Setou M. J. Mass Spectrom. Soc. Jpn. 2006;54:133. [Google Scholar]
- 44.Shimma S, Setou M. J. Mass Spectrom. Soc. Jpn. 2007;55:145. [Google Scholar]
- 45.Sugiura Y, Shimma S, Setou M. Anal. Chem. 2006;78:8227. doi: 10.1021/ac060974v. [DOI] [PubMed] [Google Scholar]
- 46.Sugiura Y, Shimma S, Setou M. J. Mass Spectrom. Soc. Jpn. 2006;54:45. [Google Scholar]
- 47.Sugiura Y, Shimma S, Moriyama Y, Setou M. J. Mass Spectrom. Soc. Jpn. 2007;55:25. [Google Scholar]
- 48.Shimma S, Sugiura Y, Hayasaka T, Zaima N, Matsumoto M, Setou M. Anal. Chem. 2008;80:878. doi: 10.1021/ac071301v. [DOI] [PubMed] [Google Scholar]
- 49.Taira S, Sugiura Y, Moritake S, Shimma S, Ichiyanagi Y, Setou M. Anal. Chem. doi: 10.1021/ac800081z. in press. [DOI] [PubMed] [Google Scholar]
- 50.Li YT, Mascos K, Chou CW, Cole RB, Li SC. J. Biol. Chem. 2003;278:35286. doi: 10.1074/jbc.M306126200. [DOI] [PubMed] [Google Scholar]
- 51.Martin RL, Brancia FL. Rapid Commun. Mass Spectrom. 2003;17:1358. doi: 10.1002/rcm.1044. [DOI] [PubMed] [Google Scholar]
- 52.Harvey DJ. Mass Spectrom. Rev. 1999;18:349. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 53.Wiegandt H. Behav. Brain Res. 1995;66:85. doi: 10.1016/0166-4328(94)00129-4. [DOI] [PubMed] [Google Scholar]
- 54.Hakomori S. Glycoconjugate J. 2000;17:627. doi: 10.1023/a:1011086929064. [DOI] [PubMed] [Google Scholar]
- 55.Venkataraman K, Futerman AH. Trends Cell Biol. 2000;10:408. doi: 10.1016/s0962-8924(00)01830-4. [DOI] [PubMed] [Google Scholar]
- 56.Gulbins E, Grassme H. Biochim. Biophys. Acta. 2002;1585:139. doi: 10.1016/s1388-1981(02)00334-7. [DOI] [PubMed] [Google Scholar]
- 57.Ii T, Ohashi Y, Nagai Y. Carbohydr. Res. 1995;273:27. doi: 10.1016/0008-6215(95)00101-x. [DOI] [PubMed] [Google Scholar]
- 58.Marbois BN, Faull KF, Fluharty AL, Raval-Fernandes S, Rome LH. Biochim. Biophys. Acta. 2000;1484:59. doi: 10.1016/s1388-1981(99)00201-2. [DOI] [PubMed] [Google Scholar]
- 59.Svennerholm L. J Neurochem. 1963;10:613. doi: 10.1111/j.1471-4159.1963.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 60.Ann Q, Adams J. Anal. Chem. 1993;65:7. [Google Scholar]